Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanoarmoring: strategies for preparation of multi-catalytic enzyme polymer conjugates and enhancement of high temperature biocatalysis†
Omkar V. Zoreab,
Paritosh Pandea,
Oghenenyerovwo Okifoa,
Ashis K. Basua,
Rajeswari M. Kasi*ab and
Challa V. Kumar*abc
aDepartment of Chemistry, University of Connecticut Storrs, CT 06269-3060, USA. E-mail: challa.kumar@uconn.edu; kasi@ims.uconn.edu; Fax: +1-860-486-4745; Tel: +1-860-486-4713
bInstitute of Materials Science, U-3136, University of Connecticut Storrs, CT 06269-3069, USA
cDepartment of Molecular and Cell Biology, University of Connecticut Storrs, CT 06269-3125, USA
First published on 6th June 2017
Abstract
We report a general and modular approach for the synthesis of multi enzyme–polymer conjugates (MECs) consisting of five different enzymes of diverse isoelectric points and distinct catalytic properties conjugated within a single universal polymer scaffold. The five model enzymes chosen include glucose oxidase (GOx), acid phosphatase (AP), lactate dehydrogenase (LDH), horseradish peroxidase (HRP) and lipase (Lip). Poly(acrylic acid) (PAA) is used as the model synthetic polymer scaffold that will covalently conjugate and stabilize multiple enzymes concurrently. Parallel and sequential synthetic protocols are used to synthesise MECs, 5-P and 5-S, respectively. Also, five different single enzyme–PAA conjugates (SECs) including GOx–PAA, AP–PAA, LDH–PAA, HRP–PAA and Lip–PAA are synthesized. The composition, structure and morphology of MECs and SECs are confirmed by agarose gel electrophoresis, dynamic light scattering, circular dichroism spectroscopy and transmission electron microscopy. The bioreactor comprising MEC functions as a single biocatalyst can carry out at least five different or orthogonal catalytic reactions by virtue of the five stabilized enzymes, which has never been achieved to-date. Using activity assays relevant for each of the enzymes, for example AP, the specific activity of AP at room temperature and 7.4 pH in PB is determined and set at 100%. Interestingly, MECs 5-P and 5-S show specific activities of 1800% and 600%, respectively, compared to 100% specific activity of AP at room temperature (RT). The catalytic efficiencies of 5-P and 5-S are 1.55 × 10−3 and 1.68 × 10−3, respectively, compared to 9.11 × 10−5 for AP under similar RT conditions. Similarly, AP relevant catalytic activities of 5-P and 5-S at 65 °C show 100 and 300%, respectively, relative to native AP activity at RT as the native AP is catalytically inactive at 65 °C. The catalytic activity trends suggest: (1) MECs show enhanced catalytic activities compared to native enzymes under similar assay conditions and (2) 5-S is better suited for high temperature biocatalysis, while both 5-S and 5-P are suitable for room temperature biocatalysis. Initial cytotoxicity results show that these MECs are non-lethal to human cells including human embryonic kidney [HEK] cells when treated with doses of 0.01 mg mL−1 for 72 h. This cytotoxicity data is relevant for future biological applications.
1. Introduction
Here an easy, novel, modular and general method to synthesize highly catalytically active and stable multi (five) enzyme poly(acrylic acid) conjugates (MECs) is presented. PAA serves as a universal scaffold and is able to chemically conjugate several enzymes of varying isoelectric points. Comprehensive catalytic activities of MEC based bioreactor are investigated and compared to single enzyme–PAA conjugates and native or unmodified enzymes.Nature utilizes a multi-enzyme catalytic complex, cellulosome, for efficient hydrolysis of cellulose.1 Using Nature as an inspiration, a major challenge is to design a bioreactor containing multiple enzymes that are covalently or chemically conjugated within a single universal scaffold. For industrially relevant applications, these multi enzyme conjugates are required to present stability, high catalytic activity, environmentally conscious features in cascade catalytic reactions or several orthogonal non-cascade catalytic reactions.2–5
Several examples of multiple enzymes physically adsorbed or encapsulated within a single scaffold are known. For instance, a set of three3 enzymes, comprised of glucose oxidase, lactate oxidase, and galactose oxidase, are non-covalently immobilized on an electrode using polyethylene glycol as a spacer and used for quantification of glucose, lactate, and galactose at millimolar concentrations.6 However, this immobilization strategy show leaching of enzymes from the electrode and displays a 20–25% reduction in catalytic activities. In another example, thirteen (13) different enzymes are physically adsorbed onto carbon nanotubes/carbon paper and used an enzymatic biofuel cascade reactor for sugar-based battery applications. While these non-immobilized multi-enzymes present good activities at room temperature, the ability to function at higher temperature and long-term storage and usage are not addressed.7 Since catalytic activities are typically enhanced at higher temperatures, strategies to stabilize multiple enzymes for catalysis at temperatures higher than 25 °C are beneficial. These limitations of enzymes can be overcome, at least in principle, by the covalent conjugation of enzymes with well-defined, suitably selected, synthetic polymers via facile but mild synthetic approaches.8 By combining proteins and polymers in a “synergistic” manner, new structures and unique materials can be designed, and properties of both components can be enhanced for specific applications.9,10
Covalent or chemical conjugation of a single enzyme within a polymer or an inorganic support is well-understood. For example, using site-specific attachment strategies, single enzymes can be conjugated to end groups of polyethylene oxide.11,12 These precisely defined single enzyme conjugates are used for biological applications.13 Single enzymes can also be randomly conjugated within polyacrylic acid (PAA) framework such that amino groups on the enzyme chemically react with carboxylic acid groups on PAA.14,15 Several such single enzyme conjugates are synthesized and display high stability and enhanced biocatalytic activities at higher temperatures and in organic solvents compared to native or unmodified enzymes.16
There is only one known example of two different enzymes covalently conjugated to a single polymer support.8 Amine groups on GOx and HRP are covalently and randomly attached to carboxylic groups on PAA catalyzed by EDC resulting in GOx–HRP–PAA bienzyme conjugates. These bienzyme conjugates present unprecedented catalytic activity and stability at 65 °C with little or no leaching of the active catalysts. Furthermore, PAA acts as a buffer and these enzymes are stabilized over pH of 2.5–7.4. While these reports suggest that PAA would be an excellent universal scaffold for conjugation of multiple enzymes, a general strategy for chemical covalent conjugation of several of different isoelectric points, amino groups and catalytic features within a universal polymer scaffold has not been achieved. The multiple enzyme polymer conjugate (MECs) platform will be very useful for cascade catalysis or in orthogonal single stream non-cascade chemical transformations.
In the present study, PAA is used a universal scaffold to covalent tether enzymes including glucose oxidase (GOx), horseradish peroxidase (HRP), lipase (Lip), acid phosphatase (AP) and lactate dehydrogenase (LDH) to form multi-catalytic enzyme complex (MEC). These five enzymes are chosen due to their widely different isoelectric points and surface availability of lysine groups (Table 1). These enzymes also used in several biocatalytic applications. For example, GOx and HRP catalyze redox transformations in sensing, catalysis, and waste-water remediation and they serve as cascade dyads.17–23 In contrast, LDH catalyzes dehydrogenation reactions for the production of enantiopure (−) lactate and 2-hydroxy acids that are precursors to many pharmaceutical drugs24,25 and for sensing of lactate.26 Hydrolases like AP are used widely for waste remediation, metal recovery,27 and determination of surfactants,28 whereas lipases are employed for the production of pharmaceutical drugs, cosmetics, leather, detergents, foods, perfumery, medical diagnostics and other synthetic materials.29–32 More importantly, LDH, GOx and HRP display significant reduction of catalytic activities at 50 °C, while the activity of Lip and AP is rapidly decreases above 40 °C.8,33–35 Methods to stabilize these enzymes so that they can function at high temperatures are needed to enhance their catalytic activities and is a challenge that should be addressed.
