Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Higher order structures involving post transcriptionally modified nucleobases in RNA†
Preethi S. P.a,
Purshotam Sharma *b and
Abhijit Mitra
*b and
Abhijit Mitra *a
*a
aCenter for Computational Natural Sciences and Bioinformatics, International Institute of Information Technology Hyderabad (IIIT-H), Gachibowli, Hyderabad, Telangana 500032, India. E-mail: abi_chem@iiit.ac.in
bComputational Biochemistry Laboratory, Department of Chemistry and Centre for Advanced Studies in Chemistry, Panjab University, Chandigarh, 160014, India. E-mail: psharma@pu.ac.in
First published on 18th July 2017
Abstract
Post transcriptionally modified nucleotides play an important structural role in RNA by providing additional chemical diversity to the architecture of the constituent nucleotides. To understand the contribution of these modified nucleotides in tertiary interactions that help in RNA folding, we present a systematic investigation of higher order hydrogen bonded structures involving modified nucleotides using bioinformatics based crystal structural database analysis and quantum chemical calculations. Our analysis reveals that 29% of the modified bases that participate in hydrogen-bonding interactions form higher order motifs, which points towards their importance as RNA building blocks. Although greater geometric variations in such motifs are observed in tRNA, they occur most commonly in rRNA, where they may play a role in RNA folding. Characterization of the optimum geometries and binding strengths of the modified motifs reveals that structures that involve either a positively charged modified nucleobase (m7G+), or a protonated nucleobase (AH+) paired with a modified base (Ψ), possess stronger binding compared to uncharged (neutral) motifs. Thus, they seem to play an important role in stabilizing complex RNA structures, possibly through enhanced electrostatic interactions. Overall, our combined statistical, geometric, energetic and contextual analysis of these modified motifs reveals their importance as stable RNA building blocks, and highlights the need for further investigations related to their functional roles.
Introduction
The role of posttranscriptional modifications in facilitating the functional diversity of RNA molecules has drawn considerable attention in contemporary biochemical literature.1–6 From a wider perspective, the characterization of such modified nucleotides as intrinsic building blocks of RNA has been a step forward towards addressing the long standing conundrum on how RNA performs a variety of functional roles, despite its limited constitutional diversity in terms of the presence of only four different nucleobases (adenine (A), cytosine (C), guanine (G) and uracil (U)). However, adequate understanding of the role of modified nucleotides in shaping the structural diversity of RNA macromolecules is yet to be achieved.The evolutionarily conserved 3D structures of folded RNA macromolecules are stabilized through a variety of non-covalent interactions between the constituent nucleotides. The most prominent among these are the stacking interactions between aromatic nucleobase rings,7 and the inter-nucleotide hydrogen bonding (IHB) interactions that constitute the base–base, base–sugar, and base–phosphate interactions.8–12 Further, in addition to the canonical (A:U and G:C) base–base interactions, ‘non canonical’ IHB interactions play unique structural roles in RNA.13,14 Such interactions can further be extended to form base triples, quadruples and other higher order structures that contribute towards RNA folding.15,16
The structures of IHB motifs have been characterized in previous studies, mainly on the basis of the general assumption that each of these motifs is an intrinsically stable recurrent building block of RNA. Specifically, two complementary techniques: structural bioinformatics based crystal structure database analysis8,14,16–19 and quantum mechanical (QM) calculations,11,20–25 have been used to characterize such motifs. In addition to analysing the occurrence frequency and the stability of IHB motifs, such studies have highlighted the differences between the “intrinsic structure” of the motif, and the structure it adopts within RNA.23,26 These structural differences point towards the role of RNA macromolecular context in shaping the geometries of IHB motifs.
There are a significant number of studies on IHB interactions involving natural nucleotides. However, though modified nucleotides play important roles in determining the structure, folding and function of RNA,16,17,19,27–30 there are only a few recent studies reporting the occurrence, structures and stabilities of IHB interactions involving modified nucleosides in RNA.31,32 Oliva et al. analysed four modified base pair and two modified triple motifs constituting the conserved tertiary interactions in tRNA using quantum chemical methods.26 Subsequent studies on one of the conserved (i.e. G15:C48) base pair motif in tRNA, established the role of magnesium binding, archaeosine modification,33 or triple formation with dihydrouridine modification present at position 20,20 in stabilization of this motif. More recently, studies have characterized the occurrence context, structures and stabilities of naturally occurring IHB pairs that involve modified nucleotides.31,32 However, a systematic analysis of structural features of higher order interactions involving modified bases in RNA is currently absent in the literature.
