Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Comparative studies on the interaction of nitrofuran antibiotics with bovine serum albumin†
Qiulan Zhanga and
Yongnian Ni *ab
*ab
aState Key Laboratory of Food Science and Technology, Nanchang University, Nanchang 330047, China. E-mail: ynni@ncu.edu.cn
bSchool of Chemistry, Nanchang University, Nanchang 330031, China
First published on 15th August 2017
Abstract
The interactions of typical nitrofuran antibiotics, nitrofurazone (NFZ) and nitrofurantoin (NFT), with bovine serum albumin (BSA) have been studied using fluorescence and FT-IR spectroscopy, circular dichroism (CD), atomic force microscopy (AFM) and molecular docking investigation. The results revealed that the fluorescence of BSA was quenched by nitrofurans with a static quenching mechanism. The conformation of BSA was changed and unfolded with the addition of nitrofurans because the alpha-helix and beta-sheet were varied to beta-turn and random structure. This study also indicated that the impact on the conformational changes of BSA caused by NFZ was more conspicuous than that of NFT. The molecular simulation studies were carried out to clearly describe the nature of the interaction between nitrofurans and BSA.
1. Introduction
The interactions between biomacromolecules and chemicals have drawn increasing interest in recent years.1–3 Serum albumin, a specific biomacromolecule substance, plays important roles in the transport and disposition of different endogenous and exogenous compounds, such as drugs, metabolites, and other biologically active substances, mostly through the formation of noncovalent complexes at typical binding sites, is the major soluble protein in the circulatory system. The ADME (absorption, distribution, metabolism, and excretion) profiles and consequently, the levels of activity and toxicity could be significantly influenced as a result of their binding to serum albumin.4 Furthermore, the conformational changes of serum albumin induced by its interaction with small molecular-weight drugs may affect the biological function of serum albumin as the carrier protein. Consequently, investigation of the binding of drugs and their derivatives to serum albumin has great toxicological and medical importance. Hence, the studies of the binding of small molecules with protein will aid the explanation of metabolism and transporting procedure of the interaction and the interpretation of the relationship between the structure and the function of the protein. Being a kind of serum albumin, bovine serum albumin (BSA) has been extensively studied, owing to its structural homology with human serum albumin and the advantages of readily availability, medical importance, and unusual ligand–binding properties.5,6Nitrofurans, such as nitrofurazone and nitrofurantoin (NFZ and NFT, see formula in Fig. 1), are widely used against common infections, which are pharmaceutically effective as bactericides, schistosomicides, fungicides and amoebicides.7 Nitrofurans have been widely used previously in livestock for therapy and precaution of various gastrointestinal infections caused by protozoa or bacteria and also as growth stimulant.8 Because of their potentially mutagenic and carcinogenic effects on human health,9 the uses of these drugs in food-producing animals are generally prohibited in many countries, except for some special applications. Therefore, the observation and investigation of nitrofurans in feeds may supply an efficient method for filtrating their contamination in feed products, which can result deleterious effect for animals taking on the contaminated materials and subsequently, people expensing products from these food of animal origin. Side-effects with superabundant doses of nitrofuran include central nervous system involvement (excitement, convulsions, tremors, and peripheral neuritis), GI (glycemic index) disturbances, poor weigh gain, and depression of spermatogenesis. Furthermore, different hypersensitivity reactions for human can also be found.10 The EU Commission's Decision on March 13, 2003 found the minimum required performance limit (MRPL) of 1.0 μg kg−1 for nitrofurans; such as NFZ and NFT, in exporting meat and poultry products.11
Although the potential poisonous effects on human health, the uses of nitrofuran antibiotics are still legal in some countries since these compounds are effective veterinaries with relatively low price. The global nitrofuran crisis which broke out during 2002–2015 exposed the frequent presence of the nitrofuran residues in aquaculture and poultry products imported to Europe and America, particularly from some developing countries.12,13
A number of analytical methods for trace nitrofuran veterinary residues in the food of animal origin were developed.14,15 In this study, the fluorescence and FT-IR spectroscopy, the CD (circular dichroism) and the AFM (atomic force microscope) were employed to explore the interactions of nitrofurans, FZD or NFT, with BSA in the simulative physiological conditions. The study supplies a quantitative and qualitative understanding of the effect of the nitrofuran antibiotics on the structure and function of BSA, which would be a useful guideline for further understanding the toxicological action for common infections in animals and humans, and also explain its binding mechanism at molecular level.
