Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Increasing the sensitivity and selectivity of a GONS quenched probe for an mRNA assay assisted with duplex specific nuclease†
Jialong Fan‡
a,
Xizhi Zhang‡a,
Yanxiang Cheng‡b,
Changhui Xiaoa,
Wei Wangc,
Xuanming Liua,
Chunyi Tong *a and
Bin Liu
*a and
Bin Liu *a
*a
aCollege of Biology, Hunan Province Key Laboratory of Plant Functional Genomics and Developmental Regulation, Hunan University, Changsha, 410082, China. E-mail: binliu2001@hotmail.com; sw_tcy@hnu.edu.cn; Fax: +86-731-89720939; Tel: +86-731-89720939
bDepartment of Obstetrics and Gynecology, Renmin Hospital of Wuhan University, Wuhan, 430060, China
cDongguan Research Center, Guangdong Medical University, Dongguan, 523808, China
First published on 19th July 2017
Abstract
The authors report a new graphene oxide nanosheet (GONS) based fluorescence method for mRNA assay with duplex-specific nuclease (DSN)-assisted signal amplification. Following hybridization between the target mRNA and its complementary DNA probe, the ssDNA in the DNA/mRNA hybrid is selectively cleaved by DSN to produce small fragments. The released mRNA molecule then initiates another cycle of hybridization and DSN digestion. In this manner, each mRNA molecule can specifically trigger many cycles of hybridization and DNA cleavages to produce numerous small DNA fragments. The short DNA fragments can exhibit strong fluorescence signals due to the weak adsorption of GONS to them. This fluorescence assay for mRNA with increased selectivity has a 1 fM detection limit under optimal conditions. Furthermore, this assay was successfully used for mRNA imaging in situ.
1 Introduction
Many studies have revealed that some genes, which possess different expression patterns between tumors and normal tissues, can act as biomarkers for early clinical diagnosis and therapy assessment.1–4 Among of them, p21, a cell cycle regulatory protein, plays a very important role in the regulation of cell proliferation and tumorigenesis.5 As an antitumor gene, the level of p21 mRNA is significantly decreased in the tumor tissues and cell lines compared with normal cells.6,7 Thus, p21 mRNA can be used as a biomarker for tumor diagnosis, clinical therapy and prognosis evaluation.Until now, several efficient methods have been established for mRNA assay including northern blotting, RT-PCR and gene chip etc.8,9 As a standard method of mRNA assay, northern blot method is complicated, time-consuming, large amount of samples demand and easy to produce false positive or negative results. Recently, several new methods based on nucleic acid amplification significantly have improved the sensitivity of mRNA detection. For example, qRT-PCR, molecular beacon detection technology (MBs), rolling ring amplification technology (RCA), etc., have been used for mRNA analysis.9–11 Although these methods improved the specificity and sensitivity in some extent, all of them introduced complicated primer or probe design with high cost. Thus, developing a simple, ultrasensitive, highly selective and cost-effective biosensing platform for mRNA assay is still of great significance for biological assays and clinical diagnostics.
Graphene oxide nanosheet (GONS) is a two-dimensional carbon material with excellent electrical, thermal and mechanical properties.12 The surface of this kind of nanomaterial contains many functional groups on the basal plane and edges, rendering it amenable to stably disperse in the water. Until now, it has been widely used for biosensor,13,14 bioimaging,15,16 cellular image and drug delivery, etc.17–19 As biosensors for molecules detection, these new systems can overcome the shortcomings of traditional methods through the following characteristics: GONS can directly adsorb long single-stranded oligonucleotides labeled with fluorophore and efficiently cause fluorescence quench through hydrophilic and π–π interaction, while the duplex formation through hybridization can reduce the adsorption capacity of GONS to DNA strands and fluorescent signal recovery through DNA strands release form GONS surface.20–22 Furthermore, GONS can be used as a high efficient carrier and protector of DNA probe for living cell image by inhibiting nucleases unspecific digestion.23–25
On the basis of the previous work, we further proposed a method based on GONS/SP-DSN (graphene oxide nanosheet/single-stranded probe-duplex specific nuclease) sensing system for mRNA assay in vitro and in vivo. At the presence of mRNA, the SP with its targets formed duplex through hybridization. Thus, the fluorescence probe detained from GONS surface and the fluorescence signal was primarily recovered. The addition of DSN triggered many cycles of hybridization and DNA cleavages that amplified the fluorescence signal to realize the ultra-sensitive and quantitative mRNA detection. By systematically studying the interaction between GONS and different DNA/RNA duplex via theoretical simulation and experiment performance, we found that GONS could bind and quench fluorophore-labeled ssDNA/RNA probe. Meanwhile, the formation of DNA/RNA duplex led to the release of the probe from GONS and fluorescence signal recovery. By using the hybridization, digestion and rehybridization manner, the GONS-based sensing platform can offer a simple and ultrasensitive method for various kinds of molecules with improved performance.
