Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mechanistic insight into the rapid one-step facile biofabrication of antibacterial silver nanoparticles from bacterial release and their biogenicity and concentration-dependent in vitro cytotoxicity to colon cells†
Suresh K. Vermaa,
Ealisha Jhab,
Babrubahan Sahoo c,
Pritam Kumar Panda
c,
Pritam Kumar Panda a,
Arun Thirumurugand,
S. K. S. Parashare and
Mrutyunjay Suar
a,
Arun Thirumurugand,
S. K. S. Parashare and
Mrutyunjay Suar *a
*a
aSchool of Biotechnology, KIIT University, Bhubaneswar, Orissa 751024, India. E-mail: msuar@kiitbiotech.ac.in; msbiotek@yahoo.com; Tel: +91 674 2725732 Tel: +91 674 2725466
bMemorial University of Newfoundland, Department of Physics and Physical Oceanography, St. John's, Newfoundland and Labrador NL A1C 5S7, Canada
cKIIT Technology Business Incubator, KIIT University, Bhubaneswar, Orissa 751024, India
dInstitute of Physics, Bhubaneswar, Orissa 751005, India
eSchool of Applied Sciences, KIIT University, Bhubaneswar, Orissa 751024, India
First published on 16th August 2017
Abstract
Progress in the research and development of green synthesis of silver nanoparticles and their applications has reached new heights in the last decade. In this study, one-step rapid facile biosynthesis of silver nanoparticles is reported, and in vitro cytotoxicity of these nanoparticles has been investigated in an HCT116 cell line. Biogenic silver nanoparticles were synthesized from the culture supernatant of Gram-positive (B. thuringiensis and S. aureus) and Gram-negative bacteria (E. coli and S. typhimurium) using UV light, termed as BTAgNP, SAAgNP, ECAgNP, and STAgNP, respectively. The synthesized silver nanoparticles were characterised by standard characterisation methods such as field emission scanning electron microscopy (FESEM), dynamic light scattering (DLS), UV-visible spectroscopy, and Fourier transform infrared spectroscopy (FTIR). An in silico investigation was performed to elucidate the mechanism of their synthesis. Uniformly distributed ECAgNP, SAAgNP, STAgNP, and BTAgNP with stable zeta potentials were synthesized with the sizes 22.6 ± 5.2 nm, 21.2 ± 4.8 nm, 23.3 ± 6.8 nm, and 29.3 ± 5.2 nm, respectively. The synthesized silver nanoparticles were found to exhibit significant antibacterial activity against their source bacteria. An in vitro assessment revealed their biogenicity and concentration-dependent cytotoxicity and genotoxicity in colon cell lines with the occurrence of morphological deformities, oxidative stress, apoptosis, and cell cycle arrest. The study provided an insight into the biogenic differences in the biological effects of silver nanoparticles.
Introduction
In recent decades, silver nanoparticles have gained significant popularity because of their impeccable application in many fields such as in electronics, engineering, textiles, paints, the food industry, cosmetics, biosensing, chronic wound treatment, and medicine.1,2 These nanoparticles have been extensively studied due to their unique physiochemical characteristics such as their optical, electronic, and catalytic activities.3,4 Significant emphasis has also been given to these nanoparticles because of their antibacterial properties. Silver nanoparticles have become a focus in biomedical technology and are considered as alternatives to conventional antibiotics and therapeutics based on combinatorial chemistry for medical purposes.5 Extensive studies and applications have increased the demands for the rapid and facile synthesis of silver nanoparticles.6 Many methods, such as chemical,7,8 biological,9,10 and physical methods,11 have been explored using different types of reducing and stabilising agents. However, the enormous growth in their exploration and synthesis has raised concerns over their environmental impact and toxicity issues. Hence, it has become essential to evaluate the biological effects of silver nanoparticles synthesized via different means.The biological effects, such as the antibacterial activity and cytotoxicity, of silver nanoparticles depend on the physiochemical parameters, such as size,12 shape,13 charge,14 and the stabilising agent15 used in their production, which are determined by their synthesis route.16 Biological synthesis route has gained a favourable reputation because it is a green method that is less cytotoxic and more biocompatible in comparison with other methods used for the synthesis of nanoparticles;17,18 this synthesis route can further be classified on the basis of the source of the reducing agent, which can be a plant19 or a bacteria.20 To date, several bacterial strains have been used in vitro and in vivo to synthesize silver nanoparticles,20 which have been proven to be beneficial and biocompatible antibacterial agents.21
It is well known that green-synthesized silver nanoparticles from a bacterial source exhibit antibacterial activity. However, it is interesting to note that the physiochemical nature of the nanoparticles depends on the strain and culture conditions. Bacteria release a number of different biomolecules during their growth in a culture medium; these biomolecules can be utilised as reducing and capping agents to synthesize silver nanoparticles although their mechanism of action still remains unknown.22,23 Das et al.24 and Fayaz et al.20 have demonstrated the syntheses of silver nanoparticles from bacterial strains, such as Bacillus and Klebsiella, and their antibacterial activity against pathogenic strains such as E. coli and S. aureus. However, the effect of the synthesized nanoparticles against their source bacterial strain still remained to be discovered. In this study, the hidden mechanism has been revealed via experimental and computational verification. Silver nanoparticles were synthesized using the supernatants of two Gram-negative (E. coli and S. typhimurium) and two Gram-positive bacterial cultures (B. thuringiensis and S. aureus) by a novel green method, and their synthesis mechanism was investigated in silico. In addition, the probable mechanism of the antibacterial activity of these synthesized silver nanoparticles was determined.
From antibacterial activity, the focus of research has shifted towards the synthesis of non-toxic and biocompatible nanomaterials. Silver ions are being used for treating burns and as an antibacterial agent in clinical prospects to fight infections.25 Previous studies have reported the size- and concentration-dependent cytotoxicity of silver nanoparticles26,27 and highlighted the mechanism of cytotoxicity as a consequence of reactive oxygen species (ROS) stress28 in normal cells as well as in cancer cell lines. Keeping these studies in consideration, it is necessary to assess the cytotoxicity of synthesized silver nanoparticles in vitro and in vivo.
The detailed assessment of the variations in the physiochemical properties and biological effects of biocompatible, rapidly synthesized silver nanoparticles from different bacterial cultures was carried out using a novel green method. We developed a protocol for the biological synthesis of silver nanoparticles within 5 min from bacterial cultures and analysed the cytotoxicity of these nanoparticles via different experimental assays in human colon cells (HCT116) taken as an in vitro model.