Addressing this void, we have carried out a comprehensive analysis of the crystal structures of different classes of RNA, to identify the full range of modified motifs and enumerate their diversity.31 Based on this search, we identified 15 different types of modified base triples and 3 different types of base quadruple modified motifs. Further, a representative structure corresponding to each modified motif was extracted from the best resolution crystal structure, which was subjected to the quantum chemical analysis based approach of energy minimization and binding energy calculations.22,34 A detailed comparison of the “experimental” (i.e. within RNA macromolecular crystal structure) and “energy minimized” structures, respectively, of each of these motifs was then carried out using standard nucleic acid motif characterization parameters.35,36 Finally, an analysis of macromolecular occurrence context of these motifs was carried out to provide clues about the associated structural and functional roles of these motifs. Overall, our detailed analysis significantly adds to the understanding of role played by higher order motifs involving modified nucleotides, in RNA structures.
Materials and methods
To identify modified motifs, the occurrence of such bases was searched in a dataset of RNA crystal structures, that was chosen according to specific search criteria (details in ESI†), and the relevant protein databank (PDB) entries, submitted till 18 July 2016 with resolution better than 3.5 Å,31 were included. BPFind software36 was used to analyse the occurrence, location and type of higher order associations containing modified bases. Geometry optimizations of the modified motifs retrieved from the RNA crystal structures, as well as their unmodified counterparts retrieved either from crystal structures or modelled from modified pairs, was carried out using the B3LYP37,38 density functional and the 6-31G(d,p) basis set using Gaussian 09.39 The geometries, optimized using this method, have been shown to compare very well with the reference RIMP2/cc-pVTZ geometries, and are sufficient for interaction energy calculations.40 Basis set superposition error41 corrected binding energies of these base–base associations were calculated at the RIMP2 level42 using the aug-cc-pVDZ basis set. The binding energies calculated at this level compare well with the reference MP2/CBS(T) values, where an underestimation of only up to 1.11 kcal mol−1 is observed in a previous reference study on RNA base pairs.43 Further, these methods were selected to maintain conformity with previous studies on natural and modified IHB motifs involving RNA.24,25,31,34,44 Additional details regarding model building and the methodology followed for binding energy calculations are provided in ESI.†Variation in geometries of the modified motifs within the crystal occurrences, between the crystal and the optimized geometries, as well as between the modified and unmodified triples or quadruples, was quantified in terms of their root mean square deviation (rmsd) values using the VMD v1.9 software.45 Such geometric variations were further analysed by comparing the base pair parameters (i.e. buckle, propeller, open angle, shear, stretch and stagger) of the motifs using upgraded version of NUPARM software.35 In addition, the relative goodness of hydrogen bonds between two different geometries of the same higher order interaction were empirically estimated using an empirical E-value parameter (detailed in ESI†) using the BPFind software.36