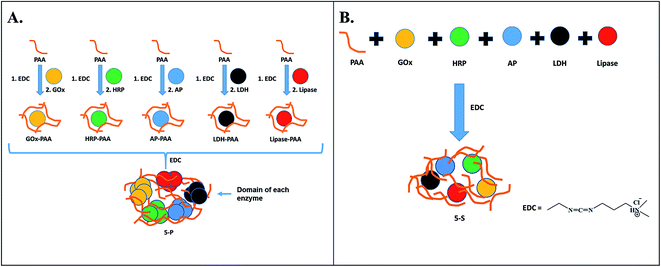
Multiple enzyme–PAA conjugates (MECs) are synthesized using two different routes (Scheme 1). In Route 1, a parallel synthesis strategy is used: (step 1) Single enzyme–PAA conjugates are synthesized using GOx (yellow), HRP (green), AP (blue), LDH (black) and lipase (red) and PAA (brown string) to form GOx–PAA, HRP–PAA, AP–PAA, LDH–PAA and Lip–PAA, (step 2) these enzyme–PAA conjugates are mixed and crosslinked using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) to form MECs (5-P, Scheme 1A). In Route 2, a sequential synthesis strategy of MEC is used: PAA and 5 enzymes are stirred together for 10–20 min and crosslinked using EDC to form MEC (5-S, Scheme 1B). The two MECs differ considerably in their microenvironment due to fundamental differences in the synthesis protocols used and, therefore, it is critical to investigate their biocatalytic behaviors. Unprecedented stability and catalytic activities of these MECs at room temperature and at 65 °C, compared to SECs and native unmodified enzymes under similar conditions, are reported.
2. Experimental section
2.1 Materials
PAA (Mv = 450![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 where Mv is viscosity average molecular weight), GOx (Aspergillus niger), lipase (Candida rugosa), hydrogen peroxide (H2O2), glucose, o-methoxyphenol, EDC, NADH, sodium pyruvate, 4-nitrophenyl acetate, 4-nitrophenyl phosphate and uranyl acetate are purchased from Sigma-Aldrich (St. Louis, MO). HRP and LDH (Sus scrofa) are purchased from Calzyme Laboratories INC, San Luis Obispo, CA. AP (Triticum aestivum) is purchased from MP Biomedicals, Solon, OH. Agarose is purchased from Molecular Biology Hoefer Inc., Allison, MA. All native enzymes are purified by dialysis. The HEK-293T cell line [ATCC CRL-11268] is purchased from ATCC (Mananas, VA).
000 g mol−1 where Mv is viscosity average molecular weight), GOx (Aspergillus niger), lipase (Candida rugosa), hydrogen peroxide (H2O2), glucose, o-methoxyphenol, EDC, NADH, sodium pyruvate, 4-nitrophenyl acetate, 4-nitrophenyl phosphate and uranyl acetate are purchased from Sigma-Aldrich (St. Louis, MO). HRP and LDH (Sus scrofa) are purchased from Calzyme Laboratories INC, San Luis Obispo, CA. AP (Triticum aestivum) is purchased from MP Biomedicals, Solon, OH. Agarose is purchased from Molecular Biology Hoefer Inc., Allison, MA. All native enzymes are purified by dialysis. The HEK-293T cell line [ATCC CRL-11268] is purchased from ATCC (Mananas, VA).
2.2 Synthesis of MECs
MECs are synthesized using five different enzymes and PAA by two different synthesis routes, which include parallel (2-steps) and sequential synthesis (1-step). Key features of the five enzymes are displayed in Table 1.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Third, the mixture is stirred for 20 min and then stock solution of GOx dissolved in PB is added dropwise such that molar ratio of enzyme
1. Third, the mixture is stirred for 20 min and then stock solution of GOx dissolved in PB is added dropwise such that molar ratio of enzyme![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PAA is 1
PAA is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and total solution volume is 10 mL and stirred for 6 h. Finally, covalent conjugation is achieved through carboxyl groups on PAA and lysine amino groups on enzymes and GOx–PAA conjugates are prepared. In a similar manner the other four enzyme–PAA conjugates are synthesized. In step 2, five different enzyme–PAA conjugate are mixed with EDC and cross-linked. The EDC
1 and total solution volume is 10 mL and stirred for 6 h. Finally, covalent conjugation is achieved through carboxyl groups on PAA and lysine amino groups on enzymes and GOx–PAA conjugates are prepared. In a similar manner the other four enzyme–PAA conjugates are synthesized. In step 2, five different enzyme–PAA conjugate are mixed with EDC and cross-linked. The EDC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) –COOH ratio is maintained at 1.5
–COOH ratio is maintained at 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PAA at 1
PAA at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and stirred for 20 min. These molar ratios are chosen based on literature reports.8,30 To this mixture, EDC is added such that the molar ratio of EDC
1 and stirred for 20 min. These molar ratios are chosen based on literature reports.8,30 To this mixture, EDC is added such that the molar ratio of EDC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) –COOH from PAA is 1.5
–COOH from PAA is 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and the mixture is stirred for 20 min. The sample is dialyzed using a 25 kDa dialysis membrane in pH 7.4 PB to remove unreacted EDC and EDC-urea byproducts. As a control experiment, all five enzymes are mixed together in the absence of PAA, which resulted in a precipitate. Thus, conjugation with PAA is necessary to prevent aggregation and precipitation of the five enzymes. Further experimental details on characterization of MECs by zeta potential, agarose gel electrophoresis, circular dichroism, transmission electron microscopy, dynamic light scattering as well as catalytic activity measurements and kinetics studies is presented in the ESI.†
1 and the mixture is stirred for 20 min. The sample is dialyzed using a 25 kDa dialysis membrane in pH 7.4 PB to remove unreacted EDC and EDC-urea byproducts. As a control experiment, all five enzymes are mixed together in the absence of PAA, which resulted in a precipitate. Thus, conjugation with PAA is necessary to prevent aggregation and precipitation of the five enzymes. Further experimental details on characterization of MECs by zeta potential, agarose gel electrophoresis, circular dichroism, transmission electron microscopy, dynamic light scattering as well as catalytic activity measurements and kinetics studies is presented in the ESI.†3. Results
In this study, a general and modular method by which GOx, HRP, AP, LDH and Lip are conjugated to PAA by parallel and sequential strategies to form MECs, is presented. In the parallel synthetic method, individual enzyme–PAA conjugates are first synthesized (GOx–PAA, HRP–PAA, Lip–PAA, AP–PAA, and LDH–PAA) and then these enzyme–PAA conjugates are further treated with EDC to produce MEC, 5-P. In the sequential synthesis route, all 5 enzymes and PAA are mixed together in specific ratios and then crosslinked covalently with EDC to form MEC, 5-S.3.1 Synthesis of MECs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) –COOH and total enzyme
–COOH and total enzyme![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PAA are maintained at 1.5
PAA are maintained at 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. Random covalent conjugation between PAA and GOx is achieved by reaction through –COOH groups on PAA and lysine NH2 groups on GOx. The conjugation reaction is carried out for 6 h followed by dialysis using 25 kDa dialysis membrane in pH 7.4 PB for 3 cycles of 5 h each. Final concentration of GOx in the GOx–PAA conjugate is 12.5 μM. Other enzyme–PAA conjugates including HRP–PAA, AP–PAA, Lip–PAA and LDH–PAA are similarly synthesized and purified by dialysis. In step 2, GOx–PAA, HRP–PAA, AP–PAA, Lip–PAA and LDH–PAA are mixed in equimolar amounts and stirred for 20 min. Then, EDC is added such that molar ratio of EDC
1, respectively. Random covalent conjugation between PAA and GOx is achieved by reaction through –COOH groups on PAA and lysine NH2 groups on GOx. The conjugation reaction is carried out for 6 h followed by dialysis using 25 kDa dialysis membrane in pH 7.4 PB for 3 cycles of 5 h each. Final concentration of GOx in the GOx–PAA conjugate is 12.5 μM. Other enzyme–PAA conjugates including HRP–PAA, AP–PAA, Lip–PAA and LDH–PAA are similarly synthesized and purified by dialysis. In step 2, GOx–PAA, HRP–PAA, AP–PAA, Lip–PAA and LDH–PAA are mixed in equimolar amounts and stirred for 20 min. Then, EDC is added such that molar ratio of EDC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) –COOH is 1.5
–COOH is 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. After 6 h, the samples are purified by dialysis to remove unreacted EDC and EDC-urea. Final concentration of each enzyme in 5-P is 6.5 μM.