Materials and methods
Microbial synthesis of silver nanoparticles
The silver nanoparticles used in this study were synthesized by a novel method using the bacterial supernatant of two Gram-positive (B. thuringiensis and S. aureus) and two Gram-negative strains (E. coli and S. typhimurium) (Fig. S1†). The bacterial strains were cultured in Luria-Bertani (LB) broth at 35 ± 2 °C using an orbital shaker incubator (New Brunswick Scientific, USA) at 160 rpm. The supernatants of overnight cultures were extracted via centrifugation of the cultures at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. Silver nitrate (AgNO3) solution (1 mM) was then mixed with the supernatant of each bacterial culture in a ratio of 1
000 rpm for 5 min. Silver nitrate (AgNO3) solution (1 mM) was then mixed with the supernatant of each bacterial culture in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, and the mixture was exposed to UV light for 3–5 min. The synthesized silver nanoparticles were then obtained by centrifugation at 10
2, and the mixture was exposed to UV light for 3–5 min. The synthesized silver nanoparticles were then obtained by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The concentrated nanoparticles were resuspended in MQ water and lyophilized. A stock solution for each analysis was prepared by suspending the nanoparticles in the corresponding medium. The solutions were characterised for their physiochemical properties by different methods. The synthesized silver nanoparticles were termed as BTAgNP, ECAgNP, SAAgNP, and STAgNP, obtained from the B. thuringiensis, E. coli, S. aureus, and S. typhimurium culture supernatants, respectively.
000 rpm for 10 min. The concentrated nanoparticles were resuspended in MQ water and lyophilized. A stock solution for each analysis was prepared by suspending the nanoparticles in the corresponding medium. The solutions were characterised for their physiochemical properties by different methods. The synthesized silver nanoparticles were termed as BTAgNP, ECAgNP, SAAgNP, and STAgNP, obtained from the B. thuringiensis, E. coli, S. aureus, and S. typhimurium culture supernatants, respectively.
Characterisation of silver nanoparticles
The synthesized silver nanoparticles were characterised for their physiochemical properties by different standard techniques. The size of the nanoparticles was determined by field emission scanning electron microscopy (FESEM) using a Zeiss EVO 60 microscope equipped with an Oxford Inca energy-dispersive X-ray spectrometer (EDS); the hydrodynamic size and zeta potential (as a measure of stability) of BTAgNP, SAAgNP, STAgNP, and ECAgNP were measured by dynamic light scattering (Zetasizer, Malvern, UK) in water, DMEM cell culture medium, and Holtfreter buffer. Fourier transform infrared (FTIR) spectroscopy measurements were performed using a Perkin Elmer RXI FTIR spectrometer with an ATR attachment. All measurements were carried out in the range of 400–4000 cm−1 at a resolution of 4 cm−1. Optical properties were determined via UV-visible spectra obtained using a UV-vis spectrophotometer (Agilent Cary 60, USA) at 200–800 nm.In silico investigation of the synthesis of silver nanoparticles
To understand the probable mechanism of the synthesis of nanoparticles from bacterial supernatants, a molecular docking approach was employed to recognize the capping of Ag nanoparticles by the Gram-positive and Gram-negative bacterial supernatant proteins. Supernatants contain the majority of biofilm proteins released by bacteria in a culture medium. Biofilm-associated protein A (bapA) of Salmonella typhimurium (3824 residues) and biofilm-associated surface protein (bap) of S. aureus (2276 residues) were considered for the molecular docking approach. Since the structures of both these proteins were unavailable, molecular modelling of these two proteins was carried out using ModBase,29 which is a database of comparative protein structure models. For bapA protein, PDB ID: 4AQ1,30 structure of the SbsB S-layer protein of S. typhimurium PV72p2 in complex with the nanobody KB6 from the region 371–1026 was chosen as a template, whereas for bap protein, PDB ID: 1D2P,31 crystal structure of two b repeating units (b1b2) of the collagen-binding protein (cna) of S. aureus was taken as a template from the region 1809–2113 for structural modelling. Furthermore, these proteins were subjected to molecular docking as receptors using AutoDock 4.2,32 whereas Ag was used as the ligand. The grid dimensions were set to 90 × 90 × 90 with a spacing of 1 Å. Lamarckian genetic algorithms (LGA) were used for the grid dimensions, and a genetic algorithm was used for docking runs using a population size of 150, with the maximum number of evaluations set to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500
500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and the maximum number of generations. Post-docking analysis was performed using AutoDock 4.2 analysis tools using conformations and clustering, and the results were visualized using Chimera.33 Further 2D plots of interactive regions were shown using LigPot+.34
000 and the maximum number of generations. Post-docking analysis was performed using AutoDock 4.2 analysis tools using conformations and clustering, and the results were visualized using Chimera.33 Further 2D plots of interactive regions were shown using LigPot+.34
Assessment of the antibacterial effect of silver nanoparticles
The antibacterial activities of the different as-synthesized AgNPs were assessed via a well diffusion assay and growth curve analysis of bacterial strains in the presence of the synthesized silver nanoparticles. The detailed protocol is described in the ESI† sheet.Analysis of morphology change in silver nanoparticle-treated bacteria