Results and discussion
Statistical overview of higher order interactions of modified bases in RNA 3D structures
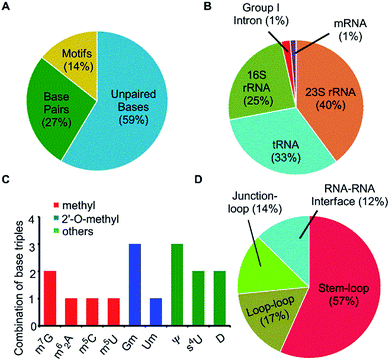
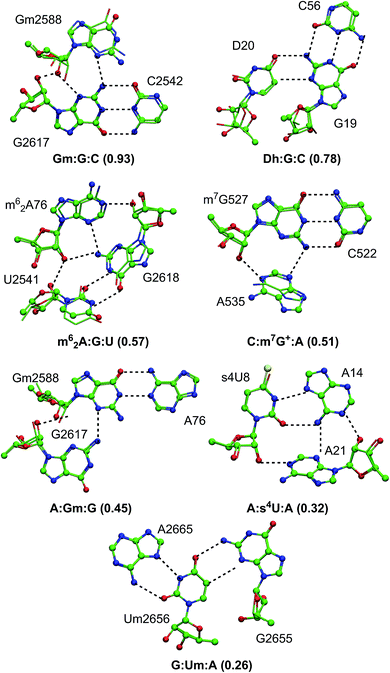
Fifteen different naturally occurring post-transcriptionally modified nucleosides that participate in RNA base pairing,31 were searched in a dataset of 207 high resolution RNA crystal structures identified in our previous study,31 to analyse their propensity to form modified motifs (Table 1). Of these, nine modified nucleosides participate in motifs (Fig. 1 and Table 1), four of which involve base-methylation (m62A, m7G+, m5C and m5U), two involve 2′-O-methylation at ribose (Gm and Um) and three involve other modifications (s4U, Ψ and D, Fig. S1–S3†). More importantly, a sizable proportion (i.e. 30%) of the base pairing occurrences involving these nine modified nucleosides occur as part of modified motifs (Fig. 2 and Table 1), which points toward their structural and functional importance in RNA.| Modified bases | Unpaired bases (631) | Base pairs (453) | Base triples (120) | Base quadruples (15) |
|---|---|---|---|---|
| N6,6-Dimethyl adenine (m62A) | — | 4 | 1 | — |
| N7-Methyl guanine (m7G+) | 14 | 55 | 31 | — |
| 2′-O-Methyl guanine (Gm) | 23 | 44 | 44 | 12 |
| C5-Methyl cytosine (m5C) | 61 | 90 | 2 | — |
| Pseudouridine (Ψ) | 231 | 79 | 26 | — |
| 5,6-Dihydrouridine (D) | 101 | 15 | 2 | — |
| C5-Methyl uracil (m5U) | 15 | 39 | 0 | 3 |
| 2′-O-Methyl uracil (Um) | 59 | 9 | 1 | — |
| 4-Thiouridine (s4U) | 1 | 23 | 13 | — |
| N1-Methyl adenine (m1A) | 53 | 21 | — | — |
| 2′-O-Methyl adenine (Am) | 12 | 12 | — | — |
| N2-Methyl guanine (m2G) | 1 | 42 | — | — |
| N2,2-Dimethyl guanine (m22G) | 25 | 16 | — | — |
| 2′-O-Methyl cytosine (Cm) | 35 | 3 | — | — |
| 2′,5-Dimethyl uracil (m5Um) | 0 | 1 | — | — |
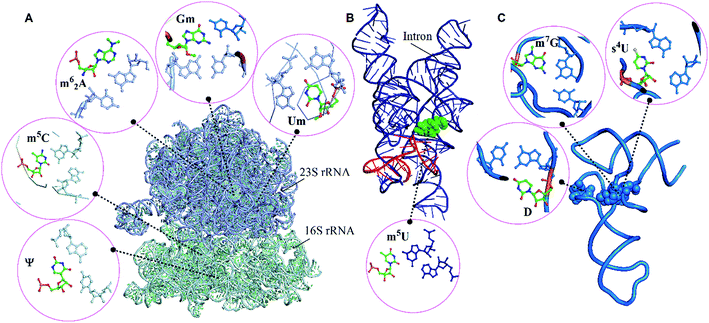 | ||
| Fig. 1 Modified motifs found in the (A) ribosomal RNA (B) group I intron/exon complex and (C and D) tRNA. The modified base in each of the motif is represented in green color. | ||
Our dataset of RNA crystal structures spanning all major RNA classes (Tables S1 and S2†) shows that modified triples and quadruples (Fig. S4†) are most commonly observed in rRNA (71%), followed by tRNA (26%) and group I intron (3%, Fig. 2). It is noteworthy that despite their high occurrence in rRNA, higher diversity of modified motifs is observed in tRNA, that spans five (i.e. m62A, m7G+, s4U, Ψ and D) of the nine modified nucleotides that form motifs.
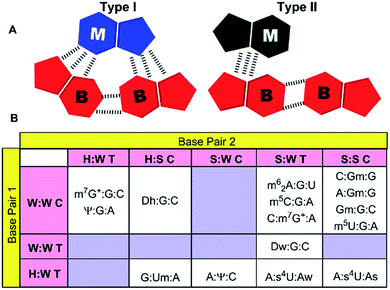
Analysis, of occurrence context of modified motifs, reveals that they are observed in all major RNA structural elements. Specifically, more than half of these interactions participate in stem–loop tertiary interactions (57%), followed by loop–loop (17%), junction–loop (14%) and RNA–RNA interface interactions (12%, Fig. 2). Additionally, ten unique modified triple associations participate in the stabilization of A/G-minor motifs46 or in dinucleotide platforms stabilization (Table 2). Overall, when classified according to the type of interacting edges (i.e. Watson–Crick edge (W), Hoogsteen edge (H) and sugar edge (S)9), our analysis reveals fifteen distinct modified triples that span nine base triple geometric families,16 and three distinct modified quadruples. Two of these modified triple motifs involve the positively charged modified base (m7G+), whereas one involves a protonated adenine (AH+) interacting with pseudouridine (Ψ, Table 2).