2 Experimental
2.1 Materials
The GO powder was purchased from Xianfeng Nanotechnology Co., Ltd (Nanjing, China). All other chemicals were of analytical grade used without further purification. MilliQ Water was used for the whole procedure. Oligonucleotides, listed in the (Table 1), were synthesized and purified by HPLC (Takara Biotechnology Inc., Dalian). All kinds of cell lines were purchased form the cell library of Xiangya Central Laboratory, Central-South University. Human gastric cancer and normal tissue samples were obtained from the Third Xiangya Hospital, Central-South University. Fluorescence measurement was performed on the Hitachi F-4500 Fluorescence spectrophotometer. AFM measurement was carried out on the Nanoscope IIIa (Digital Instrument, USA) under tapping mode, with a Pt/Ti coated tip (force constant of 4 N m−1, MicroMasch). A droplet of GONS dispersion was cast onto a freshly cleaved mica surface and the sample was maintained at room temperature for several minutes to allow water evaporation. The image was obtained at 20 °C with humidity of 30%.| Name | Sequence (5′–3′) | Base numbers |
|---|---|---|
| P1 | TTTTTTTTTAGGACACATGGGGAGCCGA-FAM | 28 |
| P2 | CACAACAGGACACATGGGGAGCCGAGTrUGTG-FAM | 31 |
| Random | TTCTTTCTTCCCTTGTTTGTT-FAM | 21 |
| MB | DABCYL-CACAACAGGACACATGGGGAGCCGAGTrUGTG-TAMRA | 31 |
| T1 | r(CUCGGCUCCCCAUGUGUCCU) | 20 |
| mT1 | r(CUCGGCUCCaCAUGUGUCCU) | 20 |
| mT2 | r(CUCGGtUCCaCAUGUGUCCU) | 20 |
| mT3 | r(CUCGGtUCCaCAUGUcUCCU) | 20 |
| P21-primer-1 | ACCTTCCAGCTCCTGTAACATACT | 24 |
| P21-primer-2 | GTCTAGGTGGAGAAACGGGAA | 21 |
| β-actin-primer-1 | TCCGTGGAGAAGAGCTACGA | 20 |
| β-actin-primer-2 | GTACTTGCGCTCAGGAGGAG | 20 |
2.2 Optimization of reaction time between oligonucleotides and GONS
The commercial graphene oxide was treated using probe sonication and bath sonication in purified water for 2 h to get a highly dispersible and homogeneous graphene oxide nanosheet. The products were stored at 4 °C for use and stability observation. The working solution of fluorescence probe was obtained by diluting the stock solution to a concentration of 100 nM using 20 mM Tris–HCl buffer (pH 8.0). To optimize the interacting time between fluorescence probe and GONS, 1 μL of the probe solution (100 nM), and 2.7 μg GONS were mixed. The mixture was diluted with Tris–HCl buffer to a final volume of 100 μL and incubated for 0, 1, 5, 10, 15, 20, 25, and 30 min at room temperature. Then, the fluorescence intensities of samples at these time points were measured with fluorescence spectrophotometer.2.3 Optimization of hybridizing time
To optimize the reaction time between DNA/mRNA hybrid and DSN, 1 μL of the probe solution (100 nM), T1 (20 nM) and 1 μL DSN solution (0.2 units) were mixed in the 47 μL buffer. The mixture solutions were incubated for 0, 5, 10, 15, 20, 25, 30, 35 and 40 min at 45 °C, respectively. Then, 2.7 μg GONS was added. The mixture was diluted with a Tris–HCl buffer to 100 μL, and incubated for 10 min at room temperature. Finally, the fluorescence intensities of these samples were measured with fluorescence spectrophotometer.2.4 Fluorescence assay
In the fluorescence quenching and hybridization assays, the fluorescent probe 1 (P1) and probe 2 (P2) with final concentration of 100 nM were hybridized with target T1 in the buffer (100 mM NaCl, 10 mM PB, 10 mM MgCl2, pH 7.4) for 10 min. In the mRNA detection methods of self-assembled graphene oxide complex of GONS/P1 and GONS/P2, GONS of 1 μL (1.5 mg mL−1) were added into hybridization buffer. After incubation for 15 min, the fluorescences of solutions were measured using Hitachi FL-4500 fluorescence spectrophotometer with excision at 450 nm and emission range from 500 to 580 nm. Spectrometer slits were set for 10 nm band-pass. In the concentration gradient assay, the fluorescent probe P1 (100 nM) were hybridized with target T1 of 1, 5, 10, 20, 50 and 100 nM, respectively for 10 min, to which GONS of 1 μL (1.5 mg mL−1) added. After incubation for 15 min, fluorescence measurements were performed with same spectrophotometer. The specificity of the new method was investigated through monitoring the signal change of the fluorescent probe (100 nM) caused by the target or base mismatch targets (50 nM). All other operation steps in the mRNA detection methods of fluorescence probes and self-assembled graphene oxide complex of GONS/P1 were similar to the above described.2.5 Cell culture, total RNA extraction and qRT-PCR