Changes in bacterial morphology were investigated by FESEM. The processing of samples is described in the ESI† sheet.Analysis of reactive oxygen species (ROS)
The analysis of the ROS production in bacterial strains upon treatment with BTAgNP, SAAgNP, STAgNP, and ECAgNP was performed by detecting the green signal of 2′,7′-dichlorodihydrofluorescein (DCF) using the BL1 filter (530/30) of a flow cytometer. The green signal corresponds to the number of DCF molecules produced by the oxidation of DCFDA dye by ROS produced by cells.35 Forward- and side-scatter dot plots were used to gate out cellular debris.Treatment of silver nanoparticles with HCT116 cells
A human colon cancer cell line (HCT116) was procured from the national animal cell repository, the National Centre for Cell Science (NCCS), Pune, India. HCT116 cells were seeded in a 24-well plate at a density of 1.0 × 104 cells per well in complete DMEM medium and allowed to adhere for 24 h. After adhesion, the medium was replaced with a medium containing silver nanoparticles with the desired concentration, and the cells were incubated for 24 h and 48 h under an atmosphere of 5% CO2 at 37 °C. The cells were observed for morphological changes using an inverted microscope.Cytotoxicity assays
To measure the cytotoxicity effect of silver nanoparticles on HCT116 cells, the MTT assay was used. Cells in the exponential growth phase were seeded at a density of 1.0 × 104 cells per well into 96-well plates in 100 μL DMEM containing 10% (vol/vol) NC serum. The cells were allowed to adhere for 24 h at 37 °C. After 24 h, the medium was replaced by fresh DMEM containing 10% (vol/vol) NC serum with the addition of silver nanoparticle samples in a series of increasing concentrations from 50 μg mL−1 to 3000 μg mL−1. After incubation for 24–48 h, the medium was removed and the wells were washed three times with 1× phosphate buffered saline (PBS). After washing, 150 μL of 0.1 mg mL−1 MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) in PBS was added to each well, and the plate was incubated for 4 h. Finally, the medium was replaced by a 150 μL MTT solvent (11 g SDS in 50 mL of 0.02 M HCl and 50 mL isopropanol) to dissolve the formazan crystals formed by live cells. The absorbance (OD value) was then obtained at 570 nm using an ELISA reader (Epoch, Biotek, Germany). The cytotoxicity was estimated by calculating the percentage of the OD value with respect to that in a control experiment without the sample. A graphical representation was then created to compare the effects of different concentrations. All the experiments were performed at least three times in triplicate.Analysis of the uptake of nanoparticles by flow cytometry
Uptake analysis was performed by flow cytometry. A brief description of the protocol is given in the ESI† sheet.Analysis of the production of reactive oxygen species (ROS) in HCT cells
The production of reactive oxygen species (ROS) was analysed by flow cytometry using 2′,7′-dichlorofluorescein diacetate (DCFDA). DCFDA is a dye that gets converted into 2′,7′-dichlorofluorescein (DCF) in the presence of ROS. Thus, via estimating the amount of DCF formed, the production of ROS induced by the nanoparticles can be determined.35 To estimate the production of ROS induced in HCT116 cells on exposure to silver nanoparticles, cells were seeded and treated as described in the treatment protocol. After the treatment, the cells were washed three times with 1× phosphate buffer solution (PBS) and scraped. The scraped cells were then stained with DCFDA (2 μg mL−1 in DMSO) for 15 min in the dark. After incubation, the cells were analysed using a attune acoustic focusing cytometer (Applied Biosystems, Life Technologies) equipped with a 488 nm argon laser. The cytometer was set up to logarithmically measure FSC and SSC. The data were processed using FCS Express 5 (DeNovo, Los Angeles, CA). The mean fluorescence intensity of each sample was measured and is presented in the form of a histogram.Acridine orange (AO)/EtBr staining for apoptosis analysis
Apoptosis analysis was performed according to the standard protocol described by Kasibhatla et al.36 The HCT116 cells were seeded in a 24-well plate at a density of 1.0 × 104 cells per well in a complete DMEM medium and allowed to adhere to the surface via incubation for 24 h under 5% CO2 at 37 °C. After incubation, the medium was replaced by a 500 μL DMEM medium containing different synthesized AgNPs with the concentrations of 50 μg mL−1 and 250 μg mL−1 (v/v), and the cells were exposed for 24 h and 48 h. After treatment, the medium was replaced by 1× PBS and the mixture was washed 3 times. After being washed, the cells were processed and stained with AO and EtBr. Images were obtained using an EVOS inverted fluorescence microscope (Thermo Scientific, USA). The images were processed in ImageJ to adjust their brightness and contrast.Cell cycle analysis
Cell cycle analysis of the HCT116 cell line was performed via propidium iodide staining of the nucleus, as mentioned in the ESI.†Results and discussion
Synthesis and physiochemical characterization of silver nanoparticles
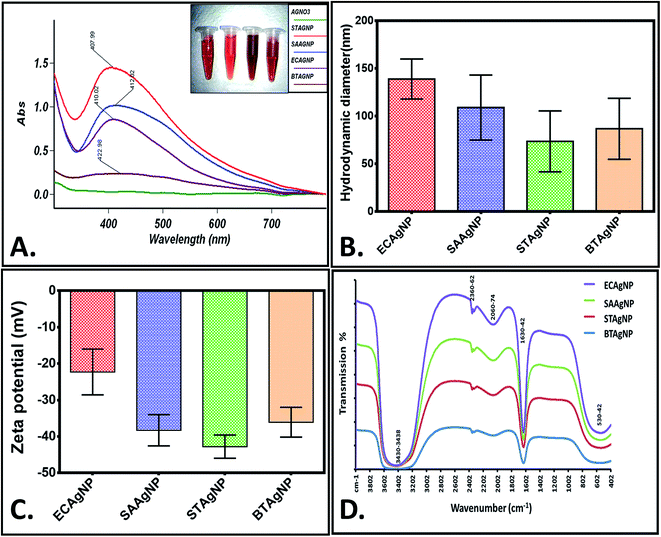