2. Experimental
2.1. Instruments and reagents
The fluorescence spectra were measured on a Perkin-Elmer LS-55 luminescence spectrometer (Perkin-Elmer Inc., USA) equipped with a thermostatic bath (Model ZC-10, Ningbo Tianheng Instruments Factory, China) and a 1.0 cm quartz cuvette. The FL Winlab software (Perkin-Elmer) was used to correct the fluorescence data. FT-IR spectra were measured on a Tensor 27 FT-IR spectrometer (Bruker, German) equipped with a deuterated triglycine sulfate (DTGS) detector in the rage of 4000–400 cm−1. Circular dichroism (CD) spectra were recorded with a Biologic MOS 450 CD spectrometer (Bio-Logic, France). For measurement in the far-UV (190–280 nm), a quartz cell with a path length of 1.0 cm was used in nitrogen atmosphere. The atomic force microscopic (AFM) images were acquired by using an AJ-III instrument (Shanghai Aijian Nanotechnology, China) with the tapping mode, which also produced simultaneously the intensity and phase data. Standard silicon cantilevers (spring constant: 0.6–6.0 N m−1) were used at their resonance frequencies (typically 60–150 kHz). All measurements were carried out at room temperature (25 ± 0.5 °C) unless stated otherwise.Stock solutions of 5 × 10−3 mol L−1 NFZ and NFT (Sigma Chemical Co., St. Louis, USA; purity – not less than 99.0%) were prepared by dissolving their crystals (9.91 × 10−3 g and 1.19 × 10−2 g, respectively) in 10 mL dimethyl formamide (DMF). BSA (2 × 10−3 mol L−1) was prepared by dissolving 1.36 g of protein (M = 66 kDa, ≥98%, lyophilized powder; purchased from Sigma-Aldrich Chemical Co. Ltd., and without further purification) in 10.0 mL 5.0 × 10−2 mol L−1 sodium chloride solution and stored at 4 °C. To confirm the purity of the prepared BSA, it was diluted to 2.0 × 10−5 mol L−1, and the measured absorbance value was (0.896) at 278 nm. This was compared with an absorbance of 0.667 from a reference of 1.0 g L−1 (1.47 × 10−5 mol L−1) pure BSA.16 All experimental solutions were adjusted with the Tris–HCl ((hydroxy methyl)amino methane–hydrogen chloride) buffer to pH 7.4. Other chemicals were Analytical grade reagents, and doubly distilled water was used throughout.
2.2. Fluorescence quenching study
Solutions used in the fluorescence experiments were prepared in a cuvette with 3.0 mL of pH 7.4 Tris–HCl buffer containing appropriate amounts of BSA and nitrofurans. BSA was kept at 6.67 × 10−8 mol L−1, NFZ or NFT, as appropriate, was added to this solution in the range of 0.00–2.13 × 10−7 mol L−1 with an interval of 2.67 × 10−8 mol L−1 (in such a way, nine samples were obtained), and the total added volume, including the BSA and nitrofurans, to cuvette was less than 0.1 mL to reduce the volume effect. The well-mixed solutions were allowed to stand for 10 min at 298 K before spectroscopic measurements. The excitation and emission slits were set at 10 nm, while the scanning rate was 1500 nm min−1. Fluorescence spectra (nine) were then measured in the range of 200–550 nm at the excitation wavelength of 280 nm. The above fluorescence experiments were repeated at another two temperatures, 302 K and 306 K, respectively.2.3. FT-IR measurements
All spectra were taken via the attenuated total reflectance (ATR) method with a resolution of 4.0 cm−1 and 60 scans. The FT-IR spectra of BSA (2.5 × 10−6 mol L−1) in the absence and presence of nitrofurans were recorded in the range of 800–2000 cm−1 at pH 7.4 Tris–HCl buffer and room temperature. The molar ratio of nitrofurans to BSA was set to 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.17 The corresponding spectra of buffer solution were measured under the same conditions and taken as blank, which were subtracted to obtain the FT-IR spectra of the sample solution. The secondary structure compositions of BSA and its nitrofuran complexes were estimated by applying the curve-fitted process to fit the Gaussian line shapes to the amide I band of the FT-IR spectral data.18
1.17 The corresponding spectra of buffer solution were measured under the same conditions and taken as blank, which were subtracted to obtain the FT-IR spectra of the sample solution. The secondary structure compositions of BSA and its nitrofuran complexes were estimated by applying the curve-fitted process to fit the Gaussian line shapes to the amide I band of the FT-IR spectral data.18
2.4. Circular dichroism (CD) and resonance light scattering (RLS) studies for the interaction
The CD spectra of BSA (2.5 × 10−7 mol L−1) in the presence of nitrofurans were recorded in the range of 200–250 nm under constant nitrogen flush. The molar ratios of nitrofurans to BSA were 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.19 Each of the obtained CD spectrum was an average of three scans and were corrected by the buffer signal. RLS spectra were obtained by synchronous scanning with the excitation and emission monochromators (Δλ = 0 nm) with a wavelength range of 200–600 nm on the LS-55 luminescence spectrometer.
1.19 Each of the obtained CD spectrum was an average of three scans and were corrected by the buffer signal. RLS spectra were obtained by synchronous scanning with the excitation and emission monochromators (Δλ = 0 nm) with a wavelength range of 200–600 nm on the LS-55 luminescence spectrometer.
2.5. Atomic force microscopy (AFM) measurements
Samples for the AFM imaging were prepared by depositing 10 μL of BSA, NFZ–BSA and NFT–BSA complexes (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on freshly cleaved mica plates separately (muscovite mica was cut into about 1.2 cm × 1.2 cm square pieces as substrate), which were dried overnight.
1) on freshly cleaved mica plates separately (muscovite mica was cut into about 1.2 cm × 1.2 cm square pieces as substrate), which were dried overnight.