Human tumor cells were cultured in DMEM medium supplemented with 10% fetal calf serum, and penicillin–streptomycin. Total cellular RNA was isolated from culture cells using TRIzol reagent (Invitrogen) and concentrations were measured by ultraviolet absorbance at 260 and 280 nm and subsequently stored in aliquots at −80 °C. Real-time PCR was performed using the LightCycler Instrument (Roche Applied Science) in a total volume of 50 μL per PCR tube. For each reaction, 1 μL of cDNA was placed in a 20 μL Platinum SYBRgreen qPCR Supermix (Takara) containing 0.1 μL of a temperature-released Taq DNA polymerase (5 U mL−1; Platinum DNA Polymerase; Takara), 0.5 μL of the primers, and 14.5 μL DEPC-treated water. The cycling protocol was identical for p21 mRNA and consisted of an initial 5 min denaturation step at 95 °C for activation of the DNA polymerase, followed by 45 cycles of denaturation at 95 °C for 15 s, annealing at 63 °C for 15 s, and extension at 72 °C for 20 s.2.6 Polyacrylamide gel electrophoresis and silver staining
15% polyacrylamide gel was prepared by adding 3 mL ddH2O, 5 mL 30% acrylamide, 2 mL × 5 TBE, 70 μL 10% ammonium persulfate and 4 μL TEMED into centrifuge tube, the mixture was quickly poured into glue. Electrophoresis was carried out at room temperature for 1 h at a constant volt of 100 V. Finally, the separated products in the gel was visualized by silver staining.3 Results and discussion
3.1 Design of a fluorescence-based method for mRNA assay
The principle of the GONS-based fluorescence assay for mRNA is illustrated in the Scheme 1. p21 mRNA as a proof-of-concept target, a single-stranded DNA probe containing a poly T tail, which was completely complementary to the p21 mRNA sequences, was firstly synthesized. The two-stage mRNA assay contains four key components of target mRNA, a FAM-labeled linear probe, single-stranded DNA specific nuclease (DSN) and water-dispersed graphene oxide nanosheet (GONS). We initially used a linear probe labeled with FAM, which forms a well characterized ‘FRET pair’ with GONS and the binding of GONS with probe in the absence of target results in fluorescence quenching of FAM fluorophore. In contrast, in the presence of p21 mRNA, the ssDNA probe hybridizes with target to form DNA/RNA heteroduplexes and produces weak signal restoration. However, the addition of DSN can selectively recognize and cleave the ssDNA in the DNA/RNA hybrid to produce very small DNA fragments. The p21 mRNA fragment will be released and initiate another round of hybridization and DSN digestion. In this manner, each target mRNA can specifically trigger various cycles of hybridization and DSN cleavage events, thus allowing sensitive detection of target mRNA using this kind of signal amplification manner.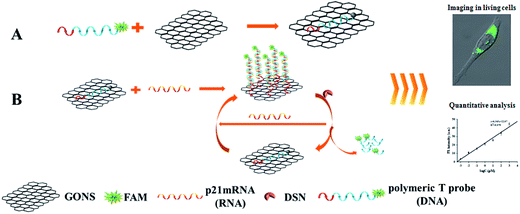 | ||
| Scheme 1 Schematic presentation of nano-MBs with GONS-enhanced quenching efficiency. The poly T tailed linear DNA has stronger infinity with GONS than hairpin or dsDNA. | ||
3.2 Characterization of graphene oxide nanosheet
After synthesizing GO using natural graphite powder by a modified Hummers method, both probe sonication and bath sonication was further used in the exfoliation of this kind of 2D layered material due to the reason that the use of only bath sonication often resulted in irregular graphene oxide nanosheet.26 Atomic force microscopy was used to directly quantify the degree of exfoliation to a single graphene sheet level after with probe and bath sonication treatment. The AFM image of the prepared GO was indicated in the Fig. 1. According to the section analysis, the thickness of the prepared was about 2–4 nm (Fig. 1A). However, the thickness of graphene oxide with probe sonication reduced to 1.5 nm (Fig. 1B), which matched well with the reported thickness of single-sheet graphene oxide. Furthermore, the obtained nanosheet graphene oxide could be dispersed in the water and TE buffer without any noticeable aggregation at room temperature for 4 days. As a control, obvious aggregation was appeared in the graphene oxide solution (Fig. 1C). From these results, it can be concluded that we have prepared a highly dispersed suspension of nanosheet graphene oxide with a lateral size of 150–200 nm and a thickness of approximately 1.5 nm by combing probe sonication with bath sonication.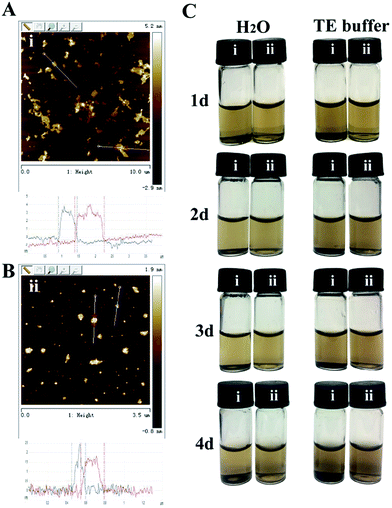 | ||