| Modified motifs | Geometry | Interaction type | Type of RNA | Position | fa | Structural element |
|---|---|---|---|---|---|---|
| a f stands for occurrence frequency of modified base triples in the dataset. | ||||||
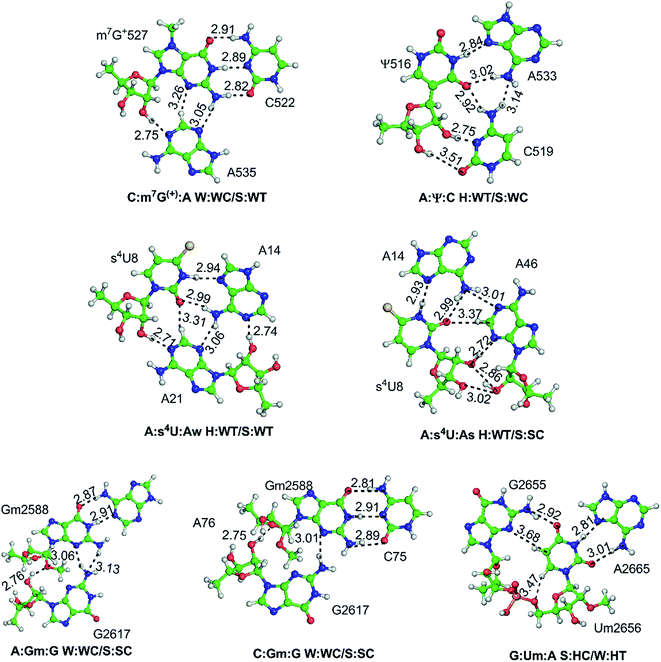
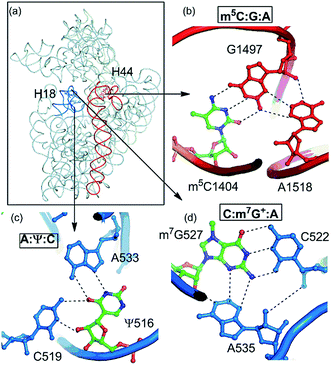
| C:m7G+:A | W:WC/S:WT | B–B/S–B | 16S rRNA | 522:527:535 | 11 | Stem–loop (A-minor) |
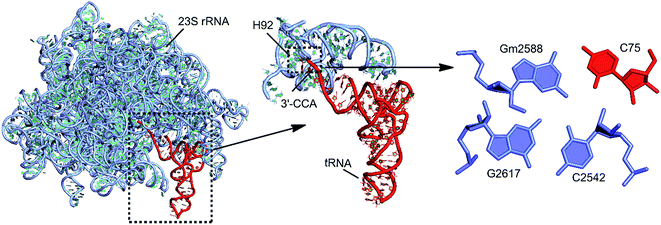
| C:Gm:G | W:WC/S:SC | B–B/S–S | tRNA:rRNA | 75:2588:2617 | 12 | RNA–RNA interface (G-minor) |
| A:Gm:G | W:WC/S:SC | B–B/S–S | tRNA:rRNA | 76:2588:2617 | 1 | RNA–RNA interface (A-minor) |
| A:s4U:Aw | H:WT/S:WT | B–B/S–B | tRNA | 14:8:21 | 12 | Junction–loop |
| A:s4U:As | H:WT/S:SC | B–B/S–S | tRNA | 14:8:46 | 1 | Junction–loop |
| A:Ψ:C | H:WT/S:WC | B–B/S–B | 16S rRNA | 533:516:519 | 22 | Loop–loop |
| G:Um:A | S:HC/W:HT | S–B/B–B | 23S rRNA | 2655:2656:2665 | 1 | Internal loop (platform) |
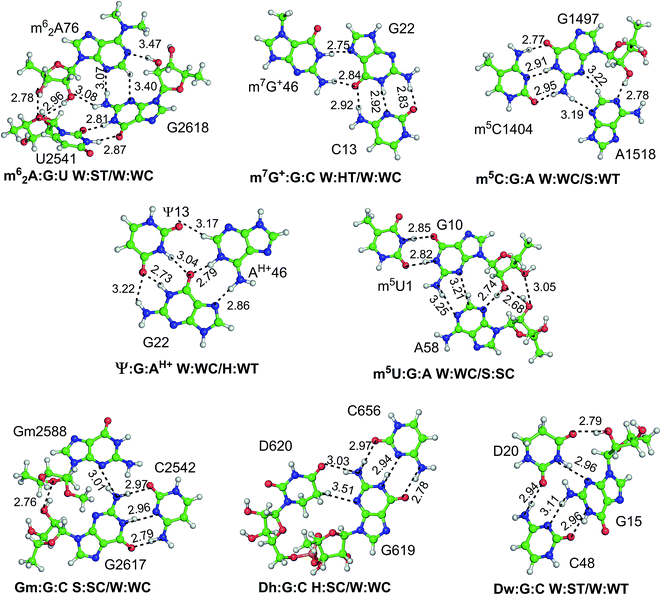
| m62A:G:U | W:ST/W:WC | B–B/S–B | tRNA:23S rRNA | 76:2618:2541 | 1 | Junction–loop (A-minor) |
| m7G+:G:C | W:HT/W:WC | B–B/B–B | tRNA | 46:22:13 | 20 | Stem–loop |
| Gm:G:C | S:SC/W:WC | S–S/B–B | 23S rRNA | 2588:2617:2542 | 30 | Stem–loop (G-minor) |
| m5C:G:A | W:WC/S:WT | B–B/S–B | 16S rRNA | 1404:1497:1518 | 2 | Stem–loop (A-minor) |
| Ψ:G:AH+ | W:WC/H:WT | B–B/B–B | tRNA | 13:22:46 | 4 | Stem–loop |
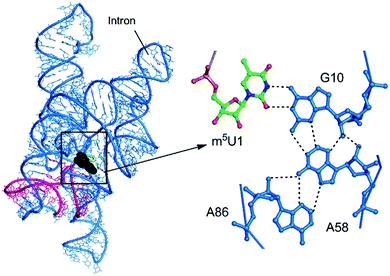
| m5U:G:A | W:WC/S:SC | B–B/S–S | Group I intron | 1:10:58 | 3 | RNA–RNA interface (A-minor) |
| Dw:G:C | W:ST/W:WT | B–S/B–B | tRNA | 20:15:48 | 1 | Loop–loop |
| Dh:G:C | H:SC/W:WC | B–S/B–B | tRNA | 20:19:56 | 1 | Loop–loop (platform) |
| C:Gm:G:C | W:WC/S:SC/W:WC | B–B/S–S/B–B | tRNA:rRNA | 75:2588:2617:2542 | 12 | RNA–RNA interface |
| A:Gm:G:C | W:WC/S:SC/W:WC | B–B/S–S/B–B | tRNA:rRNA | 76:2588:2617:2542 | 1 | RNA–RNA interface |
| m5U:G:A:A | W:WC/S:SC/H:ST | B–B/S–S/B–S | Group I intron | 1:10:58:84 | 1 | RNA–RNA interface |
Structural characteristics of higher order motifs involving modified bases
