Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Molecular mechanism of helicase on graphene-based hybridization reaction platform for microRNA detection†
Xiaojun Fana,
Yi Qia,
Zhaoyi Shia,
Yongkang Lvb and
Yujing Guo *c
*c
aCollege of Chemistry and Chemical Engineering, Taiyuan University of Technology, Taiyuan, 030024, China
bKey Laboratory of Coal Science and Technology, Ministry of Education and Shanxi Province, Taiyuan University of Technology, Taiyuan 030024, China
cInstitute of Environmental Science, Shanxi University, Taiyuan 030006, China. E-mail: guoyj@sxu.edu.cn
First published on 21st July 2017
Abstract
In this study, Escherichia coli RecQ helicase (RecQE) was introduced for the first time to demonstrate its molecular mechanism on a hybridization reaction for detecting microRNA (miRNA), using graphene oxide as a nano-quencher. In the miRNA detection system, the annealing of a “blocking” DNA strand to a fluorophore-labeled DNA probe (FDNA) effectively interfered with the non-specific binding of RecQE with FDNA. Aided by the motor force driven by adenosine triphosphate, the helicase-assisted hybridization system enhanced the hybridization of FDNA and miRNA, besides separating them. Compared with non-enzymatic detection system, the fluorescence signal of the RecQE-assisted system was increased by 17.17%. More interestingly, RecQE did not affect stability of the formed FDNA–RNA duplex. This study will greatly benefit further studies of biological sensing processes based on nucleic acid hybridization in living cells, and has great enlightening significance for designing and improving miRNA detection methods by using recombinant helicases.
1 Introduction
Nowadays molecular biology studies are revolutionizing scientific diagnosis of human diseases. Molecular diagnostics help physicians better understand each patient's clinical conditions such as disease state, disease condition prediction, drug response, and treatment prognosis.1 The essence of molecular diagnostics lies in the use of biomarkers, including proteins, carbohydrates, or nucleic acids that are associated with pathological cells.2 As a group of endogenous noncoding RNAs, microRNAs (miRNAs, 18–25 nucleotides) play significant roles in regulating the expression of target messenger RNA (mRNA).3–6 Notably, frequent aberrant expression and functional implication of miRNAs in many human diseases have lifted these small cellular components to the ranks of biomarkers.7,8Traditional methods for miRNA detection include northern blotting,9 RT-PCR,10 microarrays11 and many others. Recently, various novel methods have been developed with a special focus on sensitivity and specificity, such as nanoparticle-derived probes,12–15 isothermal amplification,16–18 electrochemical methods,19–21 etc. Among different existing nanomaterials used for miRNA detection, graphene oxide (GO) can be functionalized as an ideal transducing element thanks to high water dispersibility, stable affinity toward hydrophobic and/or π-conjugated biomolecules, and the capability of fluorescence quenching.22–24 Former studies showed that GO could bind and quench dye-labeled single-stranded DNA (ssDNA) probes, while it had less binding affinity toward dsDNA or hybrids of DNA/RNA.25,26
Helicases are enzymes conserved from bacteria to humans that use ATP-driven motor force to unwind double-stranded DNA or RNA.27,28 Such molecular activity gives helicases great potential for assisting in certain molecular diagnostic systems, especially in hybridization-dependent biosensors. However, studies on coupling helicases with GO-based nucleic acid sensors are still lacking.29–32 From this standpoint, the effects of helicases on GO-based hybridization-dependent nucleic acid detection systems need deep on-going study to enucleate the enzyme-assisted detection mechanisms in vitro and to better understand the roles of these kinds of enzymes in intracellular miRNA detection.
Methodologically, varied affinities of ssDNA and dsDNA toward GO can be explored to study molecular mechanisms of helicases on a miRNA detection system. In this work, E. coli RecQ (RecQE) was selected as a model enzyme from the RecQ helicase family for our molecular mechanism study. In the platform, BDNA is annealed to FDNA to form a FDNA–BDNA duplex for the first step (Scheme 1A). This is followed by a step of closely absorbing such duplexes onto the GO surface to form a FDNA–BDNA–GO complex (Scheme 1B) with quenched fluorescence due to noncovalent π–π stacking interactions between the hexagonal cells of graphene and the nucleobase ring structures. In the final step, RecQE and ATP are introduced to the system to form a FDNA–RNA duplex through structure switching (Scheme 1C), enabling augmentation of the fluorescent signal.