1. After 6 h, the samples are purified by dialysis to remove unreacted EDC and EDC-urea. Final concentration of each enzyme in 5-P is 6.5 μM.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PAA concentration was 1
PAA concentration was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and stirred for 20 min. Chemical conjugation is initiated by the addition of EDC and multi enzyme conjugate, 5-S is synthesized. Final concentration of each enzyme in the 5-S is 6.5 μM, similar to 5-P. Successful synthesis of enzyme–PAA, 5-P and 5-S was confirmed using agarose gel electrophoresis, zeta potential, and TEM.
1 and stirred for 20 min. Chemical conjugation is initiated by the addition of EDC and multi enzyme conjugate, 5-S is synthesized. Final concentration of each enzyme in the 5-S is 6.5 μM, similar to 5-P. Successful synthesis of enzyme–PAA, 5-P and 5-S was confirmed using agarose gel electrophoresis, zeta potential, and TEM.3.2 Agarose gel electrophoresis
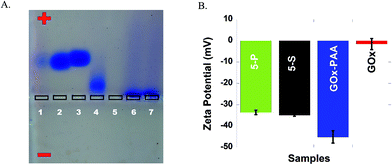
Successful conjugation of enzymes to PAA and formation of MECs is confirmed by agarose gel electrophoresis studies at pH 7.4 in 40 mM Tris acetate buffer. In Fig. 1A, lanes 1–7 are loaded with Lip, GOx, LDH, AP, HRP, 5-P and 5-S, respectively. Agarose gel data for single enzyme–PAA conjugates is shown in Fig. S1, ESI.† Electrophoretic mobility of enzymes and enzyme-conjugates will depend on the charge and the molecular weight. Native enzymes GOx, AP, Lip and LDH move towards the positive electrode due to the net negative charge on these enzymes.36 The staining of HRP in the gel is very poor and its electrophoretic properties could not be established. The MECs will have high molecular weight and may remain in the loading well without any movement through the gel. The absence of any other bands pertinent to free enzymes and single enzyme–PAA conjugates in 5-S and 5-P under electrophoresis confirms formation of MECs using PAA as a scaffold.3.3 Zeta potential studies
The charge and the electrophoretic mobility of the conjugates and native enzymes at pH 7.4 PB are established using zeta potential measurements. When enzymes conjugate with PAA to form either single enzyme–PAA conjugates or MECs, the net negative charge on these samples increase relative to unmodified enzymes due to the presence of unreacted COOH in PAA. The zeta potential of GOx and GOx–PAA, 5-P, and 5-S is determined to be −1.50 mV, −45.0, −33.5 and −35.0 mV, respectively, Fig. 1B. Similarly, HRP, AP, Lip, and LDH show zeta potentials of −2.00, −5.22, −14.0 and −20.0 mV, respectively, Fig. S2 ESI.† As expected, the potentials decrease considerably in enzyme–PAA conjugates. HRP–PAA, AP–PAA, Lip–PAA, and LDH–PAA show zeta potentials of −46.5, −42.5, −44.0 and −40.0 mV, respectively, Fig. S2 ESI.†3.4 Transmission electron microscopy (TEM)
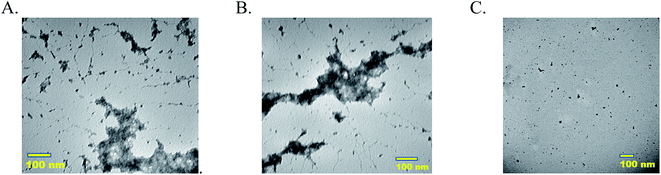
Morphologies of MECs, single enzyme–PAA and unmodified native enzymes are analyzed using TEM. Samples 5-P, 5-S, and GOx are imaged using Tecnai T20 TEM. Samples 5-P and 5-S showed extensive crosslinked nanogels whereas GOx (C) showed aggregated particles, Fig. 2(A–C). The free enzyme when drop cast tend to aggregate and precipitate easily. The nano gel morphology of GOx–PAA has been previously reported.83.5 Dynamic light scattering (DLS)
The hydrodynamic radii of MECs, enzyme–PAA, and native enzymes are determined by DLS. All samples are diluted using pH 7 PB such that the enzyme concentration in each DLS experiment is maintained at 0.5 nM. The enzymes GOx, HRP, AP, Lip, and LDH show hydrodynamic radii of 6.02, 1.84, 9.10, 1.49 and 3.44 nm, respectively. The hydrodynamic radius of GOx–PAA, HRP–PAA, AP–PAA, Lip–PA and LDH–PAA are determined to be 22.3, 26.8, 32.6, 16.7 and 16.6 nm, respectively. The MECs, 5-P and 5-S, show structures of 80–100 nm, possibly due to the random nature of conjugation and incorporation of different sized enzymes, Table S1 and Fig. S3 in ESI.†3.6 Circular dichroism (CD) studies
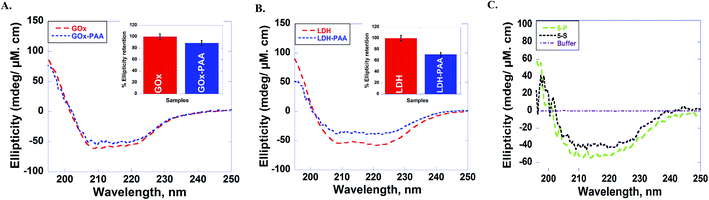
The secondary structure of enzymes before and after conjugation with PAA in pH 7.4 PB at room temperature is established using far UV circular dichroism (CD) studies. Due to the physical strain imposed on the enzyme upon PAA chemical conjugation, enzyme–PAA conjugates are expected to present reduced molar ellipticities compared to unmodified native enzymes.The CD spectra of GOx, GOx–PAA (A); LDH, LDH–PAA (B) and 5-P, 5-S (C) is shown in Fig. 3. The inset in panel A & B shows the relative % ellipticity retention of enzyme–PAA and corresponding unmodified enzyme. To calculate relative % ellipticity retention, signal intensity of samples at 222 nm is used. The signal intensity at that wavelength for unmodified enzyme is set at 100% and accordingly the relative intensity of enzyme–PAA is calculated. The CD spectra and relative % ellipticity retention of AP, AP–PAA (A), HRP, HRP–PAA (B) and Lip, Lip–PAA (C) are presented in Fig. S4, ESI.† To calculate the relative % ellipticity, retention signal intensity at lowest position in CD spectra, which is 222 nm for GOx, LDH, and AP and 208 nm for Lip and HRP. Relative % ellipticity retention of the enzymes within the single-enzyme PAA showed reduction in retention of secondary structures compared to unmodified enzyme. GOx–PAA, LDH–PAA, AP–PAA, HRP–PAA, and Lip–PAA show reduction in % relative ellipticity retention of 89, 71, 68, 66, and 4%, respectively, relative to the native enzymes' ellipticity of 100%.