2.6. Molecular docking studies
The 3D structure of NFZ/NFT as ligand was generated in Chem3D Ultra 8.0. The crystal structure of BSA with PDB ID 3VO3 was obtained from Protein data bank (http://www.rcsb.org). The docking simulation was performed with the Lamarckian genetic algorithm for 100 docking runs to find visible combination models using the MGL (molecular graphics laboratory) tools 1.5.6 matches with AutoDock software (version 4.2).203. Results and discussion
3.1. Fluorescence spectral analysis
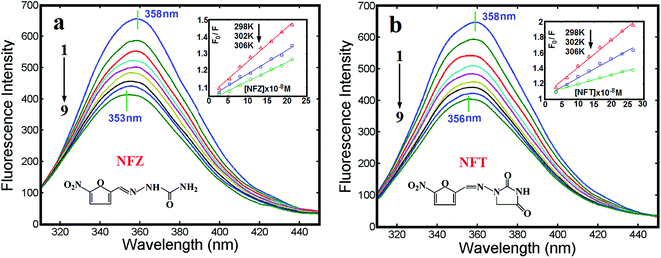
With its non-intrusive and high sensitivity measurement, fluorescence spectroscopy is generally appropriate for low concentration detection under physiological conditions. Usually, the fluorescence emission of BSA is primarily obtained from the residues of tryptophan (Trp), tyrosine (Tyr) and phenylalanine (Phe). In reality, people generally consider the fluorescence of Trp residues as the inherent fluorescence of BSA because of a relatively low quantum yield of phenylalanine and the almost completely quenched of the fluorescence of tyrosine if it is ionized or present near to an amine group, a carboxyl group or a tryptophan.21 Fig. 1 shows the fluorescence quenching spectra of BSA induced by NFZ and NFT with different concentrations in Tris–HCl buffer. The fluorescence change indicated the potentially greater toxicity to BSA at higher concentrations of nitrofurans.22,23 A blue shift of the emission peak for NFZ–BSA (5 nm, Fig. 1a) and NFT–BSA (2 nm, Fig. 1b) systems occurred, which indicated that the microenvironment around the tryptophan and tyrosine residues of BSA was changed, the polarity around the amino acid residues decreased and the hydrophobicity increased.24 Besides, NFZ was seemed to have a greater effect on the structure of BSA than that of NFT.Static and dynamic quenchings are the main processes of fluorescence quenching. The Stern–Volmer equation has been employed (eqn (1)),25 in order to find out the binding mechanism between BSA and nitrofurans:
| F0/F = 1 + Kqτ0[Q] = 1 + KSV[Q] | (1) |
| NFZ/NFT + BSA ↔ NFZ–BSA/NFT–BSA | (2) |
The following equation was employed to survey the equilibrium between the free and bound molecules and treat the experimental data on the nitrofuran–BSA systems:24
log[(F0 − F)/F] = n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka − n Ka − n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[1/([Q] − (F0 − F)[P]/F0)] log[1/([Q] − (F0 − F)[P]/F0)]
| (3) |
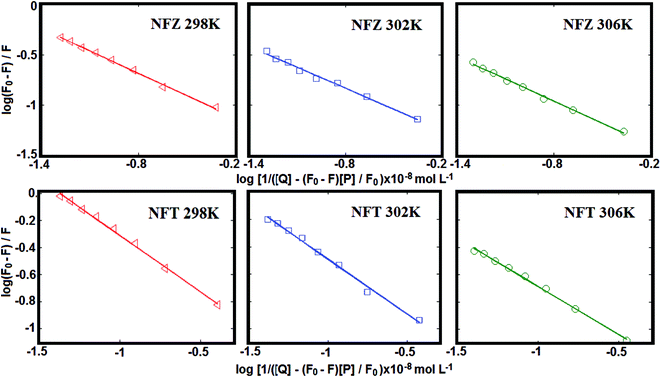 | ||
| Fig. 2 The plots of log(F0 − F)/F versus log[1/([Q] − (F0 − F)[P]/F0)] for the NFZ/NFT–BSA complex systems at 298, 302 and 306 K. | ||
| T (K) | KSV (×106 L mol−1) | R2 | Ka (×106 L mol−1) | n | ΔG (kJ mol−1) | ΔH (kJ mol−1) | ΔS (J mol−1 K−1) |
|---|---|---|---|---|---|---|---|
| NFZ | |||||||
| 298 | 2.01 | 0.9969 | 5.31 | 0.7376 | −38.37 ± 0.12 | −46.95 ± 0.23 | −29.20 ± 0.81 |
| 302 | 1.39 | 0.9959 | 4.13 | 0.7126 | −38.25 ± 0.21 | ||
| 306 | 1.11 | 0.9980 | 2.80 | 0.7375 | −37.77 ± 0.32 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| NFT | |||||||
| 298 | 3.47 | 0.9964 | 7.15 | 0.8351 | −39.10 ± 0.23 | −63.57 ± 0.35 | −81.80 ± 0.62 |
| 302 | 2.34 | 0.9932 | 5.09 | 0.8028 | −38.77 ± 0.16 | ||
| 306 | 1.18 | 0.9927 | 4.08 | 0.7044 | −38.73 ± 0.29 | ||
The synchronous fluorescence spectra of BSA in the presence of NFZ/NFT (Δλ = 60 nm) were shown in Fig. 1S, see ESI.† It was revealed that the fluorescence intensity of BSA was decreased regularly with the increasing addition of NFZ or NFT. Meanwhile, the blue shift (3 nm for NFZ–BSA, and 1 nm for NFT–BSA) for the emission peaks of tryptophan residues were observed, which indicated that the hydrophobicity surrounding tryptophan residues in BSA increased and the polarity was decreased.