2 Results and discussion
2.1 Investigating the function of a BDNA in helicase-assisted miRNA detection system
In order to obtain higher detection sensitivity, the first problem to be solved is to minimize influence of background fluorescence due to the introduction of helicase into the miRNA detection system. Using BDNA was one of many attempts that can reduce the nonspecific binding of single strand FDNA with RecQE, which was expressed in E. coli cells and purified with a nickel sepharose column (Fig. S1†), to thereby improve stability of the detection system. Before exploring the function of BDNA, the quenching ability of GO was evaluated using fluorescence spectra of FDNA–BDNA in the presence of GO. As shown in Fig. S2,† addition of 6 μg mL−1 GO resulted in complete fluorescence quenching of a 50 nM FDNA–BDNA composite probe.To investigate the fluorescence signal attenuation effect of BDNA on the RecQE-assisted system, a series of tests were conducted at different RecQE concentrations for both FDNA–GO and FDNA–BDNA–GO systems without target miRNA. As shown in Fig. 1A, curves a and b had obviously different trends in fluorescence signal enhancement with increased RecQE concentration. This phenomenon indicated that FDNA–GO system interference was easier with RecQE than the FDNA–BDNA–GO system, and the formation of FDNA–BDNA duplex could effectively reduce the nonspecific binding of RecQE to FDNA. Besides, control tests using bovine serum albumin (BSA) obtained from serum in the FDNA–BDNA–GO system produced the expected low fluorescence signal at all concentrations (Fig. 1A-c), indicating that random proteins had much lower nonspecific action with FDNA than RecQE. A small quantity of nonspecific binding of RecQE to the single stranded probe FDNA may result in dissociation of FDNA from the GO surface and fluorescence signal enhancement. The effects of ATP on both FDNA–GO and FDNA–BDNA–GO systems were similarly studied, and according to the results (Fig. 1B), the FDNA–BDNA–GO system showed a relatively better signal attenuation effect. Fluorescence resonance energy transfer (FRET) between GO and a fluorescence probe could be affected by ATP, and thereby enhance the fluorescence signal.
These results implied that the FDNA–BDNA–GO system was minimally interfered with by RecQE and other proteins.
Introducing BDNA to the miRNA detection system can accelerate formation of the FDNA–RNA duplex,33 and increase fluorescent signal generation upon addition of target miRNA. As shown in Fig. S3,† BDNA could greatly increase the fluorescent signal when the concentration of target miRNA was 25 nM, while fluorescent signal generation did not increase much when the target miRNA concentration was equal to FDNA (50 nM).
2.2 Functional exploration and concentration optimization of RecQE and ATP in the FDNA–BDNA–GO system
Although the FDNA–BDNA–GO system had certain stability, the fluorescence signal was enhanced to some extent with increasing RecQE concentration in the absence of target miRNA (Fig. 1A-b). Under such circumstances, the functions of helicase in the miRNA detection system need further studies. In this part, the hybridization reactions between FDNA–BDNA and target let-7a miRNA were tested in the presence of both RecQE and ATP inside the detection system. When the RecQE concentration was above 3 nM, signal enhancement tended to reach a stable level in the absence of ATP (Fig. 2A-a) which indicated that RecQE can anneal two complementary single-stranded nucleic acids without ATP.27 Compared with the hybridization test results with only RecQE added, fluorescence signal enhancement achieved by also adding ATP (10 μM) – the source of driving motor force – showed an overall rise (Fig. 2A-b). Results showed that the fluorescence signal of the detection system assisted by RecQE using ATP displayed a remarkable enhancement (other than attenuation) of 17.17% over an enzyme-free reaction system. In some cases, as the helicase-induced unwinding of incomplete dsDNA proceeded, the fluorescence signal decreased to some point because of the strong interaction of GO with unwound ssDNA.34 To discover the time effect of RecQE on formation of the FDNA–RNA duplex assembly, tests were deliberately designed and run to confirm the results above. As shown in Fig. 2C, fluorescence intensity changed very little from 0 to 40 min. This result further proved that RecQE could not unwind the complete FDNA–RNA duplex structure. This motivating result may be attributed to the structure of closed dsDNA without ssDNA binding sites for helicase translocation.35,362.3 Detection capability of let-7a miRNA