3.7 Activity studies
Catalytic activities of SECs and MECs are carried out at room temperature using enzyme specific substrates, assays and experimental conditions. Enzymes including GOx, HRP, AP, LDH, and Lip use glucose, o-methoxyphenol, 4-nitrophenyl phosphate, NADH, and 4-nitrophenyl acetate, respectively, as substrates to estimate catalytic features. Detailed catalytic assays are presented in the ESI.† Relative % specific activity of each enzyme conjugate is calculated by fixing the initial rate of its corresponding unmodified enzyme as 100% at room temperature.Similar trends are obtained in the case of Lip, where Lip–PAA, 5-P, and 5-S show activities of 114, 89, and 450%, respectively, relative to the native enzyme at room temperature and pH 7.4 PB. This unprecedented increase in activity of AP and Lip conjugates may be due to increase accessibility of the catalytic active site to the substrate. In general, the synthetic approach to stabilize AP and Lip with PAA, improved activities of AP in conjugates by 18-fold and activities of Lip in conjugates by 4.5-fold at room temperature relative to the native enzymes at room temperature and pH 7.4 Tris. However, in contrast, HRP and LDH present within single enzyme–PAA and MECs show decrease in catalytic activities relative to the native enzymes at room temperature and pH 7.4 PB and pH 8 Tris, respectively. For HRP conjugates, HRP–PAA, 5-P, and 5-S show activities of 60, 49, and 54%, respectively, whereas LDH conjugates, LDH–PAA, 5-P, and 5-S showed activities of 38, 57, and 40%, respectively, assuming that the native enzymes show 100% activity at room temperature and pH 8 Tris. The % specific activities of Lip, HRP and LDH are shown in Fig. S6 in ESI† and Table 2.
| Samples | % specific activity |
|---|---|
| Lip enzymatic assay | |
| 5-P | 89 |
| 5-S | 450 |
| Lip–PAA | 114 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| HRP enzymatic assay | |
| 5-P | 49 |
| 5-S | 54 |
| HRP–PAA | 60 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| LDH enzymatic assay | |
| 5-P | 57 |
| 5-S | 40 |
| LDH–PAA | 38 |
| Samples | % specific activity (65 °C) |
|---|---|
| Lip enzymatic assay | |
| 5-P | 0 |
| 5-S | 540 |
| Lip–PAA | 78 |
| Lip | 0 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| HRP enzymatic assay | |
| 5-P | 98 |
| 5-S | 138 |
| HRP–PAA | 149 |
| HRP | 47 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| LDH enzymatic assay | |
| 5-P | 0 |
| 5-S | 77 |
| LDH–PAA | 146 |
| LDH | 0 |
Catalytic activity studies at 65 °C and pH 7.4 PB are established for MECs and single enzyme conjugates. For example, while 5-P, 5-S, and GOx–PAA showed 132, 112, and 136% activity (Fig. 5A), native GOx exhibited complete deactivation. In each case, the percentage specific activity of the conjugate at 65 °C is relative to 100% activity for the corresponding native enzyme at room temperature and pH 7.4 PB. Similarly, 5-P, 5-S, and AP–PAA present 103, 298, and 106% activity (Fig. 5B), whereas unmodified AP is deactivated and unmodified AP presents 100% activity at room temperature and pH 7.4 PB. Biochemical assays at 65 °C for Lip, HRP, and LDH show that 5-S present enhanced catalytic activities compare to 5-P. These trends in catalysis may be due to microenvironment in MECs produced due to the synthetic strategy and may allow the substrate better access to active sites in 5-S compared to 5-P leading to enhanced activities in 5-S. Additionally, in 5-S, domains of similar enzymes may not form easily due to the synthetic strategy. However, cooperative effects from dissimilar enzymes may further enhance overall activities in these MECs.
The effect of the polymer scaffolding and the microenvironment of the enzymes in 5-S, 5-P, enzyme–PAA on enzymatic rate constants is evaluated using Lineweaver–Burk plots. Kinetic constants including KM, Vmax, catalytic efficiency and Kcat, are extracted and compared to the native enzymes. The data are presented below.
3.8 Kinetic studies
| 1/V = KM/Vmax[S] + 1/Vmax |
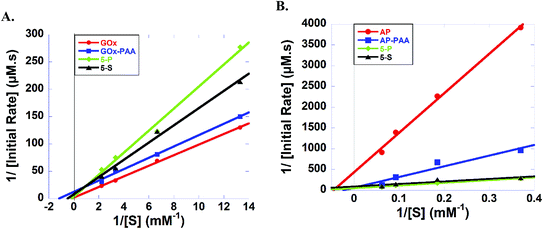
Lineweaver–Burk plots for HRP containing samples at room temperature and pH 7.4 PB are designed and KM values for 5-P, 5-S, HRP–PAA, and HRP are calculated to be 0.796, 0.330, 0.461, and 0.107 mM, respectively (Fig. S8A and Table S2†). Reduced catalytic efficiency for 5-P and 5-S are noted, 0.155 and 0.097, respectively, whereas HRP–PAA and HRP showed similar catalytic efficiency of 0.308 and 0.318, respectively. In the case of Kcat, 5-P and HRP–PAA showed similar values of 0.123 and 0.142 s−1, respectively, while, 5-S and HRP presented values of value of 0.032 and 0.034 s−1, respectively.
Kinetic parameters AP containing samples at room temperature and pH 7.4 PB are established (Fig. 6 and Table S2†). KM values of 5-P, 5-S, AP–PAA, and AP are, 12.9, 6.55, 43.0 and 21.9 mM, respectively. However, 5-P and 5-S showed highest Kcat and catalytic efficiency values. For example, Kcat values of 5-P and 5-S are 0.020 and 0.011 s−1, respectively, and catalytic efficiency values are 1.55 × 10−3 and 1.68 × 10−3, respectively. Kcat value for AP–PAA is 0.020 s−1 and catalytic efficiency is 3.96 × 10−4. In agreement with room temperature catalytic activity trends, AP shows the lowest Kcat and catalytic efficiency of 0.002 s−1 and 9.11 × 10−5, respectively.
Similarly, Lineweaver–Burk plots for LDH containing enzymes are used to derive the kinetic parameters at room temperature in pH 8 Tris (Fig. S8B and Table S2†). KM values for 5-P, 5-S, LDH–PAA, and LDH are, 0.196, 0.196, 0.073, and 0.198 mM, respectively and catalytic efficiencies are 0.036, 0.031, 0.096, and 0.364, respectively. Turn over number or Kcat values for 5-P, 5-S, and LDH–PAA are similar and show values of 0.007, 0.006, and 0.007 s−1, respectively, whereas, LDH exhibits highest Kcat of 0.072 s−1.