| Fig. 1 AFM images and the corresponding height profiles of GO with probe sonication (B) or not (A). (C) The dispersion stability of GO with probe sonication (ii) or not (i). | ||
3.3 Fluorescence quenching efficiency of GONS
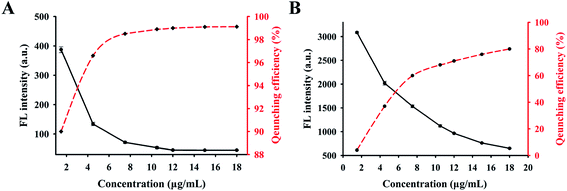
To investigate the potential use of GONS as “nanoquencher” of fluorophore and select a more sensitive probe for target assay, two kinds of pure linear and hairpin structured probe (P1 and P2) were designed and labeled with FAM or TAMRA, respectively. By comparing the fluorescence signal of these probes labeled with different fluorophore, we found that the signal of the two kinds of probe labeled with FAM fluorophore was significantly higher than those of TAMRA labeled probes, while no difference was observed when they were incubated with excess amount of GONS. Thus, FAM labeled probes (P1 and P2), which emitted strong fluorescence signals were used for the following experiment. Firstly, we compared the fluorescence quenching efficiency of P1 and P2 at the presence of different concentrations of GONS. The result indicated that quenching efficiency of the two probes gradually increased as the increasing of GONS concentration. Up to more than 99% quenching efficiency of P1 was observed when the GONS concentration was at 15 μg mL−1. However, only about 75% quenching efficiency was observed for P2 under the same condition (Fig. 2). We suspected that dramatic fluorescence difference mainly arose from the efficient differentiation ability of GONS to them, which could be accurately define the structure effect on the interaction of GONS with DNA using the ratio of Nunpaired bases/(Nunpaired bases + Nbase pairs).27 According to this ratio, the theoretic quenching efficiency of P2, a hairpin structure probe is 76%, which is very similar to the practical result (75%). In summary, these results demonstrate the highly fluorescence quenching efficiency of GONS for the linear probe rather than that hairpin structure probe.3.4 Effect of concentration of Mg2+ on the fluorescence quenching
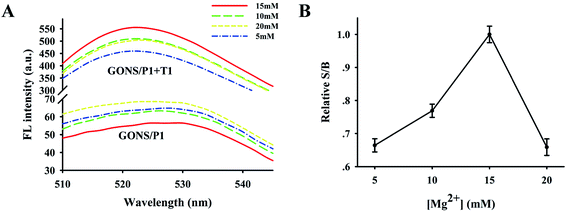
After identifying this GONS-based sensing platform for target mRNA assay, we investigated the effect of Mg2+ concentration on the fluorescence quenching of GONS and the interaction of GONS/P1 with target. From the fluorescence curves of Fig. 3A, it was found that the pure GONS/P1 signal was influenced by Mg2+ concentration and the background signal of GONS/P1 showed an inverse “V” shape upon the increase of Mg2+ from 5 to 20 mM when there was no target. We further evaluated the influence of Mg2+ on the assay by analyzing the signal to background ratio (S/B), defined as S/B = (Fhybrid − Fbuffer)/(FGONS/P1 − Fbuffer), where Fhybrid, Fbuffer and FGONS/P1 were the fluorescence intensities of the probe-target hybrid, the plain buffer solution and the pure probe, respectively. As shown in the Fig. 3, the highest S/B ratio was obtained at 15 mM Mg2+ due to the low background and increased hybrid signal of the probe. We also investigated the effect of Mg2+ on the S/B of GONS/P2 and obtained similar results (data not shown). These data suggested that the presence of stem structure could increase the background signal and thus lead to a small S/B. While the linear probe greatly improved the S/B ratio and consequently increased the sensitivity for target mRNA assay.3.5 Detection sensitivity of the assay
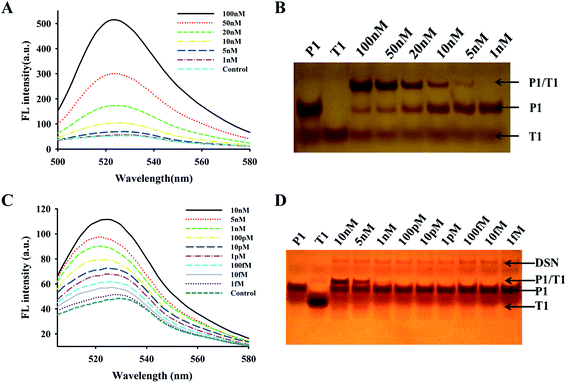
To determine the potential use of the new established system for mRNA assay, we compared the fluorescence restoration capability caused by target mRNA in the presence of DSN or not. Firstly, we employed S/B again to reflect the fluorescence restoration without DSN (Fig. S1†). As we expected, a significant fluorescence signal increase was observed after the addition of excess T1 mRNA when there was no DSN (Fig. 4A). A dramatic increase of fluorescence intensity was observed as the target concentration varied from 1 nM to 100 nM. The detection limit (determined to be three times the standard deviation in the control buffer) was 0.5 nM, which was 4-fold higher than that of the GONS-hairpin probe.27 However, the sensitivity of this approach cannot fulfill the requirement of sample quantification in the bio-lab or clinic diagnosis for the low detection limit. We further investigated the fluorescence change of the proposed approach in the presence of DSN. We firstly investigated the effect of DSN concentration on the reaction and found that the maxiumum signal was obtained under the presence of 0.2 U DSN (Fig. S2†). We then investigated the signal change by varying T1 concentration. As our expected that a dramatic increase of fluorescence intensity was observed when T1 concentration was increased from 1.0 fM to 10 nM at the presence of 0.2 U DSN (Fig. 4C). The target concentration and the corresponding fluorescence value had a good linear relationship in the range from 1.0 fM ∼ 1.0 nM, which was indicated in Fig. S3.