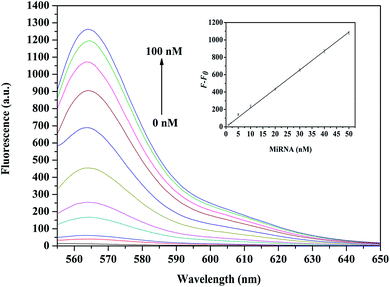
After optimizing the RecQE-assisted FDNA–BDNA–GO system, the application of such assay was investigated for various concentrations of let-7a miRNA in order to assess detection capability. As shown in Fig. 3, when the concentration of let-7a increased from 0 to 100 nM, a gradual increase in fluorescence intensity was observed, indicating dependence of fluorescence intensity upon the concentration of let-7a. By plotting the logarithmic value of let-7a concentration vs. fluorescence intensity, a fitting curve with a good correlation coefficient of 0.9985 could be obtained in the concentration range from 1 nM to 50 nM (Fig. 3, inset), and the detection limit of the method was calculated to be 0.18 nM for miRNA based on the 3σ rule. However, the fluorescence signal intensity did not show a linear relationship with let-7a concentrations in the range of 50–100 nM. The main reason might be that, when the concentration of let-7a was up to 50 nM, FDNA with fixed concentrations (say, 50 nM) was exhausted theoretically, resulting in little further increase of the fluorescence signal.There are studies on target detection sensitivity based on equivalent binding models, i.e., one target molecule can only hybridize with one fluorescent DNA probe and thereby recovers one fluorophore from the GO surface. Compared with graphene oxide-based single-layer MoS2 nanoprobes and quantum dots models,24,29 where detection limits for nucleic acids were 500 pM and 12 nM, a more sensitive detection limit was obtained in this helicase-assisted detection system. These results further suggested that RecQE had scarcely any effect on stability of the formed FDNA–RNA duplex assembly, and the proposed assay approach is appropriate for sensitive quantification of nucleic acids.
2.4 Detection specificity of target let-7a miRNA
Specificity of the proposed assay strategy was investigated by detecting let-7a, let-7f, let-7g, let-7i, and non-complementary RNA (NC) with the same concentration (50 nM) and blank control without any miRNA in reaction solutions. Fig. 4 displays fluorescence emission spectra of the RecQE-assisted FDNA–BDNA–GO system in the presence of different miRNAs. Fluorescence enhancements by the single-base-mismatched miRNA let-7f, two-base-mismatched miRNA let-7g, and four-base-mismatched miRNA let-7i were 83.23%, 63.96%, and 48.95% of that by the target let-7a, respectively. These results reveal capability of the RecQE-assisted FDNA–BDNA–GO system to detect various base-mismatched miRNAs. Moreover, this study showed that non-complementary RNA and control tests had little effect on fluorescence signal enhancement of the FDNA–BDNA–GO system, indicating high specificity of the detection system. | ||
| Fig. 4 Comparison of the fluorescence intensities produced by let-7a, let-7f, let-7g, let-7i, and NC. Control tests were run without any miRNA. | ||
2.5 Kinetics and thermodynamics
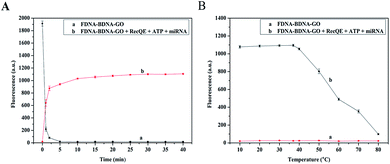
Kinetic behavior of the FDNA–BDNA–GO system assisted by RecQE and ATP was studied by monitoring fluorescence intensity as a function of time. Adsorption of the FDNA–BDNA duplex onto the GO surface was very fast at 37 °C. As shown in Fig. 5A-a, the fluorescence was effectively quenched by GO in even a shorter time than 5 min. However, the process of forming double-stranded FDNA–RNA and releasing it from GO was relatively slow (Fig. 5A-b), taking about 30 min to complete the reaction.Melting-temperature measurements were conducted to study the thermostability of FDNA–BDNA–GO and double-stranded FDNA–RNA. The FDNA–BDNA–GO system was exposed to different temperatures between 10 °C and 80 °C with 10 °C increments, allowing 30 min reaction and fluorescence signal monitoring. The FDNA–BDNA–GO complex displayed an excellent thermostability even at high temperatures, paralleling minimal change in fluorescence intensity at all the temperatures (Fig. 5B-a). However, when temperatures were above 37 °C, the formed FDNA–RNA complex started melting, resulting in a diminished fluorescence signal (Fig. 5B-b). It is worth mentioning that the highest fluorescence signal intensity of the RecQE-assisted detection system was detected at 37 °C.