Lineweaver–Burk plots for Lip are shown in Fig. S8C and Table S1.† Accordingly, KM values of 5-P, 5-S, Lip–PAA, and Lip are 0.133, 0.070, 0.122, and 0.372 mM, respectively, and catalytic efficiencies are 0.007, 0.014, 0.016, and 0.013, respectively. 5-P, 5-S, and Lip–PAA exhibited similar Kcat values of 0.001, 0.001, and 0.002, respectively, whereas Lip exhibited highest turnover number (Kcat) of 0.005 s−1.
3.9 Cellular toxicity of MECs and enzyme–PAAs in HEK-293T cells
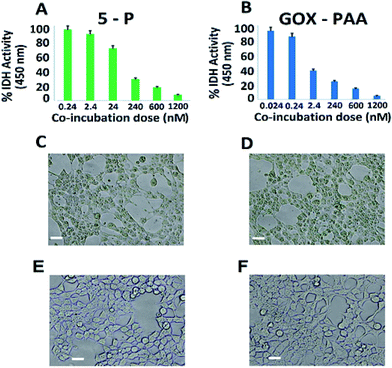
The in vitro compatibility of the enzyme–PAA conjugates with HEK-293T cells up to 16 h of co-incubation period is monitored by measuring the intracellular dehydrogenase activity, for 5-P (Fig. 7A) and GOx–PAA (Fig. 7B), which is a reliable measure for quantifying the intracellular metabolism in a mammalian cell line.38,39 Optimal intracellular dehydrogenase concentration is essential for efficient metabolism in the human cells. The reduction in dehydrogenase concentration, which is a quantitative measure of reduced intracellular metabolism, confirmed the reduction in cellular activity and eventually, the cell death. The cell viability is also examined by evaluating their extracellular morphology using phase contrast microscopy, a universal benchmark for assessing the overall physiological health of a cell,40 for 5-P (Fig. 7C), GOx–PAA (Fig. 7D) and Lip–PAA (Fig. 7E). It is evident that cells incubated with 1.2 μM Lip–PAA showed normal morphology (Fig. 7E), whereas cells incubated with 5-P and GOx–PAA at low nanomolar and picomolar levels, respectively, showed stressed cell morphology as seen in Fig. 7C and D. Fig. S9 in ESI† shows the changes in intracellular metabolism as the dose of Lip–PAA is varied from 0–1.2 μM. Similar behavior is noted for AP–PAA, HRP–PAA and LDH–PAA (data not shown).Except for the conjugates that contained GOx, no other conjugate showed any difference in intracellular metabolism or extracellular morphology relative to control cells even at the highest co-incubation dose (1.2 μM, Fig. 7D). Similar behavior is observed for the conjugates containing GOx, such as 5-P, 5-S, and GOx–PAA at low nanomolar dose (2–20 nM). In sum, the toxicity results obtained by monitoring intracellular dehydrogenase activity and light microscopy demonstrated that most enzyme–polyacrylic acid conjugates are highly biocompatible.
4. Discussion
A general and modular strategy to synthesize novel, synthetic multi-enzyme polymer chemical conjugates using different enzymes and PAA as the polymer scaffold is presented here. This strategy is developed based on the polymer's ability to conjugate one or two enzymes and protect the enzymes from stressors including temperature, aggregation and proteases as shown in previous publication.8,30,32,44 The novelty of this work arises due to the use of PAA scaffold, to conjugate multiple enzymes of varying isoelectric points, pH profiles, physical properties and catalytic properties. We also wish to investigate the temperature stability of MECs not only at room temperature but also at 65 °C compared to unmodified native enzymes. The catalytic properties presented by MECs cannot be realized from a mixture of physical mixtures of multiple enzymes, which aggregate and precipitate and lose all catalytic activity.Two distinct synthetic strategies, the parallel and sequential synthesis, are tested here. In parallel synthesis, in the first step enzyme–PAA conjugates are made via the classical covalently crosslinking of lysine amino (NH2) groups from the enzyme with carboxyl (–COOH) groups of PAA. In the second step, these individual enzyme–PAA conjugates are mixed together and further conjugated to form 5-P MEC. In the sequential one-step one-pot synthesis, all 5 enzymes and PAA are mixed together, without any visible precipitation, and crosslinked to form 5-S MEC. More importantly, microenvironments of the enzymes in 5-P and 5-S are expected to be different. In 5-P inter-penetrating domains of similar enzyme–PAA conjugates may be formed due to the synthetic protocol, while in 5-S domains of similar enzyme–PAA conjugates may not be easily attained as these are pre-mixed before crosslinking. This difference in microenvironment of 5-P and 5-S will result in very different catalytic behaviors, and this concept has never been examined before. Furthermore, the single pot strategy to covalently immobilize five different enzymes within PAA can be easily extended to thirteen or more enzymes of different pIs useful for cascade reactions including sugar to energy applications.
Successful syntheses of 5-P, 5-S, and single enzyme–PAA conjugates are confirmed using agarose gel electrophoresis, TEM, DLS, and zeta potential studies. Agarose gel electrophoresis studies are based on the size and charge of the samples. The MECs carry more negative charge due to carboxyl groups from PAA and they are larger in size compared to individual enzymes (Fig. 1A). Increased negative charge should propel the MECs towards positive electrode of the gel but the increased size of the MECs can hamper its movement. As shown in Fig. 1A, due to increase in size, the MECs were stuck in the well of the agarose gel with some streaking (lane 6 and 7). Agarose gel of the immobilized enzymes conjugated with PAA (MEC 1 and 2) indicated that all the enzymes are covalently conjugated to the polymer and hardly any unreacted enzymes or single enzyme conjugates are present in the gels. However, it is hard to estimate the concentration of the enzymes in the MECs due to their overlapping UV signatures. The concentration of the individual enzyme in the MEC is calculated based on amount of enzyme used in the synthesis protocol and quantitative crosslinking of each enzyme is expected.
TEM micrographs clearly show nanogel morphologies of 5-P and 5-S similar to reported single enzyme–PAA conjugates, Fig. 2A and B, whereas the native enzyme, GOx for example, showed discreet aggregated particles. Due to the polyvalence of the PAA as well as the proteins, chemical crosslinking of these units can result in conjugates of a distribution of sizes. The distribution and the average size of the crosslinked conjugates depends on the polymer concentration, protein concentration, EDC concentration as well as the reaction time. For example, when dilute solutions are used, smaller aggregates are expected but when high concentrations are used these smaller aggregates may undergo further cross linking and produce larger aggregates. Here reaction conditions are carefully optimized to obtain nanogels of 80–100 nm. In contrast GOx solution is drop cast onto TEM grids. During the drying process and during imaging, water is removed and GOx will form small aggregates as noted from TEM.
Zeta potential data confirms increase in negative charge after chemical conjugation with PAA, and this trend is consistent among all chemical conjugates (Fig. 1B and S1†). The average hydrodynamic radius of the MECs is determined characterized by DLS (Table S1, ESI†). Both 5-P and 5-S show increase in hydrodynamic size compared to individual enzyme–PAA conjugates as well as the free enzymes. These results are in agreement with the mobility pattern of the conjugates observed by agarose gel electrophoresis.
Retention of secondary structure of MECs is important for biocatalytic activity, which is established using CD and compared to CD spectra of free enzymes and enzyme–PAA conjugates. The CD spectra of the conjugates are compared with those of the unconjugated enzymes or native enzymes (Fig. 3 and also Fig. S2 and S4†). Except in the case of Lip–PAA, the spectra are reasonably similar in intensity as well as peak positions, but not identical. Thus, some structural changes do occur upon conjugation with the polymer. Severe loss in intensity in the case of Lip–PAA is likely due to aggregation but the Lip–PAA conjugate is nearly as active as the free enzyme. Thus, this loss in CD signal cannot be directly attributed to complete loss in enzyme secondary structure.