3.2. Thermodynamic parameters
The binding forces between ligand and protein include hydrogen bonds, van der Waals interactions, electrostatic forces and hydrophobic interaction forces, which can be determined by the thermodynamic parameters. The enthalpy change (ΔH) and the entropy change (ΔS) for the interaction between nitrofurans and BSA were calculated according to the van't Hoff equation (eqn (4)):28
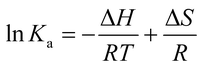 | (4) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka to 1/T (i.e. eqn (4)). To elucidate the interaction between nitrofurans and BSA, the thermodynamic parameters were then estimated from the following relationship:
Ka to 1/T (i.e. eqn (4)). To elucidate the interaction between nitrofurans and BSA, the thermodynamic parameters were then estimated from the following relationship:
ΔG = ΔH − TΔS = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka Ka
| (5) |
As shown in Table 1, the negative ΔG indicated the spontaneity of the binding interactions between nitrofurans with BSA. ΔH and ΔS were found to be −46.95 kJ mol−1 and −29.20 J mol−1 K−1 for NFZ–BSA, −63.57 kJ mol−1 and −81.80 J mol−1 K−1 for NFT–BSA, respectively. The negative ΔH and ΔS indicated that hydrogen bonds and van der Waals interactions played a major role in the binding between NFZ/NFT and BSA quenching process.
3.3. FT-IR spectra of nitrofurans–BSA complexes
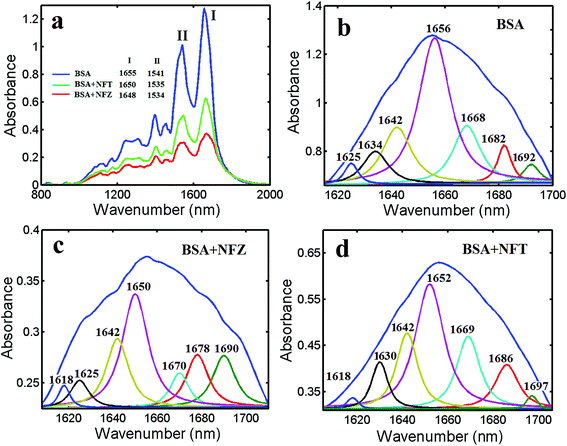
The infrared spectra of proteins are primarily constituted of amide I (1600–1700 cm−1) and amide II (1500–1600 cm−1), which are induced by the carbonyl stretching vibrations of the peptide backbone (amide I band) and the combination of N–H in-plane bending and C–N stretching vibrations of peptides groups (amide II band).29 Generally, the amide I band is more susceptive to change in protein secondary structure than the amide II band. The form of the amide I band is depended on the different secondary structure components, such as α-helix (1650–1660 cm−1), β-sheet (1610–1640 cm−1), β-turn (1660–1680 cm−1), β-antiparallel (1680–1690 cm−1) and random coil (1641–1648 cm−1).30 Conformational changes of proteins are generally occurred during ligand–protein compound formation procedure. The infrared self-deconvolution with second derivative resolution enhancement and curve-fitting procedures could be applied to determine the secondary structure of BSA. Therefore, the interaction between nitrofurans and BSA was represented by FT-IR spectroscopy. In Fig. 3a, the peak position (the amide I band) was shift from 1655 to 1648 (NFZ) and 1650 (NFT) cm−1, and that of amide II band was shift from 1541 to 1634 (NFZ) and 1635 (NFT) cm−1 along with the changed peak intensity and peak shape in accordance with the addition of NFZ and NFT to BSA. The result indicated that NFZ and NFT interacted with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–N groups of the protein polypeptides and led to the reset of the polypeptides carbonyl hydrogen-bonding network.31
O and C–N groups of the protein polypeptides and led to the reset of the polypeptides carbonyl hydrogen-bonding network.31
The quantitative analysis of the free BSA secondary structure and its nitrofurans compounds has been calculated and shown in Fig. 3b–d and Table 2. The α-helix of free protein was 46.97% (1656 cm−1), β-sheet was 11.74% (1634 and 1625 cm−1), β-turn structure was 15.91% (1668 cm−1), β-antiparallel was 8.33% (1692 and 1682 cm−1), and random coil was 17.04% (1642 cm−1). The results agreed well with the spectroscopic studies of BSA previously reported.32 Interaction with nitrofurans, an obvious decrease of α-helix from 46.97% (free BSA) to 35.94% (NFZ–BSA) and 40.40% (NFT–BSA) with a decrease in β-sheet from 11.74% (free BSA) to 9.38% (NFZ), 9.76% (NFT) was observed. An increase was measured for the β-turn structure from 15.91% (free BSA) to 21.88% (NFZ), 18.86% (NFT). Similarly, an analogical increase was also surveyed for the β-anti and random coil for NFZ–BSA and NFT–BSA. The results are also accordance with the reduction in the intensity of the protein amide I band which discussed above. The major decrease in α-helix and β-sheet structure and increase in β-turn and β-anti and random coil structures suggest a partial protein unfolding at high nitrofurans concentrations.