Of the fifteen modified triple motifs, nine associations involve multiple occurrences (Table S3†). Geometrical variations were observed within these multiple occurrences, the magnitude of which depends on the identity of the constituent nucleosides and their respective macromolecular structural context (Fig. S5†). For example, although 31 occurrences of Gm2588:G2617:C2542 triple within the crystal structures of 23S rRNA show negligible average rmsd (0.15 Å), the 13 occurrences of A14:s4U8:A21 motif in tRNA show a significant rmsd (1.07 Å). This variability may be attributed to the location of the Gm2588:G2617:C2542 triple in the stable stem–loop interaction region of 23S rRNA, which contrasts the flexible junction loop location of the A14:s4U8:A21 in tRNA structures. Overall, the average rmsd ranges between 0 Å and 1 Å within different crystal occurrences of modified motifs (Table S4†). Further, analysis of base pair parameters of modified motifs reveals that associations involving only base–base (B–B) interactions show little deviation in the base pair parameters, and exhibits up to 12° of average of buckle, propeller and open values. Within the motifs involving base–sugar (B–S) and/or sugar–sugar (S–S) interactions, largest buckle is observed in m62A:G:U, mainly due to deviation in pairing geometry on methylation of A. Similarly, Dw:G:C motif exhibits largest propeller twist, due to inherent puckering of the dihydrouridine ring. However, owing to the constraints arising out of satisfying planarity requirements, triples that stabilize the platform interactions (i.e. G:Um:A and Dh:G:C) display small deviations in the structural parameters. Overall, when the distribution of base pair parameters with respect to the identity of the modified base is considered, motifs involving D or m62A exhibit the most nonplanar geometries with large (up to 40°) deviation in the buckle and propeller parameters. It may however be noted that, due to limited resolution of RNA crystal structures, many of the observed geometrical deformations, in crystal structure occurrences of modified triples or quadruples, may be artefacts arising out of crystal data refinement errors. Thus, to characterize optimum geometries of these motifs in the absence of crystal environment, we carried out quantum chemical energy minimizations.