† According to the standard curve, we obtained a regression equation of y = 6.365x + 22.07, the corresponding regression coefficient (R2) is 0.979, where y represents the fluorescence intensity of 522 nm, x indicates the value of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (C shows different T1 concentration in pM). The detection limit was 1.0 fM (estimated from three times the standard deviation in the blank solution). Comparing with the above direct assay system, the sensitivity increased about 5 orders magnitude with the aid of DSN. This data suggested that the proposed approach was appropriate for sensitive quantification of mRNA. In order to confirm the reliability of the assay, we further detected the products of the assay using PAGE method. From Fig. 4B and D, it was found that the duplex of DNA/RNA increased as the upregulation of target concentration, while smear bands were only produced with the presence of DSN (Fig. 4D). This result further confirmed the reliability of the fluorescence detection system.
C (C shows different T1 concentration in pM). The detection limit was 1.0 fM (estimated from three times the standard deviation in the blank solution). Comparing with the above direct assay system, the sensitivity increased about 5 orders magnitude with the aid of DSN. This data suggested that the proposed approach was appropriate for sensitive quantification of mRNA. In order to confirm the reliability of the assay, we further detected the products of the assay using PAGE method. From Fig. 4B and D, it was found that the duplex of DNA/RNA increased as the upregulation of target concentration, while smear bands were only produced with the presence of DSN (Fig. 4D). This result further confirmed the reliability of the fluorescence detection system.
3.6 Selectivity investigation of GONS-quenched probe for mRNA assay
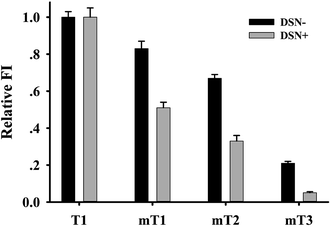
The high specificity is the major advantage of hairpin structure probe.28,29 We also studied whether the proposed dual amplification approach had similar ability to discriminate the perfect and mismatch targets. The complementary target (T1) and the mismatch targets containing 1, 2 and 3 mismatch bases (mT1, mT2 and mT3) were used as detection objects. Fig. S4† and 5 displayed relative fluorescence intensity of GONS-P1 in the presence of these targets with or without DSN, respectively. From the two figures, it was found that all targets differentially caused the fluorescence increase of GONS/P1, while the fluorescence enhancement by the mT1, mT2 and mT3 were 75%, 50% and 20% of that caused by the complimentary T1, respectively. This data indicates that the linear probe can discriminate base mismatch. It has been reported that the existence of mismatch base pair in the duplex can significantly reduce the efficiency of DSN digestion.30 We also found that the fluorescence signal caused by mT1 and mT2 was only 50%, 25% of that by T1, nearly no fluorescence increase was observed for mT3 after introducing DSN for the assay (Fig. 5). These data clearly demonstrated that the introduction of DSN significantly improved base mismatch discrimination ability and accordingly increased selectivity of the assay.3.7 p21 mRNA detection in complicated biosamples
The origin of tumor cell had been shown to be a potent regulator of p21 mRNA expression level.31,32 We wanted to determine whether this proposed approach could accurately quantify p21 mRNA levels with different cell origin. Four different tumor cell lines were selected and total RNA extracted from these cells was subjected to the GONS-probe based assay and the result was indicated in Fig. S5.† From this figure, it was found that the level of p21 mRNA decreased according to the sequence of BT-549, SMMC-7721, QBC-939 and HCT-116, respectively. Among of them, the quantitative test showed that the level of BT-549 cell was about 4-fold higher than that of HCT-116 in (Table 2).| Tumor cell lines | p21 mRNA (10−13 mol μg−1) |