| Compound | α-Helix (%) | β-Sheet (%) | β-Turn (%) | β-Anti (%) | Random coil (%) |
|---|---|---|---|---|---|
| BSA | 46.97 | 11.74 | 15.91 | 8.33 | 17.04 |
| NFZ–BSA | 35.94 | 9.38 | 21.88 | 14.06 | 18.75 |
| NFT–BSA | 40.40 | 9.76 | 18.86 | 12.79 | 18.18 |
3.4. CD spectra analysis
In the period of the interaction of nitrofurans with BSA, the changed in intramolecular or intermolecular forces, which remain the secondary or tertiary structures, might be affected, thus leading to further structure variations.33 CD was conducted to predict the conformational changes in BSA induced through the binding of the nitrofurans.Usually, in order to study the stereo structure of proteins, CD spectroscopy is prevalent in molecular biology study, particularly for secondary structure and variations of their conformation. CD spectroscopy supplies rapid evaluation of the conformational changes and measurements of protein secondary structure induced by ligand addition.34 A slight change in the near UV-CD spectra (250–320 nm) of BSA in the presence of NFZ/NFT was also observed (Fig. 2S, ESI†). This suggested that NFZ/NFT induced the changes in the secondary and tertiary structure of the protein. In order to ascertain the secondary structural changes induced by NFZ/NFT, CD measurements in the far UV-CD region (200–250 nm) were carried out. Upon binding of ligand, intermolecular forces responsible for sustaining the secondary and tertiary structure may get rearranged, resulting in conformational alteration in the protein. Fig. 4 shows the CD spectra of free BSA and nitrofurans–BSA complex, and it demonstrated two negative bands at wavelengths of 208 nm and 222 nm in the UV region, which were both assigned to n → π* shifts of the peptide bonds in the α-helix structure. The two wavelength points are the characteristic peaks of α-helix structures and both of them can be used to calculate its content. Although the intensity of CD spectra was reduced with the addition of nitrofurans, the peaks did not shift obviously. If the CD result was manifested in line with MRE (mean residue ellipticity) in deg cm−2 dmol−1, the following formula could be used:
| MRE = θobs/(10nlcp) | (6) |
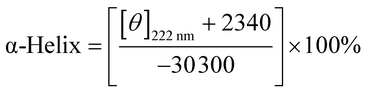 | (7) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 is the MRE of pure α-helix at 222 nm. Through the addition of nitrofurans, the intensities of the protein negative bands at 208 and 222 nm reduced gradually. The explanation may be that the stretched polypeptide structures of BSA caused by the combination of NFZ/NFT resulted in the exposure of the perturbation of the microstructures and hydrophobic cavities around the aromatic amino acid residues. Furthermore, the α-helix content of BSA reduced with the additions of NFZ/NFT when the molar ratio of BSA/nitrofuran was 1
300 is the MRE of pure α-helix at 222 nm. Through the addition of nitrofurans, the intensities of the protein negative bands at 208 and 222 nm reduced gradually. The explanation may be that the stretched polypeptide structures of BSA caused by the combination of NFZ/NFT resulted in the exposure of the perturbation of the microstructures and hydrophobic cavities around the aromatic amino acid residues. Furthermore, the α-helix content of BSA reduced with the additions of NFZ/NFT when the molar ratio of BSA/nitrofuran was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, which was in accordance with the results of the fluorescence and FT-IR spectra. From eqn (6) and (7), the quantitative analysis consequences of the α-helix content were gained. A decrease of the α-helix from 55.6% (free BSA) to 45.0% (NFZ–BSA 4
4, which was in accordance with the results of the fluorescence and FT-IR spectra. From eqn (6) and (7), the quantitative analysis consequences of the α-helix content were gained. A decrease of the α-helix from 55.6% (free BSA) to 45.0% (NFZ–BSA 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex) and 50.3% (NFT–BSA 4
1 complex) and 50.3% (NFT–BSA 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex) was obtained, indicating the reduction of α-helix due to the interaction.
1 complex) was obtained, indicating the reduction of α-helix due to the interaction.