3 Conclusions
For the first time, this study reports the molecular mechanism of helicase on a GO-based nucleic acid sensor with equivalent hybridization models in vitro. Comprehensive results showed that helicases had an important role in the detection process of a GO-based nucleic acid sensor, and the enzyme did not affect stability of the formed double-stranded FDNA–RNA duplex. In addition, BDNA can weaken the nonspecific interaction between RecQE and FDNA. Since helicases have an extensive applicability with hybridization reactions of nucleic acids, this work will benefit further studies of biological sensing processes based on nucleic acid hybridization.4 Experimental section
4.1 Materials and apparatus
HPLC-purified DNA and miRNAs were synthesized and purified by Sangon Biotech Co., Ltd. (Shanghai, China). Sequences of the oligonucleotide probes and miRNAs are listed in Table 1. Diethylpyrocarbonate (DEPC)-treated deionized water and ribonuclease inhibitor were purchased from Solarbio Biotechnology Co., Ltd. (Beijing, China). GO was synthesized from natural graphite according to the classic protocol with a modification of Hummers' method.37,38 pET24a-RecQE recombinant plasmid was constructed and presented by Hua Ren (Department of Biomedical Sciences, School of Life Science, East China Normal University, Shanghai 200241, China). Unless otherwise noted, all other chemicals were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). DEPC-treated deionized water was used in all experiments.| Name | Sequences (5′-3′) |
|---|---|
| a Note: different bases in let-7 were underlined. | |
| FDNA | Cy3-AACTATACAACCTACTACCTCA |
| BDNA | TGAGGTA |
| Let-7a | UGAGGUAGUAGGUUGUAUAGUU |
| Let-7f | UGAGGUAGUAGAUUGUAUAGUU |
| Let-7g | UGAGGUAGUAGUUUGUACAGUU |
| Let-7i | UGAGGUAGUAGUUUGUGCUGUU |
| Non-complementary RNA | UUGUACUACACAAAAGUACUGG |
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a DYY-10C Electrophoresis Apparatus (Liuyi, Beijing, China). Ultrapure water was purified by a Simplicity UV Water Purification System (Millipore SAS, France). Fluorescence spectra were obtained using a Hitachi F-2700 Fluorescence Spectrophotometer (Hitachi, Japan).
4.2 FDNA–BDNA–GO system
The FDNA–BDNA–GO system contains 50 nM FDNA and 50 nM BDNA. Prior to reaction, FDNA and BDNA were both incubated in 1 × SPSC buffer (0.75 M NaCl and 50 mM Na2HPO4, pH 7.4) at 37 °C to fold the FDNA–BDNA duplex assembly. After that, GO (6 μg mL−1) was added to the solution and incubated for 30 min at 37 °C.4.3 Optimization of RecQE and ATP on FDNA–BDNA–GO system
Optimization of RecQE concentration in the FDNA–BDNA–GO system was performed in 1 mL of a FDNA–BDNA–GO system containing 50 nM let-7a, 10 μM ATP, 40U RNase inhibitor, and different concentration levels of RecQE (1, 2, 3, 4, 5, and 6 nM). The final mixture was incubated at 37 °C for 30 min.For optimizing ATP concentration in the FDNA–BDNA–GO system, the reaction was performed in 1 mL of a FDNA–BDNA–GO system containing 50 nM let-7a, 5 nM RecQE, 40U RNase inhibitor, and different concentrations of ATP (2, 4, 6, 8, 10, and 12 μM). The final mixture was incubated at 37 °C for 30 min.