|---|---|
| BT-549 | 5.18 |
| QBC-939 | 3.83 |
| SMMC-7721 | 2.64 |
| HCT-116 | 1.28 |
| Tumor cell lines | p21 mRNA (10−13 mol μg−1) | |
|---|---|---|
| Normal | Gastric cancer | |
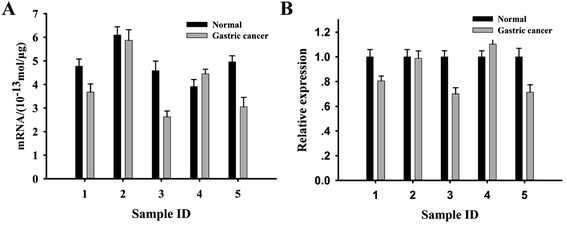
| 1 | 4.78 | 3.67 |
| 2 | 6.09 | 5.87 |
| 3 | 4.59 | 2.62 |
| 4 | 3.91 | 4.45 |
| 5 | 4.96 | 3.05 |
It had been reported that p21 mRNA was generally lower in the tumor tissues than that of normal control.33 We then investigated whether the new approach could be used to discriminate the difference of p21 mRNA in tissue samples. 5 pair gastric tissues were selected and the total RNA extracted from these tissue samples were subjected to the fluorescence assay. The result in Fig. 6A showed that a different extent decrease of p21 mRNA in the 4 patients (80%) compared with the corresponding adjacent tissues. Only one patient didn't show fluorescence signal change between the tumor and adjacent tissue. Meanwhile, the result also confirmed that p21 mRNA level was related with the personal origin tissue. According to the standard curve (Fig. S3†), we obtained p21 mRNA level of the investigated cell lines and tissue samples quantitatively (Table 2). We further measured p21 mRNA levels of these tissue samples using qRT-PCR method, the results are indicated in Fig. 6B. By comparing the results in Fig. S5† and Fig. 6A and B, it was found that the p21 mRNA levels of these tissue samples observed by the two methods are consistent. However, the new proposed method showed the significant advantages of simplicity, high speed and low cost comparing with classical RT-PCR method.
3.8 p21 mRNA image in situ using GONS/P1-DSN system
It has been reported that GONS, a highly efficient carrier for delivering DNA into cells, can strongly protect DNA from nuclease digestion.24,25,34,35 In order to further wide the capability of the new method, we used it to monitor p21 mRNA level in vivo by using BT-549 cells as a model. After incubation of BT-549 cells with GONS/P1 for 4 h under 45 °C, we observed a weak green fluorescence signal in tumor cells, which indicated the successful entry of GONS/P1 into the cells and hybridization with p21 mRNA. Almost no fluorescence signal was observed in the control sample. However, GONS/P1-DSN co-incubating with BT549 cells showed stronger fluorescence signal comparing with that of cells without DSN (Fig. 7K vs. 7I). This result clearly indicated that DSN effectively amplified the fluorescence signal in the cell, which is urgent for mRNA assay and early diagnosis in vivo, especially for the identification of cells infected with low abundance RNA virus.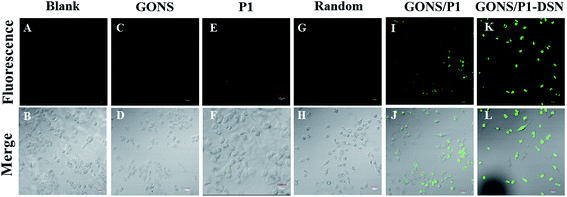 | ||
| Fig. 7 p21 mRNA image in situ BT-549 cells were incubated with GONS/P1 at the presence of DSN or not. | ||
4 Conclusions
In summary, this study designed a simple, high sensitive and specific method for mRNA assay using GONS labeled single-stranded DNA probe and DSN, a specific enzyme amplification cascade cycle. Based on the GONS/P1-DSN sensing system, the purpose of mRNA sensitive detection was achieved, the detection limit reached 1 fM. In addition, the sensing system can not only detect mRNA in living cells specifically, but also is hopeful for monitoring the dynamic change of mRNA in situ. Based on these characteristics, this assay system will be helpful for enhancing the understanding of mRNA function in a wide range of biological processes, as well as for the early diagnosis of disease and new drug development.Acknowledgements
This work was partially supported by the Natural Science Foundation of Hunan Province (h14JJ2049), the Natural Science Foundation of China (81374062, 85673179 and 31672457), the Science Foundation for Outstanding Young Scholars of Hunan Province (2015JJ1007) and the Fundamental Research Funds for the Central Universities of China (2015JCA03).References
- D. Sidransky, Nucleic acid-based methods for the detection of cancer, Science, 1997, 278(5340), 1054–1058 CrossRef CAS PubMed.
- H. Schwarzenbach, D. S. Hoon and K. Pantel, Cell-free nucleic acids as biomarkers in cancer patients, Nat. Rev. Cancer, 2011, 11(6), 426–437 CrossRef CAS PubMed.