The secondary structure of each enzyme within 5-P and 5-S could not be compared with the single enzyme conjugates or native enzymes due to difficulty in resolving and deconvoluting the spectra of each enzyme in the MECs. The spectral shapes of 5-P and 5-S are comparable to the single enzyme–PAA conjugates, which may indicate at least partial retention of secondary structure. In the absence of concrete structural retention data from UV and CD studies, enzymatic activities of 5-P and 5-S are investigated and compared with single enzyme–PAA conjugates and free enzymes and is an indirect form of structural retention investigation.
The catalytic activity of one enzyme in the presence of other enzymes within MECs and the effect of method of MEC synthesis (parallel vs. sequential addition) have never been explored before. The interpenetrating domains of a single enzyme–PAA conjugates within the 5-P MEC and the domains in 5-S MEC where the enzymes are mixed together and then conjugated (Scheme 1) could impact enzyme conformational states. The polymer scaffold may also sequester or expel the substrate and play an active role in the catalysis, as demonstrated before.41
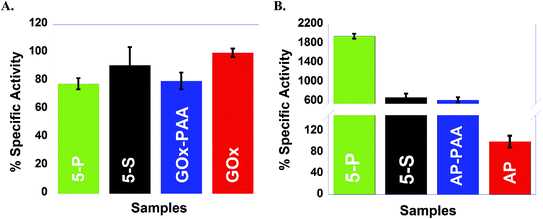
Catalytic activities are determined at room temperature at pH 7.4 PB are established using five different enzymatic assays where each assay is designed to interrogate one of the five enzymes in the MECs. For example, with GOx based assays, 5-P and 5-S show activities similar to GOx–PAA and GOx at room temperature and pH 7.4 PB. In contrast, for HRP and LDH based assays, ∼50% loss in activity is noticed for 5-P and 5-S when compared to pristine enzymes at room temperature and pH 7.4 PB. In another example, enzymatic assays for AP show unprecedented increase in activity for 5-P, 5-S and AP–PAA relative to native AP. However, enzymatic assays for Lip showed moderate increase in catalytic activity for 5-S and Lip–PAA relative to native Lip at room temperature and pH 7.4 PB. These improvements in catalytic activities of AP and Lip conjugates may be attributed to the exposure of the active sites to the substrate.42,43 Contribution of volumetric changes to the catalytic activity profile after conjugation to polymer in single and multi-enzyme fashion is minimal due to the excessive volume of the buffer used during catalysis. The total volume of PAA and all enzymes is small when compared to the volume of the buffer.
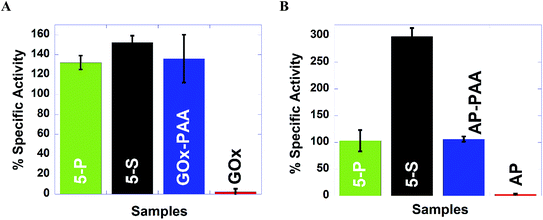
To test the robustness of these MECs, activity studies are performed at 65 °C at pH 7.4 PB. MECs and enzyme–PAA conjugates present improved stability and activity retention compared to the native unmodified enzyme. For example, GOx activity assay indicates more than 100% activity retention for 5-P, 5-S and GOx–PAA at 65 °C at pH 7.4 PB compared to GOx activity at room temperature and pH of 7.4 PB% since free enzyme at 65 °C did not present any catalytic activity. Similar observations are noted for AP and HRP activity assays where 5-P, 5-S, AP–PAA, and HRP–PAA 65 °C at pH 7.4 PB retain more than 100% of the activity relative to the activity of the unconjugated/free enzyme at room temperature and pH 7.4 PB. Both native enzymes AP and HRP present negligible activity at 65 °C. The activity of 5-S in the case of AP and HRP activity assay increase to 300 and 200%, respectively, at 65 °C compared to native enzymes at room temperature. Increase in these activities is only observed for 5-S but not for 5-P. The domain formation in the case of 5-P seems to be an impediment for augmentation of activity at 65 °C. However, domain formation did not negatively impact the activity of 5-P in the case of GOx, AP, or HRP based catalytic assays.
In the case of LDH and Lip activity assays, 5-P did not show any activity at 65 °C and pH 8 Tris and pH 7.4 PB, respectively. However, 5-S in the case of LDH and Lip activity assay showed 77% and 540% of activity, which is significantly better compared than 5-P. The microenvironment in 5-S is different from 5-P and could significantly impact observed difference in catalytic activities for LDH and Lip based assays at 65 °C and pH 7.4 PB. Thus 5-S may be exploited for high temperature catalytic applications as it is easier to synthesize, more robust and catalytic more active at 65 °C compared to 5-P.
To further understand the activity enhancement at room temperature and pH 7.4 PB (GOx, HRP, AP and Lip) and pH 8 Tris (LDH), enzyme kinetics are carried out and systematic changes in KM, Vmax, Kcat, and catalytic efficiencies of 5-P, 5-S, single enzyme–PAA conjugates and native unmodified enzymes are obtained using Lineweaver–Burk plots. For example, in GOx containing samples, KM values of GOx–PAA and 5-S are low compared to KM values of unmodified GOx. In contrast, 5-P shows slightly higher KM compared to GOx. Enzyme samples with lower KM have higher affinity for the substrate. However, lower KM values observed for some of the conjugates may partly explain the retention or enhancement of activity at 65 °C compared to native GOx, which loses its catalytic activity completely at 65 °C. In another example, huge enhancement of activity using AP activity assay for AP–PAA, 5-P, and 5-S can be explained based on KM and catalytic efficiency. At room temperature and pH 7.4 PB, AP–PAA and 5-S had 6 times higher activity and 5-P had 20 times higher activity compared to AP. These correlate well with the catalytic efficiencies of these samples. Also, 5-S and 5-P show higher substrate affinity as their KM values are lower than that of AP at room temperature. These nanogel type enzyme conjugates may pre-concentrate the substrate around the enzyme, which directly enhances substrate affinity and reaction rate.28
Reduction in activity of LDH and HRP containing samples after polymer conjugation and formation of MECs can be explained via changes in KM and catalytic efficiency. Catalytic efficiency decreases in the case of HRP–PAA, 5-P, and 5-S to 0.308, 0.155, and 0.097 from 0.318 of HRP, which directly relates to the reduction in activity of HRP (100%) to (45–60%) for HRP–PAA, 5-P, and 5-S. A similar trend is noted in the case of LDH containing enzyme conjugates. Catalytic efficiency decreases from 0.364 in LDH to 0.03–0.09 in the case of LDH–PAA, 5-P, and 5-S. Similarly, activity retention of LDH–PAA, 5-P, and 5-S is reduced by 40–60% when compared to LDH.