| Higher order interaction | Modified | Unmodified |
|---|---|---|
| C:m7G+:A | −59.4 | −45.9 |
| C:Gm:G | −45.7 | −42.8 |
| A:Gm:G | −34.1 | −39.4 |
| A:s4U:Aw | −28.1 | −29.5 |
| A:s4U:As | −35.2 | −34.9 |
| A:Ψ:C | −34.2 | −35.6 |
| G:Um:A | −25.6 | −26.6 |
| m62A:G:U | −30.5 | −31.4 |
| m7G+:G:C | −38.0 | −20.2 |
| Gm:G:C | −19.3 | −25.9 |
| m5C:G:A | −29.7 | −29.1 |
| Ψ:G:AH+ | −24.1 | −23.1 |
| m5U:G:A | −17.2 | −16.8 |
| Dw:G:C | −20.3 | −21.1 |
| Dh:G:C | −7.7 | −10.4 |
| m5U:G:A:A | −17.1 | −16.7 |
| C:Gm:G:C | −43.1 | −38.4 |
Within the fifteen motifs that undergo modification at the nucleobase portion of the nucleotide, two motifs (C:m7G+A and m7G+:GC) exhibit significant enhancement (by 14 and 28 kcal mol−1 respectively) in binding energy due to introduction of positive change on the nucleobase architecture. However, rest of the twelve motifs undergo little (i.e. within 2 kcal mol−1) change in binding energy on modification (Table 3).
Structurally and functionally important modified base triples and quadruples in RNA
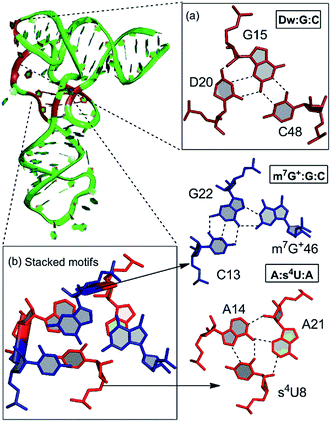
Our detailed analysis of the occurrence contexts of modified triples and quadruples revealed the presence of such motifs within unique structural contexts in different RNA classes, which indicate their putative functional roles. Here we outline the RNA structural environment around each of these motifs, and project the conclusions derived from our quantum chemical studies on isolated motifs onto the wider biochemical context of their occurrence in RNA structures.In addition to structure stabilization, our analysis reveals that the presence of modified bases in tRNA tertiary interactions may also play a role in stabilization of alternate conformations, which may in turn enhance tRNA flexibility. Specifically, depending on the sequence, the 8:14 pair can form two different triple motifs. The first motif observed in E. coli involves the s4U8:A14:A21 triple that stacks with the 13:22:46 triple (Fig. 7). However, in the second motif present in T. aquaticus, A21 is flipped out, and the resulting s4U8:A14:A46 triple stacks over only the 13:22 pair. The associated loss of hydrogen bonding between 13:22 and 46 in the second motif, contrasting the scenario in the first motif, is partially compensated by enhanced binding (by 7 kcal mol−1) within the s4U8:A14:A46 triple compared to the s4U8:A14:A21 motif. This intricate trade-off, between competing tertiary interactions, may help in stabilization of the alternate local tRNA conformations on the one hand, and provide local flexibility on the other.
Another example of involvement of modified bases, in stabilization of alternate tertiary interaction motifs, is observed on comparison of tRNA crystal structures of E. coli and T. thermophilus. Whereas the D20 residue, with its flexible χ-torsion interacts with the conserved G15:C48 pair in T. thermophilus to form a D:G:C modified motif that connects the D-loop and the V-arm, it interacts with the G19:C56 pair in E. coli to form a D20:G19:C56 triple containing a dinucleotide platform,20 that connects the D-loop with the TΨC loop (Fig. 7). In 20:15:48 modified motif, the binding of D20 to the intrinsically unstable G15:C48 Levitt base pair20 provides an extra stabilization of 21 kcal mol−1 to the motif. On the other hand, binding of D20 to the G19:C56 W:W cis pair results in the formation of a stable platform, which provides an additional 20 kcal mol−1 extra stabilization. Overall, our analysis demonstrates how the presence of modified motifs provides enhanced stabilization, and conformational variability to tRNA structures.