For the time effect of RecQE on detection of the FDNA–RNA duplex assembly, FDNA (50 nM) and let-7a (50 nM) were first incubated together at 37 °C for 1 h to perfectly fold into a FDNA–RNA duplex assembly, and then, GO (6 μg mL−1) was added to the system and incubated for 30 min at 37 °C. Subsequently, RecQE (5 nM) and ATP (10 μM) were mixed with the solution, and the fluorescence signal was measured every five minutes (0–40 min).
4.4 Target miRNA detection
The hybridization reaction was performed in 1 mL of the FDNA–BDNA–GO system containing 5 nM RecQE, 10 μM ATP, 40U RNase inhibitor, and let-7a in different concentrations. Subsequently, the mixture was incubated at 37 °C for 30 min.4.5 Fluorescence spectra
Fluorescence of a Cy3-labeled DNA probe was excited at 540 nm, and the resulting spectra were recorded between 555 nm and 650 nm. The fluorescence emission maximized at 564 nm. Spectrometer slits were set for 5 nm band pass and operating voltage at 700 V.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported the Natural Science Foundation of Shanxi Province (No. 201603D321094) and the Young “Sanjin” Scholar Foundation of Shanxi Province, 2016. The authors thank Lihui Bai, Min Li and Lu Li for their help with some of the experiments.References
- J. Hoggatt, Mol. Diagn. Ther., 2011, 15, 53–55 CrossRef PubMed.
- A. B. Chinen, C. M. Guan, J. R. Ferrer, S. N. Barnaby, T. J. Merkel and C. A. Mirkin, Chem. Rev., 2015, 115, 10530 CrossRef CAS PubMed.
- L. He, J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon and S. M. Hammond, Nature, 2005, 435, 828 CrossRef CAS PubMed.
- S. M. Johnson, H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K. L. Reinert, D. Brown and F. J. Slack, Cell, 2005, 120, 635–647 CrossRef CAS PubMed.
- J. J. Rossi, Cell, 2009, 137, 990–992 CrossRef CAS PubMed.
- A. Ventura and T. Jacks, Cell, 2009, 136, 586–591 CrossRef CAS PubMed.
- N. Chiorazzi, K. R. Rai and M. Ferrarini, N. Engl. J. Med., 2005, 352, 804–815 CrossRef CAS PubMed.
- A. Esquelakerscher and F. J. Slack, Nat. Rev. Cancer, 2006, 6, 259–269 CrossRef CAS PubMed.
- A. Válóczi, C. Hornyik, N. Varga, J. Burgyán, S. Kauppinen and Z. Havelda, Nucleic Acids Res., 2004, 32, e175 CrossRef PubMed.
- C. Chen, D. A. Ridzon, A. J. Broomer, Z. Zhou, D. H. Lee, J. T. Nguyen, M. Barbisin, N. L. Xu, V. R. Mahuvakar, M. R. Andersen, K. Q. Lao, K. J. Livak and K. J. Guegler, Nucleic Acids Res., 2005, 33, e179 CrossRef PubMed.
- W. Li and K. Ruan, Anal. Bioanal. Chem., 2009, 394, 1117–1124 CrossRef CAS PubMed.
- F. Degliangeli, P. Kshirsagar, V. Brunetti, P. P. Pompa and R. Fiammengo, J. Am. Chem. Soc., 2014, 136, 2264–2267 CrossRef CAS PubMed.
- D. W. Hwang, I. C. Song, D. S. Lee and S. Kim, Small, 2010, 6, 81–88 CrossRef CAS PubMed.
- Z. Lu, L. Zhang, Y. Deng, S. Li and N. He, Nanoscale, 2012, 4, 5840–5842 RSC.
- Y. P. Zeng, G. Zhu, X. Y. Yang, J. Cao, Z. L. Jing and C. Y. Zhang, Chem. Commun., 2014, 50, 7160–7162 RSC.
- H. Dong, J. Zhang, H. Ju, H. Lu, S. Wang, S. Jin, K. Hao, H. Du and X. Zhang, Anal. Chem., 2012, 84, 4587–4593 CrossRef CAS PubMed.