Lip containing samples showed an increase in catalytic efficiencies after polymer conjugation, for example, Lip–PAA and 5-S while 5-P showed a decrease. Catalytic efficiency of Lip (0.013) is increased for Lip–PAA (0.016) and 5-S (0.014), indicating 114% and 450% increase in activity of Lip–PAA and 5-S, respectively. Structure of the lipase active site and its conformation are known to regulate its activity.31 If the enzyme is trapped in its active state during the conjugation, then it is likely that the conjugate will be more active and vice versa. Similarly, the decrease in activity of 5-P (89%) is related to its decrease in catalytic efficiency (0.007) where the enzyme is locked into a less active confirmation.31
There are several examples of multi-enzyme systems that are physically adsorbed or physically encapsulated within a support, which showed enhancement or retention in catalytic activity in cascade or non-cascade catalysis.47–54 While, recyclability of these physically encapsulated system has been optimized and these enzymes can last a few cycles, their catalytic performance at higher temperatures has not been investigated. Catalytic activity is typically enhanced at higher temperatures and strategies to stabilize multiple enzymes at different temperatures are beneficial. In sharp contrast, 5-S and 5-P show significant activity retention or even catalytic activity enhancement in non-cascade or orthogonal biocatalysis at room temperature and at 65 °C compared to native enzymes at room temperature and pH 7.4 PB. Our previous work has shown that chemical confinement of the single enzyme and bi-enzymes by PAA will reduce the overall conformational entropy of the system and results in structure retention while improving thermal stability and catalytic performance at higher temperatures.8 But However, optimization of crosslink densities, molecular weight of PAA, and concentration of enzymes need to be investigated to recycle these enzyme conjugates. Here recyclability studies of MECs are not evaluated but methods to optimize recyclability of these MECs will be investigated in the near future.
To extend the use of these MECs for implantable bioreactor applications in animals, bio-toxicities are tested. The toxicity of enzyme–polyacrylic acid conjugates on HEK-293T cells is evaluated for four to forty-eight hours by quantifying the intracellular dehydrogenase activity and monitoring the cell morphology using phase-contrast optical microscopy. The results show no toxic effects of enzyme–PAA conjugates in HEK-293T cells up to 1.2 μM dose except for GOx containing samples like GOx–PAA, 5-P, and 5-S, which show toxicity at low nanomolar levels and MECs (5-P and 5-S) which showed toxicity at low picomolar levels. The toxicity of GOx containing samples can be attributed to the glucose oxidase mediated conversion of D-glucose (4.5 g L−1) present in the cell culture media into H2O2 and D-glucose-1,5-lactone which gets further hydrolyzed to D-gluconic acid. The catalytic generation of H2O2 and gluconic acid significantly lowers the pH of cell culture media from its normal physiological range (pH 7.4–7.6, blood red) to the acidic range (pH 6.0–6.8, yellow) resulting in metabolic acidosis in HEK-293 cells. Due to the excellent stability of enzyme–PAA conjugates, the GOx containing PAA-conjugates and MECs are able to completely overwhelm the intrinsic bicarbonate buffer system present in the cell culture media. This, in turn, resulted in an acidic environment adversely affecting the cell survivability.
To our knowledge, this is the first study that reports the biocompatibility of enzyme–PAA and MECs in a well-established human cell line for 48 hours. Our results strongly indicate that not only these complexes are highly stable in the complex biological growth medium, but also they are non-toxic. These conjugates, therefore, may be considered for implantable devices.
5. Conclusions
This is the first report in which 5 enzymes with different pI values and catalytic properties are stabilized using functional polymer to form multi-catalytic enzyme complexes (MECs) using two distinct strategies. This is a general and modular strategy that can be extended to many enzymes as most enzymes contain amino groups that can covalently conjugate with hydrophilic PAA, which serves as a universal scaffold. In the parallel synthesis protocol (5-P), individual enzyme–PAA complexes are synthesized first and these were further crosslinked by EDC chemistry. In sequential method of synthesis (5-S), all 5 enzymes are mixed with PAA and then crosslinked using EDC. In 5-S, the enzyme structures are locked-in by the polymer shroud before exposing to other enzyme–polymer conjugates while in 5-P, each enzyme has the potential to interact with other enzymes as well as PAA during the conjugation. Thus, in 5-P microdomains of each-enzyme polymer networks are formed which are interpenetrated during the second step. In 5-S, these microdomains are less likely to form, unless certain enzymes aggregate individually or in the presence of specific partners, aggregation/precipitation is absent, and hence, enzymes in 5-S may be more homogeneously dispersed. These MECs are used for room and high temperature catalysis. Room temperature activity studies showed that MEC formation enhanced the AP and Lip activities of 5-P and 5-S by 5–20 folds when compared to the activities of corresponding unmodified enzymes. In contrast, MEC formation had no effect on GOx activity of 5-P and 5-S. At high temperature, all unmodified enzymes nearly lost all their activities, except HRP. In contrast, 5-P and 5-S showed 1 to 3 fold increase in activity for AP, GOx and HRP, whereas LDH and Lip showed peculiar behavior where 5-P completely lost activity and 5-S retained 70% in the case of LDH and 540% activity in the case of Lip. In principle, the enzymatic activities are tunable by controlling the affinity of the substrates to the polymer scaffolding surrounding the enzymes, which can promote substrate binding by increasing the local concentrations of the substrates near the enzyme. In the case of cellulosome, the cellulose-binding domain enhances the catalytic activities by increasing the affinity of the multi-catalytic complex with cellulose. Our studies are only preliminary in this context, but they point to possibilities for tuning the enzymatic activities by controlling the synthesis method and composition of the MECs in a rational manner. The general method of making robust, active, artificial multi-catalytic complexes of multiple enzymes of different pI values opens up new avenues for high temperature catalysis and low-toxicity biocatalysts for implantable biofuel cells.Acknowledgements
C. V. K acknowledges NSF EAGER grant (DMR-1441879) for financial support of the work. C. V. K and R. M. K thank the University of Connecticut Research Foundation, Research Excellence Program Award 2015. This research was also partially funded by NIEHS grant ES023350 to AKB.References
- C. M. Fontes and H. J. Gilbert, Annu. Rev. Biochem., 2010, 79, 655–681 CrossRef CAS PubMed.
- P. A. Santacoloma, G. Sin, K. V. Gernaey and J. M. Woodley, Org. Process Res. Dev., 2011, 15, 203–212 CrossRef CAS.
- Bio Based Economy, http://www.bio-economy.net/applications/applications_enzymes.html, accessed February 25, 2016.
- N. Gurung, S. Ray, S. Bose and V. Rai, BioMed Res. Int., 2013, 2013, 1–18 CrossRef PubMed.
- S. Li, X. Yang, S. Yang, M. Zhu and X. Wang, Comput. Struct. Biotechnol. J., 2012, 2, 1–10 Search PubMed.
- W. J. Sung and Y. H. Bae, Sens. Actuators, B, 2006, 114, 164–169 CrossRef CAS.
- Z. Zhu, T. K. Tam, F. Sun, C. You and Y. H. P. Zhang, Nat. Commun., 2014, 5, 1–8 CrossRef.
- H. Klok, J. Polym. Sci., Part A: Polym. Chem., 2005, 43, 1–17 CrossRef CAS.
- C. Boyer, X. Huang, M. R. Whittaker, V. Bulmus and T. P. Davis, Soft Matter, 2010, 7, 1599–1614 RSC.
- K. Heredia and H. D. Maynard, Org. Biomol. Chem., 2007, 5, 45–53 CAS.
- A. Abuchowski, T. Van Es, N. Palczuk and F. Davis, J. Biol. Chem., 1977, 252, 3578–3581 CAS.
- D. Bontempo, K. L. Heredia, B. A. Fish and H. D. Maynard, J. Am. Chem. Soc., 2004, 126, 15372–15373 CrossRef CAS PubMed.
- F. Veronese and G. Pasut, Drug Discovery Today, 2005, 10, 1451–1458 CrossRef CAS PubMed.