The rapid one-pot biosynthesis of silver nanoparticles was achieved using supernatants of overnight cultures of two Gram-positive (B. thuringiensis and S. aureus) and two Gram-negative (E. coli and S. typhimurium) bacterial strains, as shown in Fig. S1.† UV radiation was used to reduce silver nitrate, whereas biomolecules released by the bacteria in the supernatant were used to stabilize the silver nanoparticles. The synthesis of silver nanoparticles was confirmed via a change in colour. As determined by FESEM (Fig. 1), the average sizes of ECAgNP, SAAgNP, STAgNP, and BTAgNP were found to be 22.6 ± 5.2 nm, 21.2 ± 4.8 nm, 23.3 ± 6.8 nm, and 29.3 ± 5.2 nm, respectively. EDS analysis showed the presence of silver nanoparticles with a small amount of chlorine, which may be due to the presence of sodium chloride in the bacterial culture medium (LB broth). A small amount of carbon was also detected by EDS, which can be attributed to the small amount of bacterial biomass present in the solution. The synthesized silver nanoparticles (ECAgNP, SAAgNP, STAgNP, and BTAgNP) were characterised for their optical properties via UV-vis spectroscopy. UV-vis spectroscopy is the most widely known technique for characterising nanoparticles via quantification of their light absorption phenomenon. As shown in Fig. 2A, the absorption spectra of all four nanoparticles displayed a surface plasmon resonance (SPR) peak at 400–420 nm, which was attributed to silver nanoparticles in previous reports.37 The presence of this peak in the spectra of all four nanoparticles confirmed the presence of silver nanoparticles. The hydrodynamic diameters of ECAgNP and SAAgNP were found to be 139.2 ± 21.0 and 109.1 ± 34.0 nm, respectively, whereas those of STAgNP and BTAgNP were 73.6 ± 32.0 and 86.8 ± 32.0 nm, respectively (Fig. 2B and Table 1). The differences in the diameters of the nanoparticles, as determined by FESEM and DLS, can be ascribed to the presence of water molecules around the particles in solution.38 The hydrodynamic diameter varied in different media used for biological assays (Table 1). The zeta potential of the silver nanoparticles was measured in different media to determine the degree of repulsion among the particles in different media.39 As shown in Fig. 2C, STAgNP and SAAgNP exhibited lower zeta potentials of −42.8 ± 3.2 mV and −38.3 ± 4.3 mV as compared to ECAgNP and BTAgNP (−22.3 ± 6.3 mV and −36.1 ± 4.1 mV), respectively. Thus, the results indicated the dispersed suspension of silver nanoparticles in all the types of media. However, the variation in values in different media can be attributed to the interaction of nanoparticles with different types of molecules present in the media.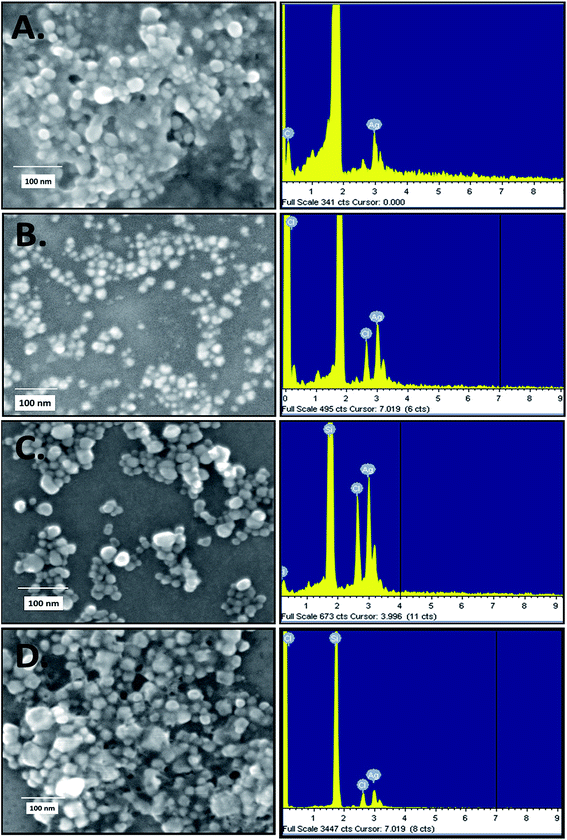 | ||
| Fig. 1 Size determination by FESEM and EDS analysis of silver nanoparticles: (A) ECAgNP, (B) SAAgNP, (C) STAgNP, and (D) BTAgNP. | ||
| Nanoparticles | Size (nm) by FESEM | Hydrodynamic diameter (nm) | Zeta potential (mV) | SPR peak (nm) | ||||
|---|---|---|---|---|---|---|---|---|
| Aq. | PBS | DMEM (complete) | Aq. | PBS | DMEM (complete) | |||
| ECAgNP | 22.6 ± 5.2 | 139.2 ± 21 | 215.3 ± 22 | 230.1 ± 11 | −22.3 ± 6.3 | −26.5 ± 5.3 | −19.3 ± 7.1 | 410 |
| SAAgNP | 21.2 ± 4.8 | 109.1 ± 34 | 116.1 ± 51 | 109.5 ± 43 | −38.3 ± 4.3 | −28.3 ± 4.2 | −23.1 ± 5.4 | 412 |
| STAgNP | 23.3 ± 6.8 | 73.6 ± 32 | 113.6 ± 16 | 110.9 ± 17 | −42.8 ± 3.2 | −46.9 ± 3.3 | −27.2 ± 4.6 | 407 |
| BTAgNP | 29.3 ± 5.2 | 86.8 ± 32 | 152.1 ± 36 | 208.6 ± 13 | −36.1 ± 4.1 | −43.8 ± 2.3 | −28.3 ± 3.4 | 422 |
Fig. 2D shows the FTIR spectra of the biosynthesized silver nanoparticles. Distinct peaks were observed at 530–542 cm−1, 1630–1642 cm−1, 2060–2074 cm−1, 2360–2362 cm−1, and 3430–3438 cm−1. This complexity of the peaks may be due to the presence of different biomolecules released by the bacteria. The broad peak observed at 3430–3438 cm−1 corresponds to the O–H and N–H bond stretching vibrations. The small peaks at 2360–2362 cm−1 and 2060–2074 cm−1 correspond to the stretching vibrations of the compounds containing C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bonds. A sharp peak was found at 1630–1642 cm−1, which may be due to the presence of amides. The small peak at 530–542 cm−1 may be due to the leaching of organic compounds present in the supernatant of the bacterial cultures.40,41 With reference to the analysis of the results of synthesis and characterisation, a uniform and well-dispersed formation of silver nanoparticles can be confirmed.
N bonds. A sharp peak was found at 1630–1642 cm−1, which may be due to the presence of amides. The small peak at 530–542 cm−1 may be due to the leaching of organic compounds present in the supernatant of the bacterial cultures.40,41 With reference to the analysis of the results of synthesis and characterisation, a uniform and well-dispersed formation of silver nanoparticles can be confirmed.
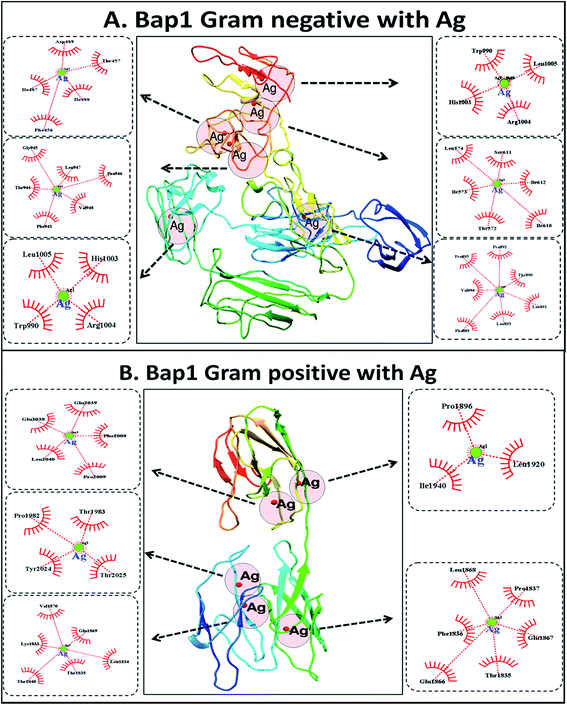
To understand the probable mechanism of synthesis of the silver nanoparticles, an in silico approach was employed. Molecular docking was performed using Ag as the ligand and Bap1 proteins from Gram-positive (S. aureus) and Gram-negative (S. typhimurium) bacterial strains. Bap1 protein has been recognised as a biofilm protein released by bacteria into their matrix during their growth. Fig. S2† shows the structure of Bap1 proteins in S. aureus and S. typhimurium. As shown in Fig. 3, reduced silver ions from silver nitrate were predicted to be incorporated into the matrix protein in six possible conformations in Gram negative bacterial protein and five possible conformations in Gram positive bacterial protein. In Gram-negative matrix biofilm protein, the amino acid residues found to interact with Ag were aspartic acid, threonine, isoleucine, and phenylalanine, whereas in some clusters, valine, leucine, arginine, and histidine were also found to play a role. In the Gram-positive bacterial matrix, along with other residues, proline was found to play a significant role. Although in both cases the interactions occurred through hydrophobic bonds, polar amino acid residues were predicted to interact with silver in the proteins of Gram-negative bacteria. With reference to these results, it can be speculated that the synthesis of silver nanoparticles from the bacterial supernatants was influenced by the biogenicity and structural as well as functional aspects of the bacterial strains. In the reaction setup, silver nitrate salt was reduced to silver and nitrate radicals in the presence of reducing agents. Silver atoms were further capped by interactions with amino acids in the matrix proteins for stabilisation. Hence, the physiochemical nature of the synthesized silver nanoparticles varies due to variations in matrix protein amino acid residues of Gram-positive and Gram-negative bacterial strains. The reaction can be represented as follows:
| AgNO3 + reducing agent (matrix biomolecules) → Ag+ + NO3− |
| Ag+ + capping agent (matrix protein amino acids) → AgNP |
Antibacterial activity of ECAgNP, SAAgNP, STAgNP, and BTAgNP silver nanoparticles
The antibacterial activities of the biosynthesized ECAgNP, SAAgNP, STAgNP, and BTAgNP were first examined by the well diffusion method to observe the formation of an inhibition zone against S. typhimurium and E. coli. Fig. S3† shows the inhibition zones exhibited by all four silver nanoparticles with E. coli and S. typhimurium. The observation of inhibition zones revealed the significant antibacterial activity of the synthesized silver nanoparticles. The objective of this study was to specify the antibacterial activities of the synthesized nanoparticles against their source bacterial cultures. To achieve this, growth curves were determined by incubating ECAgNP, SAAgNP, STAgNP, and BTAgNP with E. coli, S. aureus, S. typhimurium, and B. thuringiensis in a culture medium at different concentrations. As shown in Fig. S4(a)–(d),† all nanoparticles inhibited the growth of their corresponding bacterial strains with the increasing concentrations, among which ECAgNP was most effective. The MIC values of the nanoparticles varied considerably against their source bacterial strain. The growth curves for S. aureus, S. typhimurium, and B. thuringiensis became gradually steeper with an increase in concentration, whereas slope declination was highest in E. coli against ECAgNP, depicting its highest efficacy. The variation in the activity of the nanoparticles against their corresponding bacterial strains can be attributed to the released biomolecules, the nature of the amino acid residues utilised in their synthesis, and the difference in the structural composition of each bacterial culture.42To understand the mechanism of action of the synthesized nanoparticles on their corresponding source bacteria, FESEM was performed to visualise the bacterial cell morphology after treatment. All the untreated bacterial cells were found to display normal morphology after treatment for 4 h, as shown in Fig. 4, whereas those treated with the corresponding silver nanoparticles were found to lose their membrane integrity and exhibited damaged shapes. The attachment of silver nanoparticles was confirmed by EDS. The results indicated the firm attachment of nanoparticles to the surface of bacteria that led to their death. These results were in accordance with the reports focusing on reciprocity between AgNPs and bacterial cell membranes.43,44 These reports referred to the zeta potential, size, and shape of silver nanoparticles as causes of the accumulation and destruction of membranes. ECAgNP, SAAgNP, STAgNP and BTAgNP had sizes of around 100 nm and negative charges, which therefore facilitated their attachment to cell membranes, exerting the antibacterial effects. Moreover, the attachment of silver nanoparticles has been attributed to a change in the membrane permeability of bacterial cells owing to the interactions with membrane proteins45 and histidine-rich periplasmic proteins.46 This acute change may cause the release and degradation of membrane proteins; this leads to the formation of fissures and perforations in the cell membrane. The formation of fissures and perforation leads to the leakage of cytoplasmic contents from the cell; this increases the death consequences. Because the capping materials of the synthesized silver nanoparticles were derived from the supernatants of their corresponding bacterial cultures, it can be argued that the biomolecules increased their probability of attachment to the corresponding bacteria owing to their similar chemical natures.
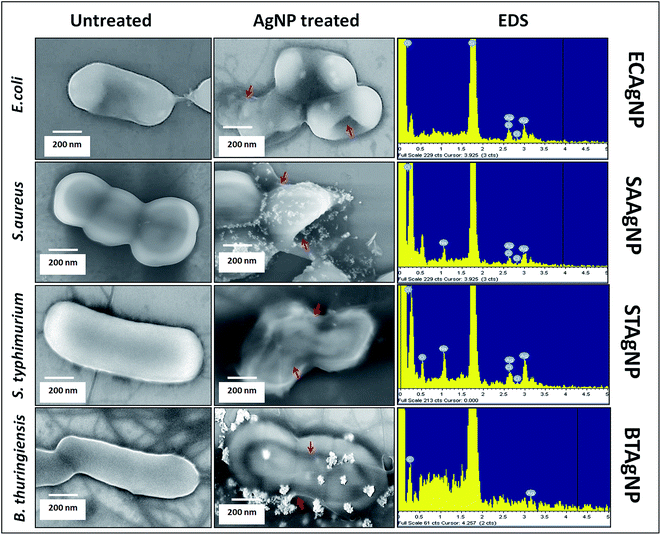 | ||
| Fig. 4 Antibacterial activity of AgNPs: morphological analysis of bacterial strains with EDS analysis of bacterial surfaces treated with the corresponding synthesized biogenic silver nanoparticles. | ||
Another well-known reason for the bactericidal activity of silver nanoparticles is the induction of oxidative stress in bacterial cells upon exposure to nanoparticles.47,48 Reasonable effects of the interactions of the synthesized silver nanoparticles with the corresponding bacterial strains were investigated via the fluorescence intensity of DCFDA in bacterial cells. As shown in Fig. S5,† the production of ROS was found to increase with the incubation time (4 h) in a concentration-dependent manner. Although each type of nanoparticle induced an enhancement in the production of ROS in the corresponding bacterial strain, the intensity was highest in the case of ECAgNP with E. coli, followed by STAgNP, BTAgNP, and SAAgNP. The difference may be correlated with an enhancement in the interaction and internalisation of ECAgNP nanoparticles as compared to that of others.
These results suggested the variable antibacterial efficiency of biosynthesized silver nanoparticles from different bacterial cultures against their corresponding bacteria. Moreover, the results indicate the interesting fact that the components of bacterial supernatants can be used as a weapon against the bacteria themselves.