- W. Li, T. Hou, M. Wu and F. Li, Talanta, 2016, 148, 116–121 CrossRef CAS PubMed.
- H. Liu, L. Li, L. Duan, X. Wang, Y. Xie, L. Tong, Q. Wang and B. Tang, Anal. Chem., 2013, 85, 7941–7947 CrossRef CAS PubMed.
- A. Erdem and G. Congur, Talanta, 2014, 118, 7–13 CrossRef CAS PubMed.
- S. P. Jonstrup, J. Koch and J. Kjems, RNA, 2006, 12, 1747–1752 CrossRef CAS PubMed.
- F. Li, J. Peng, J. Wang, H. Tang, L. Tan, Q. Xie and S. Yao, Biosens. Bioelectron., 2014, 54, 158–164 CrossRef CAS PubMed.
- J. N. Lekitima, K. I. Ozoemena, C. J. Jafta, N. Kobayashi, Y. Song, D. Tong, S. Chen and M. Oyama, J. Mater. Chem. A, 2013, 1, 2821–2826 CAS.
- Z. Wang, J. Zhang, P. Chen, X. Zhou, Y. Yang, S. Wu, L. Niu, Y. Han, L. Wang, P. Chen, F. Boey, Q. Zhang, B. Liedberg and H. Zhang, Biosens. Bioelectron., 2011, 26, 3881–3886 CrossRef CAS PubMed.
- C. Zhu, Z. Zeng, H. Li, F. Li, C. Fan and H. Zhang, J. Am. Chem. Soc., 2013, 135, 5998–6001 CrossRef CAS PubMed.
- S. He, B. Song, D. Li, C. Zhu, W. Qi, Y. Wen, L. Wang, S. Song, H. Fang and C. Fan, Adv. Funct. Mater., 2010, 20, 453–459 CrossRef CAS.
- W. Qiang, W. Li, X. Li, X. Chen and D. Xu, Chem. Sci., 2014, 5, 3018–3024 RSC.
- Y. Wu, J. Nucleic Acids, 2012, 2012, 140601 Search PubMed.
- D. L. Croteau, V. Popuri, P. L. Opresko and V. A. Bohr, Annu. Rev. Biochem., 2014, 83, 519–552 CrossRef CAS PubMed.
- H. Dong, W. Gao, F. Yan, H. Ji and H. Ju, Anal. Chem., 2010, 82, 5511–5517 CrossRef CAS PubMed.
- C. H. Lu, H. H. Yang, C. L. Zhu, X. Chen and G. N. Chen, Angew. Chem., Int. Ed., 2009, 121, 4879–4881 CrossRef.
- L. Li, J. Feng, H. Liu, Q. Li, L. Tong and B. Tang, Chem. Sci., 2016, 7, 1940–1945 RSC.
- L. Yang, C. Liu, W. Ren and Z. Li, ACS Appl. Mater. Interfaces, 2012, 4, 6450–6453 CAS.
- J. Song, P. S. Lau, M. Liu, S. Shuang, C. Dong and Y. Li, ACS Appl. Mater. Interfaces, 2014, 6, 21806–21812 CAS.
- H. Jang, Y. K. Kim, H. M. Kwon, W. S. Yeo, D. E. Kim and D. H. Min, Angew. Chem., Int. Ed., 2010, 122, 5839–5843 CrossRef.
- S. X. Dou, P. Y. Wang, H. Q. Xu and X. G. Xi, J. Biol. Chem., 2004, 279, 6354–6363 CrossRef CAS PubMed.
- H. Q. Xu, E. Deprez, A. H. Zhang, P. Tauc, M. M. Ladjimi, J. C. Brochon, C. Auclair and X. G. Xi, J. Biol. Chem., 2003, 278, 34925–34933 CrossRef CAS PubMed.
- W. S. Hummers Jr and R. E. Offeman, J. Am. Chem. Soc., 1958, 80, 1339 CrossRef.
- Y. Guo, S. Guo, J. Ren, Y. Zhai, S. Dong and E. Wang, ACS Nano, 2010, 4, 4001–4010 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra06203f |
| This journal is © The Royal Society of Chemistry 2017 |