- V. Thilakarathne, V. A Briand, R. M. Kasi and C. V. Kumar, Langmuir, 2011, 27, 7663–7671 CrossRef CAS PubMed.
- V. Mudhivarthi, K. Cole, M. Novak, K. Westley, I. Deshapriya, Y. Zhou, R. M. Kasi and C. V. Kumar, J. Mater. Chem., 2012, 22, 20423–20433 RSC.
- O. Zore, P. J. Lenehan, C. V. Kumar and R. M. Kasi, Langmuir, 2014, 30, 5176–5184 CrossRef CAS PubMed.
- O. V. Zore, A. Pattammattel, S. Gnanaguru, C. V. Kumar and R. M. Kasi, ACS Catal., 2015, 5, 4979–4988 CrossRef CAS.
- X. Cao, Y. Li, Z. Zhang, J. Yu, J. Qian and S. Liu, Analyst, 2012, 137, 5785–5791 RSC.
- A. Heller and B. Feldman, Chem. Rev., 2008, 108, 2482–2505 CrossRef CAS PubMed.
- C. Huber, M. Preis, P. J. Harvey, S. Grosse, T. Letzel and P. Schroder, Chemosphere, 2016, 146, 435–441 CrossRef CAS PubMed.
- J. Gong, C. S. Lee, E. J. Kim, Y. Y. Chang and Y. S. Chang, J. Hazard. Mater., 2016, 310, 135–142 CrossRef CAS PubMed.
- Y. Zhang, Y. Yong, J. Ge and Z. Liu, ACS Catal., 2016, 6, 3789–3795 CrossRef CAS.
- X. Ji, Z. Su, P. Wang, G. Ma and S. Zhang, ACS Catal., 2014, 4, 4548–4559 CrossRef CAS.
- N. G. Karaguler, R. B. Sessions and B. Binay, Biochem. Soc. Trans., 2007, 35, 1610–1615 CrossRef CAS PubMed.
- M. J. Kim and G. M. Whitesides, J. Am. Chem. Soc., 1988, 110, 2959–2964 CrossRef CAS.
- A. Chaubey, M. Gerard, R. Singhal, V. S. Singh and B. D. Malhotra, Electrochim. Acta, 2001, 46, 723–729 CrossRef.
- M. Claire, Bacterial acid phosphatase and its application to waste remediation and metal recovery, PhD thesis, University of Birmingham, UK, 2011.
- H. Tanaka, Y. Horiuchi and K. Konishi, Anal. Biochem., 1975, 66, 489–497 CrossRef CAS PubMed.
- N. N. Gandhi, Applications of Lipase, J. Am. Oil Chem. Soc., 1997, 74, 621–634 CrossRef CAS.
- C. J. Gray, J. S. Narang and S. A. Barker, Enzyme Microb. Technol., 1990, 12, 800–807 CrossRef CAS PubMed.
- S. Negishi, S. Shirasawa, Y. Arai, J. Suzuki and S. Mukataka, Enzyme Microb. Technol., 2003, 32, 66–70 CrossRef CAS.
- R. Fernandez-Lafuente, J. Mol. Catal. B: Enzym., 2010, 62, 197–212 CrossRef CAS.
- J. Huang, Y. Huang, X. Wu, W. Du, X. Yu and X. Hu, Parasitol. Res., 2009, 104, 287–293 CrossRef PubMed.
- L. Nguyen, T. Dao, T. Zivkovic, M. Fehrholz, W. Schaefer and S. Salomon, Enzyme Microb. Technol., 2010, 46, 479–486 CrossRef CAS PubMed.
- G. Q. Zhang, Q. J. Chen, J. Sun, H. X. Wang and C. H. Han, J. Basic Microbiol., 2013, 53, 868–875 CAS.
- G. Terlecki, E. Czapinska and K. Hotowy, Cell Mol. Biol. Lett., 2007, 12, 378–395 CAS.
- T. Akiyama, H. Uchimiya and H. Suzuki, Plant Cell Physiol., 1981, 22, 1023–1028 CAS.
- Y. C. Liou, A. G. Marangoni and R. Y. Yada, Food Res. Int., 1998, 31, 243–248 CrossRef CAS.
- A. Ghimire, O. V. Zore, V. K. Thilakarathne, V. A. Briand, P. J. Lenehan, Y. Lei, R. M. Kasi and C. V. Kumar, Sensors, 2015, 15, 23868–23885 CrossRef CAS PubMed.
- O. V. Zore, P. J. Lenehan, C. V. Kumar and R. M. Kasi, Langmuir, 2014, 30, 5176–5184 CrossRef CAS PubMed.
- Y. Liu, Y. Luo, J. Wu, Y. Wang, X. Yang, R. Yang, B. Wang, J. Yang and N. Zhang, Sci. Rep., 2013, 3, 1–8 Search PubMed.
- A. P. Pattammattel, C. L. Williams, P. Pande, A. K. Basu and C. V. Kumar, RSC Adv., 2015, 5, 59364 RSC.
- B. Ekwall, V. Silano, A. Paganuzzi-Stammati and F. Zucco, Toxicity tests with mammalian cells, in Short-term Toxicity Tests for Non-genotoxic Effects, ed. P. B., John Wiley and Sons Ltd., 1990, ch. 7 Search PubMed.
- C. M. Riccardi, K. S. Cole, K. R. Benson, J. R. Ward, K. M. Bassett, Y. Zhang, O. V. Zore, B. Stromer, R. M. Kasi and C. V. Kumar, Bioconjugate Chem., 2014, 25, 1501–1510 CrossRef CAS PubMed.
- B. Abel and K. Aslan, J. Colloid Interface Sci., 2014, 415, 133–142 CrossRef CAS PubMed.
- E. H. Lee, T. Tsujimoto, H. Uyama, M. H. Sung, K. Kim and S. Kuramitsu, Polym. J., 2010, 42, 818–822 CrossRef CAS.
- Y. Zhang, Y. Yong, J. Ge and Z. Liu, ACS Catal., 2016, 6, 3789–3795 CrossRef CAS.
- Q. Menga, Y. Zhanga, X. Jua, C. Maa, H. Maa, J. Chena, P. Zhenga, J. Suna, J. Zhua, Y. Maa, X. Zhaob and T. Chenb, J. Biotechnol., 2016, 226, 8–13 CrossRef PubMed.
- W. Jiang and B. Fang, Sci. Rep., 2016, 6, 30462 CrossRef CAS PubMed.
- Q. Jia, B. Wanga, J. Tanb, L. Zhua and L. Lia, Process Biochem., 2016, 51, 1193–1203 CrossRef.
- A. Alshammari, M. G. Posner, A. Upadhyay, F. Marken, S. Bagby and A. Ilie, ACS Appl. Mater. Interfaces, 2016, 8, 21077–21088 CAS.
- S. Kale, G. Ulas, J. Song, G. W. Brudvig, W. Furey and F. Jordan, Proc. Natl. Acad. Sci. U. S. A., 2008, 4, 1158–1163 CrossRef PubMed.
- P. A. Santacoloma, G. Sin, K. V. Gernaey and J. M. Woodley, Org. Process Res. Dev., 2011, 15, 203–212 CrossRef CAS.
- L. Betancor, M. Fernández, K. J. Weissman and P. F. Leadlay, ChemBioChem, 2008, 9, 2962–2966 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supporting information file contains experimental details and supplemental data as noted in the text. See DOI: 10.1039/c7ra05666d |
| This journal is © The Royal Society of Chemistry 2017 |