In vitro cytotoxicity of AgNPs against HCT116 colon cancer cell line
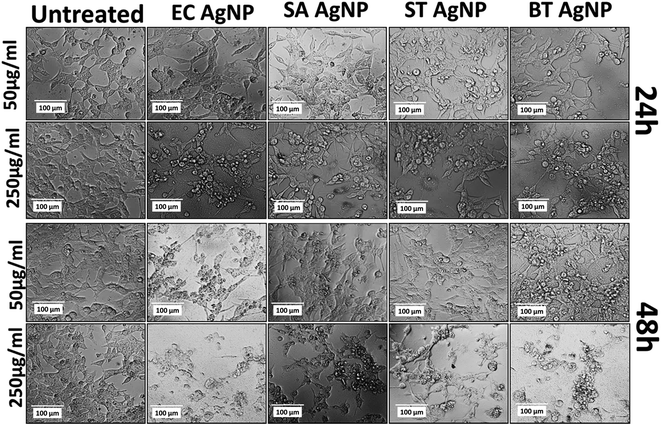
The cytotoxicity of the synthesized silver nanoparticles (AgNPs) was investigated in an HCT116 colon cancer cell line via standard established assays. Viability was determined by the MTT assay. The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay is based on the determination of the mitochondrial activity of living cells via estimation of the conversion of MTT into formazan crystals by them.49 The LC50 value of a compound can be determined thereafter by comparing the viability with that of untreated cells that display 100% viability. As shown in Fig. S6A,† STAgNP was most cytotoxic followed by ECAgNP, SAAgNP, and BTAgNP. This observation was further supported by determining the LC50 values of the nanoparticles, as indicated in Fig. S6B.† The difference in the cytotoxicity of these nanoparticles can be attributed to variations in their interaction mechanism owing to the presence of different biomolecules on their surface.50 A further investigation of the mechanism of cytotoxicity was carried out via an analysis of the morphological changes in AgNP-treated cells by bright-field microscopy. Fig. 5 shows the morphology of the HCT116 cells treated with 50 μg mL−1 and 250 μg mL−1 ECAgNP, SAAgNP, STAgNP, and BTAgNP after exposure for 24 h and 48 h. After exposure for 24 h, a change was observed in the morphology of cells exposed to a low concentration (50 μg mL−1) of nanoparticles in comparison with that of the untreated cells, which was significant after exposure for 48 h. Cells were found to be detached from the surface and had lost their original morphology. The intensity increased upon treatment with a high concentration (250 μg mL−1) after exposure for both 24 h and 48 h; moreover, after 48 h, the cell viability decreased. This observation corresponded to the results of the MTT assay. The images also illustrate the accumulation of nanoparticles on the surfaces of cells; this is considered to be the first step of the mechanism of cell cytotoxicity.51,52The mechanism of the cytotoxicity of silver nanoparticles has been described in many reports.26,52–54 It has been stated that the cytotoxic effect basically proceeds in three stages: nanoparticles accumulate on the surfaces of cells and interact with the cell membrane; this leads to changes in the charge and permeability of the membrane.51,52 The interaction is followed by the internalization of nanoparticles inside the cells via endocytosis and pores in the cell membrane.55 After the internalisation of nanoparticles, alterations in cellular physiological processes such as the generation of ROS56,57 and other metabolic processes58 occur with changes at the nuclear level;59,60 this lead to abnormalities and, ultimately, cell death. To determine whether ECAgNP, SAAgNP, STAgNP, and BTAgNP follow the same mechanism and elucidate the differences between their activities, further experimental analysis of the uptake of nanoparticles and alterations in physiological processes was carried out. The analysis of the uptake of AgNPs inside the HCT116 cells was performed via a flow cytometer by measuring the changes in cell granularity, as described by previous reports.61 As shown in Fig. S7,† exposure time- and concentration-dependent changes in cell granularity were observed with all four types of AgNP; this indicated the uptake of nanoparticles. Interestingly, ECAgNP was taken up by cells to a greater extent after 48 h, followed by STAgNP, SAAgNP, and BTAgNP. Speculated consequences of the uptake of AgNPs were further investigated by an analysis of the oxidative stress, apoptosis, and abnormalities in the cell cycle in treated cells. Fig. S8† shows the concentration- and time-dependent changes in the fluorescence of DCFDA, which indicate an increase in the production of ROS in all cells treated with AgNPs. However, AgNPs synthesized from Gram-positive bacterial sources induced the production of less ROS in comparison with AgNPs synthesized from Gram-negative bacterial sources. The investigation of apoptosis in treated cells was performed by dual staining with acridine orange/EtBr. Acridine orange is a vital dye that stains both live and dead cells. EtBr only stains dead cells that have lost their membrane integrity. Hence, early apoptotic cells can be identified by their green colour and bright green nuclei; late apoptotic cells display an orange colour, whereas necrotic cells exhibit an orange colour with a nuclear morphology that resembles that of the live cells.36 As shown in Fig. 6, ECAgNP and STAgNP-treated cells were found to be in the late apoptotic phase after exposure for 24 h and 48 h to both low and high concentrations, whereas in the cases of treatment with SAAgNP and BTAgNP, early apoptosis was found at low concentrations. However, treatment with a high concentration (250 μg mL−1) led to the rapid onset of late apoptosis. The dynamic basis of the cell cycle is crucial in cells in any physiological condition62 as it determines abnormal cell proliferation and cancer. To understand the effects of the biogenic AgNPs on the stages of the cell cycle, the phase of the cell cycle was determined by PI staining analysis at low and high concentrations. As shown in Fig. S9,† cells were found to undergo arrest in the G0/G1 phase upon exposure. Interestingly, the amount of cells treated with SAAgNP and BTAgNP also increased in the S phase. These results suggested a mechanism of in vitro cytotoxicity in the HCT116 colon cells. Similar cytotoxic effects have been reported by different groups with regard to the cytotoxicity of biogenic silver nanoparticles63,64 in different cell lines. Many studies have also mentioned the cytotoxic effects of silver nanoparticles synthesized by different methods on normal cell lines and reported acute cytotoxicity and genotoxicity as results of the regulation of oxidative stress, apoptosis, and cell cycle arrest.65 Moreover, cytotoxicity against both normal and cancer cell lines has been reported to depend on the capping agent.66 With reference to previous studies and experimental data obtained, it can be concluded that the cytotoxicity of silver nanoparticles depends on the capping agent. However, an interesting point that emerged from this investigation is the variation in cytotoxicity due to the biogenicity of silver nanoparticles. Silver nanoparticles synthesized from Gram-positive bacterial supernatants were less cytotoxic than those synthesized from Gram-negative bacteria. This variation can be attributed to the variation in bacterial biomolecules released into their supernatant due to their physiological and structural differences.42