Conclusions
Our crystal structure analysis reveals the importance of higher order associations involving modified base pairs, where ∼30% of such base pairs are present as constituents of triples. The significant frequency exhibited by these higher order interactions point towards their important structural and functional roles in RNA macromolecules. Although our previous study reveals that, tRNA contains greater percentage of modified bases and base pairs compared to other RNA classes, the proportion of higher order associations containing modified bases is comparatively greater in 23S rRNA. Further, the fact that the hydrogen bonding pattern within the crystal occurrences of such motifs does not change on optimization, indicates that such motifs are intrinsically stable well-defined building blocks of RNA.Analysis of stability of higher order modified motifs reveals that those modifications, which introduce positive charge within the motifs, impart greater stabilization compared to other modifications, mainly due to enhanced electrostatic interactions (Table S10†). Further analysis of detailed structural environment, around these motifs, also indicates that these motifs are present in structurally and functionally important regions within various RNA structures. This includes the tRNA, tRNA:rRNA interface, rRNA and the intron–exon complex. These findings underscore the need for further investigations towards unravelling the hitherto unknown functional roles of such motifs, in the context of many biological processes involving RNA.
Conflict of interest
The authors declare no competing financial interest.Acknowledgements
PS thanks the Department of Science and Technology (DST) and University Grants Commission (UGC), New Delhi for financial support through the DST INSPIRE (IFA14-CH162) and the UGC FRP (F.4-5(176-FRP/2015(BSR))) programs, respectively. AM thanks DBT, Government of India project BT/PR-14715/PBD/16/903/2010 for partial financial support.References
- P. F. Agris, EMBO Rep., 2008, 9, 629–635 CrossRef CAS PubMed.
- C. S. Chow, T. N. Lamichhane and S. K. Mahto, ACS Chem. Biol., 2007, 2, 610–619 CrossRef CAS PubMed.
- B. El Yacoubi, M. Bailly and V. de Crécy-Lagard, Annu. Rev. Genet., 2012, 46, 69–95 CrossRef CAS PubMed.
- M. Helm, Nucleic Acids Res., 2006, 34, 721–733 CrossRef CAS PubMed.
- Y. Motorin and M. Helm, Wiley Interdiscip. Rev.: RNA, 2011, 2, 611–631 CrossRef CAS PubMed.
- S. Raychaudhuri, J. Conrad, B. G. Hall and J. Ofengand, RNA, 1998, 4, 1407–1417 CrossRef CAS PubMed.
- J. Černý and P. Hobza, Phys. Chem. Chem. Phys., 2007, 9, 5291–5303 RSC.
- N. B. Leontis, J. Stombaugh and E. Westhof, Nucleic Acids Res., 2002, 30, 3497–3531 CrossRef CAS PubMed.
- N. B. Leontis and E. Westhof, RNA, 2001, 7, 499–512 CrossRef CAS PubMed.
- J. Stombaugh, C. L. Zirbel, E. Westhof and N. B. Leontis, Nucleic Acids Res., 2009, 37, 2294–2312 CrossRef CAS PubMed.
- M. Zgarbová, P. Jurecka, P. Banáš, M. Otyepka, J. E. Sponer, N. B. Leontis, C. L. Zirbel and J. I. Šponer, J. Phys. Chem. A, 2011, 115, 11277–11292 CrossRef PubMed.
- C. L. Zirbel, J. E. Šponer, J. Šponer, J. Stombaugh and N. B. Leontis, Nucleic Acids Res., 2009, 37, 4898–4918 CrossRef CAS PubMed.
- T. Hermann and E. Westhof, Chem. Biol., 1999, 6, R335–R343 CrossRef CAS PubMed.
- E. Westhof and V. Fritsch, Struct., 2000, 8, R55–R65 CrossRef CAS.
- P. B. Moore, Annu. Rev. Biochem., 1999, 68, 287–300 CrossRef CAS PubMed.
- A. S. Abu Almakarem, A. I. Petrov, J. Stombaugh, C. L. Zirbel and N. B. Leontis, Nucleic Acids Res., 2012, 40, 1407–1423 CrossRef CAS PubMed.