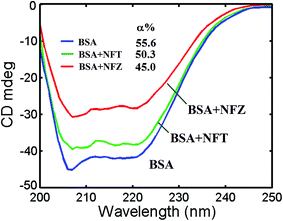 | ||
Fig. 4 Far UV-CD spectra of BSA alone and in the presence of NFZ and NFT (cBSA = 2.5 × 10−7 mol L−1, the molar ratio of NFZ/NFT to BSA is 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1). | ||
The results reveal that the impact of NFZ on the serum albumin conformational change is more remarkable than that of NFT, which prompts BSA unfold and adjust to a more incompact conformational status. The diverse effects of the two compound on the α-helix structure of BSA could be associated with the configuration associative binding affinities. The NFZ/NFT complexation could change the polypeptide structures of BSA that would then generate the beneficial spatial holes for the binding of BSA with NFZ/NFT. In addition, the CD spectra of BSA in the absence and presence of the nitrofurans were similar in shape, which indicating that the basic structure of BSA is conspicuously α-helix even after bonding.35
3.5. RLS and AFM analysis of the nitrofurans–BSA complex
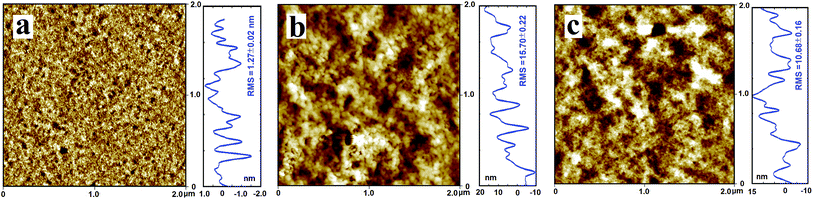
RLS is a sensitive method for investigation of aggregation of chromophores on biomacromolecules, and the RLS spectra of the nitrofurans–BSA system in Tris–HCl buffer (pH 7.4) are shown in Fig. 3S.† It can be seen that both free BSA and free nitrofurans exhibits very vulnerable RLS signals, but when a settled concentration of NFZ and NFT was added with different amounts of BSA, the intensity of RLS were conspicuously increased. Basing on the theory of RLS,36 it can be deduced that RLS intensity is correlated to the dimension of the shaped particle and directly proportional to the square of molecular volume, and Chamani et al.37 have indicated that the larger particles could induce stronger light scattering signals. Therefore, the added NFZ and NFT possibly interacted with BSA in solution, and the formation of NFZ/NFT–BSA compound led to an increase of the RLS signal.AFM was also executed in order to verdict the particle morphology and dimensions of the NFZ/NFT–BSA conjugates. BSA was observed to be adsorbed on the mica, the RMS (root mean square) roughness of the individual BSA molecule was found to be 1.27 ± 0.02 nm. The BSA molecule particle observed to be looser on the mica substrate upon interaction with NFZ/NFT (Fig. 5). The RMS roughness of the molecules were changed to be 15.70 ± 0.22 nm for the NFZ–BSA interaction and 10.68 ± 0.16 nm for the NFT–BSA interaction, respectively. The change of the NFZ–BSA structure is greater than that of the NFT–BSA, which indicates that NFZ is more toxic than NFT.
The greater hydrodynamic radius of the conjugate particles indicates the formation of larger molecular weight polymers. In order to reduce the number of unpleasant factors to constitute a stable structure, the BSA molecular lessened the surface area in contact with water by molecular aggregation.38 Hence, on the mica substrate we can see larger molecules, and this consequence also displayed that there were hydrophobic interactions between BSA and NFZ/NFT.39 The AFM result was accordance with the found of the FT-IR and CD and SFS spectra and confirmed the deduction from RLS experiment.
3.6. Molecular docking studies
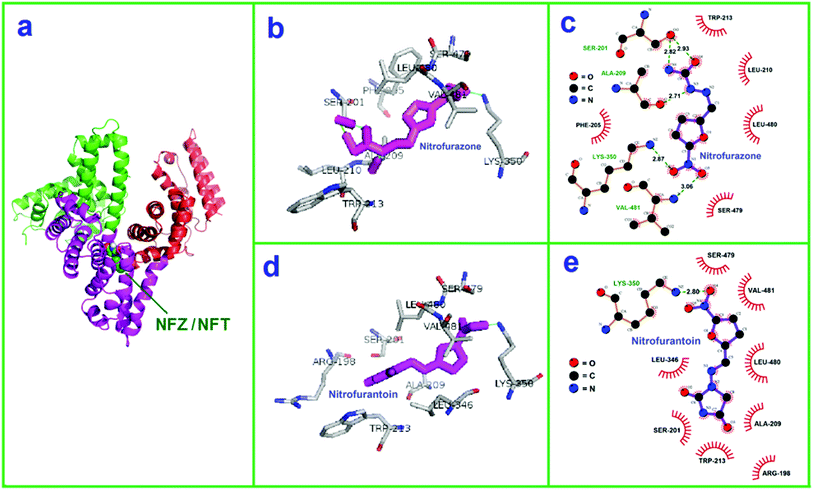
BSA is a spherical protein constituting of a single peptide chain of 582 amino acids. It is composed of three analogical domains, each domain can be divided into two subdomains, A and B, which assembles to form heart shaped. The primary binding region on BSA is located in hydrophobic cavities in subdomains IIA, which corresponds to the site I.40 According to Sudlow et al., the warfarin binds specifically to the site I whereas ibuprofen binds to site II of serum albumin.41 Hence, in order to identify the binding site of NFZ/NFT on BSA, some extra experiments were done in this study. The concentration of BSA and warfarin/ibuprofen were kept constant at 6.67 × 10−8 mol L−1 and NFZ/NFT was added in the range of 0.00–2.13 × 10−7 mol L−1, the binding constants (obtained from the double logarithmic equation) of NFZ–BSA/NFT–BSA were varied in presence of warfarin, which was less influenced by ibuprofen (Fig. 4S, Table 1S, in ESI†). The binding constant of NFZ–BSA and NFT–BSA decreased from 4.27 × 106 L mol−1 to 3.17 × 106 L mol−1 and 5.11 × 106 L mol−1 to 2.80 × 106 L mol−1 in the present of warfarin, respectively, which suggests the binding of NFZ/NFT to the site I.The molecular modeling study was carried out to clearly describe the bioactivity mechanism and help us to better visual understanding the binding site on molecular level about the interaction between NFZ/NFT and BSA. See Fig. 6a, there is a large hydrophobic cavity in subdomain IIA to accommodate the NFZ/NFT, which indicates that the pattern of interaction is hydrophobic in essence because the binding site is located deeply inside the subject of protein instead of being on the surface of the BSA.42 The results revealed that there have hydrophobic amino acids Trp213, Leu210, Leu480, Phe205, Ala209, Val481 for NFZ and Leu480, Val481, Leu346, Ala209, Trp213 for NFT, indicating the existence of hydrophobic interaction between NFZ/NFT and BSA (Fig. 6b–e). There are hydrogen bonds interactions between the nitryl on the furan ring of NFZ/NFT and terminal amino group of the amino acids Lys-350. However, there are more hydrogen bonds interactions between NTZ and BSA, which further showed that NFZ is more vivacious than NFT when transported by BSA. What is more, there are also a number of weak electrostatic interactions between charged amino acids Ser-479, Ser-201, Lys-350 with NFZ, Ser-479, Ser-201, Arg-198, Lys-350 with NFT, respectively. The results of molecular modeling manifested that the binding between NFZ/NFT and BSA was hydrogen binding and hydrophobic and electrostatic synergy, which was consistent with the above results.