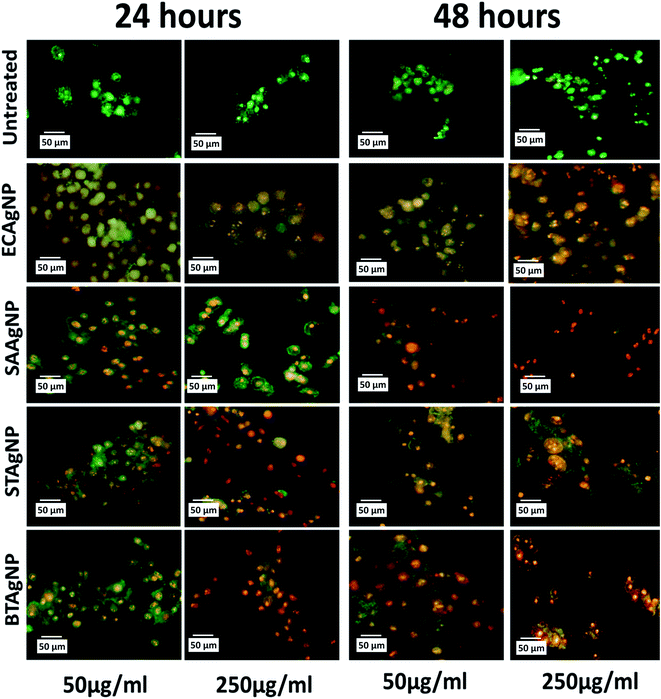 | ||
| Fig. 6 Apoptosis of HCT116 cells after treatment with different types of silver nanoparticle for 24 h and 48 h determined by staining with acridine orange/EtBr. | ||
Conclusion
In summary, the successful and facile synthesis of well-characterised biogenic silver nanoparticles (AgNPs) was achieved from the culture supernatants of two Gram-positive (S. aureus and B. thuringiensis) and two Gram-negative (E. coli and S. typhimurium) bacterial strains, and the products were termed as SAAgNP, BTAgNP, ECAgNP, and STAgNP, respectively. The antibacterial efficacy of the synthesized AgNPs was determined against their source bacteria, and their in vitro cytotoxicity was investigated with a colon cancer cell line (HCT116). The analysis of their antibacterial activity confirmed their efficacy against their source bacteria in a concentration-dependent manner due to damage to membranes and the effects of oxidative stress. The assessment of their cytotoxicity demonstrated their biogenicity and concentration-dependent in vitro cytotoxic effects in colon cancer cell lines. AgNPs that originated from Gram-negative strains were shown to exhibit higher cytotoxicity than AgNPs that originated from Gram-positive strains and induced greater oxidative stress, morphological changes, apoptosis, and cell cycle arrest in the G0/G1 phase. The overall study concludes that the biological effects of silver nanoparticles synthesized from different bacterial culture supernatants depend on their biogenicity, concentration, and exposure time. It is believed that variations in the biomolecules utilised as stabilizing agents in the synthesis of AgNPs present in the bacterial culture supernatant with variations in the bacterial culture strain determined the physiochemical properties, antibacterial activity, and cytotoxicity of the silver nanoparticles. An in silico molecular-level investigation predicted and confirmed this fact, in particular, in the context of proteomics. Moreover, pathological outcome, such as DNA damage and alterations to specific proteins and enzymes responsible for different physiological processes, of the biogenic AgNPs at the molecular level need further investigation. With the help of the study reported herein, future work can be directed towards studying the difference in the roles of Ag+ ions released by biogenic AgNPs and their in vitro and in vivo biological consequences. This study has pointed out the need of right selection of green biogenic materials and methodologies for the synthesis of silver nanoparticles for different purposes with respect to the environment and human health.Conflicts of interest
There are no conflicts to declare.References
- Q. H. Tran, V. Q. Nguyen and A.-T. Le, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2013, 4, 33001 CrossRef.
- L. Wei, J. Lu, H. Xu, A. Patel, Z. Chen and G. Chen, Drug Discovery Today, 2015, 1–7 Search PubMed.
- K. M. M. Abou El-Nour, A. Eftaiha, A. Al-Warthan and R. A. A. Ammar, Arabian J. Chem., 2010, 3, 135–140 CrossRef CAS.
- M. Nasrollahzadeh, F. Babaei, S. Mohammad Sajadi and A. Ehsani, Spectrochim. Acta, Part A, 2014, 132, 423–429 CrossRef CAS PubMed.
- D. R. Monteiro, L. F. Gorup, A. S. Takamiya, A. C. Ruvollo-Filho, E. R. de Camargo and D. B. Barbosa, Int. J. Antimicrob. Agents, 2009, 34, 103–110 CrossRef CAS PubMed.
- M. Brust and C. J. Kiely, Colloids Surf., A, 2002, 202, 175–186 CrossRef CAS.
- B. Wiley, T. Herricks, Y. Sun and Y. Xia, Nano Lett., 2004, 4, 1733–1739 CrossRef CAS.
- N. Jana and L. Gearheart, Chem. Commun., 2001, 617–618 RSC.
- K. N. Thakkar, S. S. Mhatre and R. Y. Parikh, Nanomed. Nanotech. Biol. Med., 2010, 6, 257–262 CrossRef CAS PubMed.
- V. K. Sharma, R. A. Yngard and Y. Lin, Adv. Colloid Interface Sci., 2009, 145, 83–96 CrossRef CAS PubMed.
- J. Zhu, S. Liu, O. Palchik, Y. Koltypin and A. Gedanken, Langmuir, 2000, 16, 6396–6399 CrossRef CAS.
- M. Rai, A. Yadav and A. Gade, Biotechnol. Adv., 2009, 27, 76–83 CrossRef CAS PubMed.
- S. Pal, Y. K. Tak and J. M. Song, J. Biol. Chem., 2015, 290, 1712–1720 CrossRef PubMed.
- A. M. El Badawy, R. G. Silva, B. Morris, K. G. Scheckel, M. T. Suidan and T. M. Tolaymat, Environ. Sci. Technol., 2011, 45, 283–287 CrossRef CAS PubMed.
- P. Raveendran, J. Fu and S. L. Wallen, J. Am. Chem. Soc., 2003, 125, 13940–13941 CrossRef CAS PubMed.
- Y. G. Sun and Y. N. D. A.-D. Xia, ST–Shape–controlled synthesis of gold, Science, 2002, 298, 2176–2179 CrossRef CAS PubMed.
- R. Rajan, K. Chandran, S. L. Harper, S. Il Yun and P. T. Kalaichelvan, Ind. Crops Prod., 2015, 70, 356–373 CrossRef CAS.
- A. M. Schrand, L. K. Braydich-Stolle, J. J. Schlager, L. Dai and S. M. Hussain, Nanotechnology, 2008, 19, 235104 CrossRef PubMed.
- J. Yong, S. Æ. Beom and S. Kim, Production, 2009, 79–84 Search PubMed.
- A. Fayaz, K. Balaji, M. Girilal and R. Yadav, Biol. Med., 2010, 6, 103–109 CAS.
- Y. H. Hsueh, K. S. Lin, W. J. Ke, C. Te Hsieh, C. L. Chiang, D. Y. Tzou and S. T. Liu, PLoS One, 2015, 10(12), e0144306 Search PubMed.
- S. Shivaji, S. Madhu and S. Singh, Process Biochem., 2011, 46, 1800–1807 CrossRef CAS.
- A. Nanda and M. Saravanan, Nanomed. Nanotech. Biol. Med., 2009, 5, 452–456 CrossRef CAS PubMed.
- V. L. Das, R. Thomas, R. T. Varghese, E. V. Soniya, J. Mathew and E. K. Radhakrishnan, 3 Biotech, 2013, 4, 121–126 CrossRef PubMed.
- J. Fong and F. Wood, Int. J. Nanomed., 2006, 1, 441–449 CrossRef CAS PubMed.
- A. Gliga, S. Skoglund, I. Odnevall Wallinder, B. Fadeel and H. Karlsson, Part. Fibre Toxicol., 2014, 11, 11 CrossRef PubMed.
- X. Guo, Y. Li, J. Yan, T. Ingle, M. Y. Jones, N. Mei, M. D. Boudreau, C. K. Cunningham, M. Abbas, A. M. Paredes, T. Zhou, M. M. Moore, P. C. Howard and T. Chen, Nanotoxicology, 2016, 10, 1373–1384 CrossRef CAS PubMed.
- J. W. Han, S. Gurunathan, J.-K. Jeong, Y.-J. Choi, D.-N. Kwon, J.-K. Park and J.-H. Kim, Nanoscale Res. Lett., 2014, 9, 459 CrossRef PubMed.