- S. Li, R. Li, M. Dong, L. Zhang, Y. Jiang, L. Chen, W. Qi and H. Wang, High-throughput, selective, and sensitive colorimetry for free microRNAs in blood via exonuclease I digestion and hemin-G-quadruplex catalysis reactions based on a “self-cleaning” functionalized microarray, Sens. Actuators, B, 2016, 222, 198–204 CrossRef CAS.
- Y. Si, Z. Sun, N. Zhang, W. Qi, S. Li, L. Chen and H. Wang, Ultrasensitive electroanalysis of low-level free microRNAs in blood by maximum signal amplification of catalytic silver deposition using alkaline phosphatase-incorporated gold nanoclusters, Anal. Chem., 2014, 86(20), 10406–10414 CrossRef CAS PubMed.
- A. L. Gartel and S. K. Radhakrishnan, Lost in transcription: p21 repression, mechanisms, and consequences, Cancer Res., 2005, 65(10), 3980–3985 CrossRef CAS PubMed.
- S. K. Rayala, P. R. Molli and R. Kumar, Nuclear p21-activated kinase 1 in breast cancer packs off tamoxifen sensitivity, Cancer Res., 2006, 66(12), 5985–5988 CrossRef CAS PubMed.
- S. Roy, M. Gu, K. Ramasamy, R. P. Singh, C. Agarwal, S. Siriwardana, R. A. Sclafani and R. Agarwal, p21/Cip1 and p27/Kip1 are essential molecular targets of inositol hexaphosphate for its antitumor efficacy against prostate cancer, Cancer Res., 2009, 69(3), 1166–1173 CrossRef CAS PubMed.
- G. S. Pall, C. Codony-Servat, J. Byrne, L. Ritchie and A. Hamilton, Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot, Nucleic Acids Res., 2007, 35(8), e60 CrossRef PubMed.
- H. D. VanGuilder, K. E. Vrana and W. M. Freeman, Twenty-five years of quantitative PCR for gene expression analysis, Biotechniques, 2008, 44(5), 619 CrossRef CAS PubMed.
- L. Qiu, C. Wu, M. You, D. Han, T. Chen, G. Zhu, J. Jiang, R. Yu and W. Tan, A targeted, self-delivered, and photocontrolled molecular beacon for mRNA detection in living cells, J. Am. Chem. Soc., 2013, 135(35), 12952–12955 CrossRef CAS PubMed.
- M. M. Ali, F. Li, Z. Zhang, K. Zhang, D. K. Kang, J. A. Ankrum, X. C. Le and W. Zhao, Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine, Chem. Soc. Rev., 2014, 43(10), 3324–3341 RSC.
- H. Wang, S. Li, Y. Si, N. Zhang, Z. Sun, H. Wu and Y. Lin, Platinum nanocatalysts loaded on graphene oxide-dispersed carbon nanotubes with greatly enhanced peroxidase-like catalysis and electrocatalysis activities, Nanoscale, 2014, 6(14), 8107–8116 RSC.
- H. S. Jung, M. Y. Lee, W. H. Kong, I. H. Do and S. K. Hahn, Nano graphene oxide–hyaluronic acid conjugate for target specific cancer drug delivery, RSC Adv., 2014, 4(27), 14197–14200 RSC.
- T. H. Kim, I. Song, D. H. Lee and J. W. Choi, Live cell biosensing platforms using graphene-based hybrid nanomaterials, Biosens. Bioelectron., 2017, 94, 485–499 CrossRef CAS PubMed.
- H. Y. Kim, F. Li, J. Y. Park, D. Kim, J. H. Park, H. S. Han, J. W. Byun, Y. S. Lee, J. M. Jeong and K. Char, In vivo visualization of endogenous miR-21 using hyaluronic acid-coated graphene oxide for targeted cancer therapy, Biomaterials, 2017, 121, 144–154 CrossRef PubMed.
- C. Zhao, J. Fan, L. Peng, L. Zhao, C. Tong, W. Wang and B. Liu, An end-point method based on graphene oxide for RNase H analysis and inhibitors screening, Biosens. Bioelectron., 2017, 90, 103–109 CrossRef CAS PubMed.
- W. Xu, L. Peng, B. Li, Z. Xie, C. Tong and B. Liu, A real time S1 assay at neutral pH based on graphene oxide quenched fluorescence probe, Sens Biosensing Res., 2016, 7, 42–47 CrossRef.
- L. Peng, J. Fan, C. Tong, Z. Xie, C. Zhao, X. Liu, Y. Zhu and B. Liu, An ultrasensitive fluorescence method suitable for quantitative analysis of mung bean nuclease and inhibitor screening in vitro and vivo, Biosens. Bioelectron., 2016, 83, 169–176 CrossRef CAS PubMed.
- J. Chen, J. Ge, L. Zhang, Z. Li and L. Qu, Poly(styrene sulfonate) and Pt bifunctionalized graphene nanosheets as an artificial enzyme to construct a colorimetric chemosensor for highly sensitive glucose detection, Sens. Actuators, B, 2016, 233, 438–444 CrossRef CAS.