It was found that the nitrofurans with a furan ring could interact with protein's hydrophobic residues. Generally, an electrostatic interaction could not be ruled out with respect to present findings, because BSA with an isoelectric point of 5.8, is negatively charged in Tris–HCl buffer of pH 7.4. For NFT, there is one N–H and two carbonyl groups, and two five-membered heterocyclics, the greater steric hindrance from the structures, the less the possibility of hydrophobic interaction. Nitryl on the furan ring of NFZ/NFT was found to interact with the proteins hydrophobic residues to form hydrogen bonding. As a result, the apparent interaction between NFZ/NFT and BSA may be hydrogen binding.
4. Conclusions
The binding of two nitrofurans (NFZ and NFT) to BSA, has been systematically studied using fluorescence, FT-IR, CD spectroscopy and AFM. It was discovered that BSA recognized NFZ and NFT with different binding affinity, spectral response and structural perturbation. The fluorescence experimental results manifest that the quenching mechanism of BSA by NFZ/NFT is a static quenching procedure. The molecular simulation indicates that the hydrophobic forces and electrostatic interactions play dominant role during the ligand–protein binding process and effectively proves the binding of NFZ/NFT to BSA. The gained binding constants for NFZ/NFT with BSA are 106 L mol−1, and the relatively high binding affinities manifest that the binding of NFZ/NFT to BSA might be specific. It has also found that the binding affinity of NFT to BSA was stronger than that of NFZ due to their diverse molecular structure. The FI-IR, CD and AFM showed that NFZ/NFT interact with both C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–N groups of BSA, leading to the loss of α-helix structure and the corresponding changes of some other secondary structures with different influence degrees. The impact on the conformational changes of BSA caused by NFZ is more conspicuous than that of NFT, which illustrates that NFZ is more toxic than NFT.
O and C–N groups of BSA, leading to the loss of α-helix structure and the corresponding changes of some other secondary structures with different influence degrees. The impact on the conformational changes of BSA caused by NFZ is more conspicuous than that of NFT, which illustrates that NFZ is more toxic than NFT.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support of this study by the National Natural Science Foundation of China (NSFC-21665017), the State Key Laboratory of Food Science and Technology of Nanchang University (SKLF-KF-201606 and SKLF-ZZA-201612).References
- F. Kuralay and A. Erdem, Analyst, 2015, 140, 2876–2880 RSC.
- M. M. Marshall, J. Ruzicka, O. K. Zahid, V. C. Henrich, E. W. Taylor and A. R. Hall, Langmuir, 2015, 31, 4582–4588 CrossRef CAS PubMed.
- A. Basu and G. S. Kumar, Food Chem., 2015, 175, 137–142 CrossRef CAS PubMed.
- A. Frostell-Karlsson, A. Remaeus, H. Roos, K. Andersson, P. Borg, M. Hamalainen and R. Karlsson, J. Med. Chem., 2000, 43, 1986–1992 CrossRef CAS PubMed.
- S. Bhuiya, A. B. Pradhan, L. Haque and S. Das, J. Phys. Chem. B, 2016, 120, 5–17 CrossRef CAS PubMed.
- S. Tunc, O. Duman, I. Soylu and B. K. Bozoglan, J. Hazard. Mater., 2014, 273, 36–43 CrossRef CAS PubMed.
- P. Thongsrisomboon, B. Liawruangrath, S. Liawruangrath and S. Satienperakul, Food Chem., 2010, 123, 834–839 CrossRef CAS.
- K. M. Cooper, J. V. Samsonova, L. Plumpton, C. T. Elliott and D. G. Kennedy, Anal. Chim. Acta, 2007, 592, 64–71 CrossRef CAS PubMed.
- N. Taokaenchan, T. Tangkuaram, P. Pookmanee, S. Phaisansuthichol, S. Kuimalee and S. Satienperakul, Biosens. Bioelectron., 2015, 66, 231–237 CrossRef CAS PubMed.
- S. E. Aiello, The Merck veterinary manual, Whitehouse Station, New Jersey, 2005 Search PubMed.