- U. Pieper, B. M. Webb, G. Q. Dong, D. Schneidman-Duhovny, H. Fan, S. J. Kim, N. Khuri, Y. G. Spill, P. Weinkam, M. Hammel, J. A. Tainer, M. Nilges and A. Sali, Nucleic Acids Res., 2014, 42, D336–D346 CrossRef CAS PubMed.
- E. Baranova, R. Fronzes, A. Garcia-Pino, N. Van Gerven, D. Papapostolou, G. Péhau-Arnaudet, E. Pardon, J. Steyaert, S. Howorka and H. Remaut, Nature, 2012, 487, 119–122 CAS.
- C. C. S. Deivanayagam, R. L. Rich, M. Carson, R. T. Owens, S. Danthuluri, T. Bice, M. Höök and S. V. L. Narayana, Structure, 2000, 8, 67–78 CrossRef CAS PubMed.
- G. Morris and R. Huey, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- A. C. Wallace, R. A. Laskowski and J. M. Thornton, Protein Eng., 1995, 8, 127–134 CrossRef CAS PubMed.
- E. Eruslanov and S. Kusmartsev, Methods Mol. Biol., 2010, 594, 57–72 CAS.
- S. Kasibhatla, Cold Spring Harb. Protoc., 2006, 2006, 4493 Search PubMed.
- K. B. Mogensen and K. Kneipp, J. Phys. Chem. C, 2014, 118, 28075–28083 CAS.
- J. Jiang, G. Oberdörster and P. Biswas, J. Nanopart. Res., 2009, 11, 77–89 CrossRef CAS.
- R. Xu, Particuology, 2008, 6, 112–115 CrossRef CAS.
- A. R. Binupriya, M. Sathishkumar, K. Vijayaraghavan and S. I. Yun, J. Hazard. Mater., 2010, 177, 539–545 CrossRef CAS PubMed.
- P. S. Pimprikar, S. S. Joshi, A. R. Kumar, S. S. Zinjarde and S. K. Kulkarni, Colloids Surf., B, 2009, 74, 309–316 CrossRef CAS PubMed.
- R. Y. Parikh, S. Singh, B. L. V Prasad, M. S. Patole, M. Sastry and Y. S. Schouche, ChemBioChem, 2008, 9, 1415–1422 CrossRef CAS PubMed.
- B. Salopek-Sondi and I. Sondi, J. Colloid Interface Sci., 2004, 275, 177–182 CrossRef PubMed.
- O. Choi, K. K. Deng, N.-J. Kim, L. Ross Jr, R. Y. Surampalli and Z. Hu, Water Res., 2008, 42, 3066–3074 CrossRef CAS PubMed.
- M. Jose Ruben, E. Jose Luis, C. Alejandra, H. Katherine, B. K. Juan, R. R. Jose Tapia and Y. Miguel Jose, Nanotechnology, 2005, 16, 2346 CrossRef PubMed.
- L. Mirolo, T. Schmidt, S. Eckhardt, M. Meuwly and K. M. Fromm, Chem.–Eur. J., 2013, 19, 1754–1761 CrossRef CAS PubMed.
- M. Stevanović, V. Uskoković, M. Filipović, S. D. Škapin and D. Uskoković, ACS Appl. Mater. Interfaces, 2013, 5, 9034–9042 Search PubMed.
- M. Stevanović, B. Kovačević, J. Petković, M. Filipič and D. Uskoković, Int. J. Nanomed., 2011, 6, 2837–2847 CrossRef PubMed.
- Assay Guidance Manual [Internet], ed. G. S. Sittampalam, N. P. Coussens, K Brimacombe, et al., Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda, MD, 2004, pp. 26–270 Search PubMed.
- W. Jiang, A. Saxena, B. Song, B. B. Ward, T. J. Beveridge and S. C. B. Myneni, Langmuir, 2004, 20, 11433–11442 CrossRef CAS PubMed.
- S. M. Hussain, K. L. Hess, J. M. Gearhart, K. T. Geiss and J. J. Schlager, Toxicol. In Vitro, 2005, 19, 975–983 CrossRef CAS PubMed.
- B. D. Chithrani and W. C. W. Chan, Nano Lett., 2007, 7, 1542–1550 CrossRef CAS PubMed.
- E. J. Park, J. Yi, Y. Kim, K. Choi and K. Park, Toxicol. In Vitro, 2010, 24, 872–878 CrossRef CAS PubMed.
- R. Foldbjerg, D. A. Dang and H. Autrup, Arch. Toxicol., 2010, 3, 279–290 Search PubMed.
- A. Lankoff, W. J. Sandberg, A. Wegierek-Ciuk, H. Lisowska, M. Refsnes, B. Sartowska, P. E. Schwarze, S. Meczynska-Wielgosz, M. Wojewodzka and M. Kruszewski, Toxicol. Lett., 2012, 208, 197–213 CrossRef CAS PubMed.
- S. Kim, J. E. Choi, J. Choi, K.-H. Chung, K. Park, J. Yi and D.-Y. Ryu, Toxicol. In Vitro, 2009, 23, 1076 CrossRef CAS PubMed.
- A. Manke, L. Wang and Y. Rojanasakul, BioMed Res. Int., 2013, 2013, 942916 Search PubMed.
- R. Foldbjerg, P. Olesen, M. Hougaard, D. A. Dang, H. J. Hoffmann and H. Autrup, Toxicol. Lett., 2009, 190, 156–162 CrossRef CAS PubMed.
- S. Hackenberg, A. Scherzed and M. Kessler, Toxicol. Lett., 2011, 201, 27–33 CrossRef CAS PubMed.
- P. V. AshaRani, G. L. K. Mun, M. P. Hande and S. Valiyaveettil, ACS Nano, 2009, 3, 279–290 CrossRef CAS PubMed.
- E. J. Zucker Massaro, K. M. Sanders, L. L. Degn and W. K. Boyes, Cytometry, Part A, 2010, 77, 677–685 CrossRef PubMed.
- C. Gerard and A. Goldbeter, Interface Focus, 2014, 4, 20130075 CrossRef PubMed.
- R. Vivek, R. Thangam, K. Muthuchelian and P. Gunasekaran, Process Biochemistry, 2012, 47, 2405–2410 CrossRef CAS.
- R. Sukirtha, K. M. Priyanka, J. J. Antony, S. Kamalakkannan, R. Thangam, P. Gunasekaran, M. Krishnan and S. Achiraman, Process Biochem., 2012, 47, 273–279 CrossRef CAS.
- P. V. AshaRani, G. L. K. Mun, M. P. Hande and S. Valiyaveettil, ACS Nano, 2009, 3, 279–290 CrossRef CAS PubMed.
- P. Netchareonsirisuk, S. Puthong, S. Dubas, T. Palaga and K. Komolpis, J. Nanopart. Res., 2016, 18, 322 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra05943d |
| This journal is © The Royal Society of Chemistry 2017 |