- C. Zhang, Y. Yuan, S. Zhang, Y. Wang and Z. Liu, Biosensing platform based on fluorescence resonance energy transfer from upconverting nanocrystals to graphene oxide, Angew. Chem., Int. Ed., 2011, 50(30), 6851–6854 CrossRef CAS PubMed.
- Y. Wang, L. Tang, Z. Li, Y. Lin and J. Li, In situ simultaneous monitoring of ATP and GTP using a graphene oxide nanosheet-based sensing platform in living cells, Nat. Protoc., 2014, 9(8), 1944–1955 CrossRef CAS PubMed.
- J. Chen, J. Ge, L. Zhang, Z. Li, J. Li, Y. Sun and L. Qu, Reduced graphene oxide nanosheets functionalized with poly(styrene sulfonate) as a peroxidase mimetic in a colorimetric assay for ascorbic acid, Microchim. Acta, 2016, 183(6), 1847–1853 CrossRef CAS.
- H. Zhang, H. Zhang, A. Aldalbahi, X. Zuo, C. Fan and X. Mi, Fluorescent biosensors enabled by graphene and graphene oxide, Biosens. Bioelectron., 2017, 89(Pt 1), 96–106 CrossRef CAS PubMed.
- C. H. Lu, C. L. Zhu, J. Li, J. J. Liu, X. Chen and H. H. Yang, Using graphene to protect DNA from cleavage during cellular delivery, Chem. Commun., 2010, 46(18), 3116–3118 RSC.
- S. R. Ryoo, J. Lee, J. Yeo, H. K. Na, Y. K. Kim, H. Jang, J. H. Lee, S. W. Han, Y. Lee and V. N. Kim, Quantitative and multiplexed microRNA sensing in living cells based on peptide nucleic acid and nano graphene oxide (PANGO), ACS Nano, 2013, 7(7), 5882–5891 CrossRef CAS PubMed.
- D. C. Marcano, D. V. Kosynkin, J. M. Berlin, A. Sinitskii, Z. Sun, A. Slesarev, L. B. Alemany, W. Lu and J. M. Tour, Improved synthesis of graphene oxide, ACS Nano, 2010, 4(8), 4806–4814 CrossRef CAS PubMed.
- F. Li, Y. Huang, Q. Yang, Z. Zhong, D. Li, L. Wang, S. Song and C. Fan, A graphene-enhanced molecular beacon for homogeneous DNA detection, Nanoscale, 2010, 2(6), 1021–1026 RSC.
- C. H. Lu, J. Li, J. J. Liu, H. H. Yang, X. Chen and G. N. Chen, Increasing the sensitivity and single-base mismatch selectivity of the molecular beacon using graphene oxide as the “nanoquencher”, Chem. – Eur. J., 2010, 16(16), 4889–4894 CrossRef CAS PubMed.
- D. M. Zhou, Q. Xi, M. F. Liang, C. H. Chen, L. J. Tang and J. H. Jiang, Graphene oxide-hairpin probe nanocomposite as a homogeneous assay platform for DNA base excision repair screening, Biosens. Bioelectron., 2013, 41, 359–365 CrossRef CAS PubMed.
- W. Lu, Y. Chen, Z. Liu, W. Tang, Q. Feng, J. Sun and X. Jiang, Quantitative Detection of MicroRNA in One Step via Next Generation Magnetic Relaxation Switch Sensing, ACS Nano, 2016, 10(7), 6685–6692 CrossRef CAS PubMed.
- T. Abbas and A. Dutta, p21 in cancer: intricate networks and multiple activities, Nat. Rev. Cancer, 2009, 9(6), 400–414 CrossRef CAS PubMed.
- F. Q. Nie, M. Sun, J. S. Yang, M. Xie, T. P. Xu, R. Xia, Y. W. Liu, X. H. Liu, E. B. Zhang and K. H. Lu, Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression, Mol. Cancer Ther., 2015, 14(1), 268–277 CrossRef CAS PubMed.
- K. F. Macleod, N. Sherry, G. Hannon, D. Beach, T. Tokino, K. Kinzler, B. Vogelstein and T. Jacks, p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage, Genes Dev., 1995, 9(8), 935–944 CrossRef CAS PubMed.
- Y. Wang, Z. Li, D. Hu, C. T. Lin, J. Li and Y. Lin, Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells, J. Am. Chem. Soc., 2010, 132(27), 9274–9276 CrossRef CAS PubMed.
- Y. Wang, Z. Li, T. J. Weber, D. Hu, C. T. Lin, J. Li and Y. Lin, In situ live cell sensing of multiple nucleotides exploiting DNA/RNA aptamers and graphene oxide nanosheets, Anal. Chem., 2013, 85(14), 6775–6782 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra05656g |
| ‡ These authors contributed to the work equally and should be regarded as co-first authors. |
| This journal is © The Royal Society of Chemistry 2017 |