- H. Y. Zhai, L. C. Zhang, Y. F. Pan, K. S. Yuan, L. Huang and X. Yu, Chromatographia, 2015, 78, 551–556 CAS.
- J. Points, D. T. Burris and M. J. Walker, Food Control, 2015, 50, 92–103 CrossRef CAS.
- A. Kaufmann, P. Butcher, K. Maden, S. Walker and M. Widmer, Anal. Chim. Acta, 2015, 862, 41–52 CrossRef CAS PubMed.
- D. Kim, B. Kim, S. W. Hyung, C. H. Lee and J. Kim, J. Food Compos. Anal., 2015, 40, 24–31 CrossRef CAS.
- Y. B. Zhang, H. O. Qiao, C. Chen, Z. L. Wang and X. D. Xia, Food Chem., 2016, 192, 612–617 CrossRef CAS PubMed.
- T. Peter, All about albumin: Biochemistry Genetic and Medical Applications, Academic Press, San Diego, CA, 1996, pp. 9–75 Search PubMed.
- B. Tang, Y. M. Huang, X. L. Ma, X. X. Liao, Q. Wang, X. N. Xiong and H. Li, Food Chem., 2016, 212, 434–442 CrossRef CAS PubMed.
- Y. P. Wang, G. W. Zhang and L. H. Wang, J. Agric. Food Chem., 2015, 63, 75–84 CrossRef CAS PubMed.
- D. Wu, J. Yan, J. Wang, Q. Wang and H. Li, Food Chem., 2015, 170, 423–429 CrossRef CAS PubMed.
- X. Peng, G. W. Zhang, Y. J. Liao and D. M. Gong, Food Chem., 2016, 190, 207–215 CrossRef CAS PubMed.
- A. Sulkowska, J. Mol. Struct., 2002, 614, 227–232 CrossRef CAS.
- S. Huang, H. N. Qiu, S. Y. Lu, F. W. Zhu and Q. Xiao, J. Hazard. Mater., 2015, 285, 18–26 CrossRef CAS PubMed.
- T. T. Chen, X. T. Zhu, Q. Chen, M. Ge, X. P. Jia, X. Wang and C. W. Ge, Food Chem., 2015, 186, 292–297 CrossRef CAS PubMed.
- P. Mandal and T. Ganguly, J. Phys. Chem. B, 2009, 113, 14904–14913 CrossRef CAS PubMed.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Kluwer Academic/Plenum Publishers, New York, 2nd edn, 1999 Search PubMed.
- J. R. Lakowicz, Principles of fluorescence spectroscopy, Plenum Press, New York, 3rd edn, 2006, p. 277 Search PubMed.
- T. P. Lakshmi, M. Mondal, K. Ramadas and S. Natarajian, Spectrochim. Acta, Part A, 2017, 183, 90–102 CrossRef PubMed.
- P. D. Ross and S. Subramanian, Biochemistry, 1981, 20, 3096–3102 CrossRef CAS PubMed.
- D. M. Charbonneau and H. A. Tajmir-Riahi, J. Phys. Chem. B, 2010, 114, 1148–1155 CrossRef CAS PubMed.
- J. A. Pierce, R. S. Jackson, K. W. Vanevery, P. R. Griffiths and H. J. Gao, Anal. Chem., 1990, 62, 477–484 CrossRef CAS.
- S. N. Khan, B. Islam, R. Yennamalli, A. Sultan, N. Subbarao and A. U. Khan, Eur. J. Pharm. Sci., 2008, 35, 371–382 CrossRef CAS PubMed.
- P. Bourassa, C. D. Kanakis, P. Tarantilis, M. G. Pollissiou and H. A. Tajmir-Riahi, J. Phys. Chem. B, 2010, 114, 3348–3354 CrossRef CAS PubMed.
- A. S. Sharma and M. Ilanchelian, J. Phys. Chem. B, 2015, 119, 9461–9476 CrossRef PubMed.
- Q. M. Sun, J. He, H. Q. Yang, S. S. Li, L. D. Zhao and H. Li, Food Chem., 2017, 233, 190–196 CrossRef CAS PubMed.
- Y. Liu, M. M. Chen, L. G. Jiang and L. Song, Food Chem., 2015, 181, 170–178 CrossRef CAS PubMed.
- J. Yang, X. D. Chen, R. W. Fu, W. A. Luo, Y. B. Li and M. Q. Zhang, Phys. Chem. Chem. Phys., 2010, 12, 2238–2245 RSC.
- H. Iranfar, O. Rajabi, R. Salari and J. Chamani, J. Phys. Chem. B, 2012, 116, 1951–1964 CrossRef CAS PubMed.
- G. W. Zhang, L. Wang and J. H. Pan, J. Agric. Food Chem., 2012, 60, 2721–2729 CrossRef CAS PubMed.
- D. Y. Kim and W. S. Shin, Food Chem., 2015, 173, 1–6 CrossRef CAS PubMed.
- A. Bujacz, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 1278–1289 CrossRef CAS PubMed.
- G. Sudlow, D. J. Birkett and D. N. Wade, Mol. Pharmacol., 1976, 12, 1052–1061 CAS.
- M. Sarkar, S. S. Paul and K. K. Mukherjea, J. Lumin., 2013, 142, 220–230 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra05570f |
| This journal is © The Royal Society of Chemistry 2017 |