- S. Bhattacharya, S. Mittal, S. Panigrahi, P. Sharma, S. P. Preethi, R. Paul, S. Halder, A. Halder, D. Bhattacharyya and A. Mitra, Database, 2015, bav011 CrossRef PubMed.
- J. Sponer, J. E. Šponer, A. I. Petrov and N. B. Leontis, J. Phys. Chem. B, 2010, 114, 15723–15741 CrossRef CAS PubMed.
- M. Sarver, C. L. Zirbel, J. Stombaugh, A. Mokdad and N. B. Leontis, J. Math. Biol., 2008, 56, 215–252 CrossRef PubMed.
- M. Chawla, S. Abdel-Azeim, R. Oliva and L. Cavallo, Nucleic Acids Res., 2014, 42, 714–726 CrossRef CAS PubMed.
- M. Chawla, P. Sharma, S. Halder, D. Bhattacharyya and A. Mitra, J. Phys. Chem. B, 2011, 115, 1469–1484 CrossRef CAS PubMed.
- P. Sharma, M. Chawla, S. Sharma and A. Mitra, RNA, 2010, 16, 942–957 CrossRef CAS PubMed.
- P. Sharma, A. Mitra, S. Sharma, H. Singh and D. Bhattacharyya, J. Biomol. Struct. Dyn., 2008, 25, 709–732 CAS.
- J. Šponer, P. Jurečka and P. Hobza, J. Am. Chem. Soc., 2004, 126, 10142–10151 CrossRef PubMed.
- J. E. Šponer, N. a. Špačková, P. Kulhánek, J. Leszczynski and J. Šponer, J. Phys. Chem. A, 2005, 109, 2292–2301 CrossRef PubMed.
- R. Oliva, L. Cavallo and A. Tramontano, Nucleic Acids Res., 2006, 34, 865–879 CrossRef CAS PubMed.
- W. A. Cantara, P. F. Crain, J. Rozenski, J. A. McCloskey, K. A. Harris, X. Zhang, F. A. P. Vendeix, D. Fabris and P. F. Agris, Nucleic Acids Res., 2011, 39, D195–D201 CrossRef CAS PubMed.
- S. Dunin-Horkawicz, A. Czerwoniec, M. J. Gajda, M. Feder, H. Grosjean and J. M. Bujnicki, Nucleic Acids Res., 2006, 34, D145–D149 CrossRef CAS PubMed.
- F. Jühling, M. Mörl, R. K. Hartmann, M. Sprinzl, P. F. Stadler and J. Pütz, Nucleic Acids Res., 2009, 37, D159–D162 CrossRef PubMed.
- D. Piekna-Przybylska, W. A. Decatur and M. J. Fournier, Nucleic Acids Res., 2008, 36, D178–D183 CrossRef CAS PubMed.
- P. P. Seelam, P. Sharma and A. Mitra, RNA, 2017, 23, 847–859 CrossRef PubMed.
- M. Chawla, R. Oliva, J. M. Bujnicki and L. Cavallo, Nucleic Acids Res., 2015, 43, 9573 CrossRef CAS PubMed.
- R. Oliva, A. Tramontano and L. Cavallo, RNA, 2007, 13, 1427–1436 CrossRef CAS PubMed.
- P. Sharma, J. E. Šponer, J. Šponer, S. Sharma, D. Bhattacharyya and A. Mitra, J. Phys. Chem. B, 2010, 114, 10234 CrossRef CAS.
- S. Mukherjee, M. Bansal and D. Bhattacharyya, J. Comput.-Aided Mol. Des., 2006, 20, 629–645 CrossRef CAS PubMed.
- J. Das, S. Mukherjee, A. Mitra and D. Bhattacharyya, J. Biomol. Struct. Dyn., 2006, 24, 149–161 CAS.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B, 1988, 37, 785–789 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, N. J. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian, Inc., Wallingford, CT, USA, 2009.
- J. Šponer, P. Jurečka and P. Hobza, J. Am. Chem. Soc., 2004, 126, 10142–10151 CrossRef PubMed.
- S. F. Boys and F. Bernardi, Mol. Phys., 1970, 19, 553–566 CrossRef CAS.
- F. Weigend, M. Häser, H. Patzelt and R. Ahlrichs, Chem. Phys. Lett., 1998, 294, 143–152 CrossRef CAS.
- A. Mládek, P. Sharma, A. Mitra, D. Bhattacharyya, J. Šponer and J. E. Šponer, J. Phys. Chem. B, 2009, 113, 1743–1755 CrossRef PubMed.
- J. Šponer, M. Zgarbová, P. Jurečka, K. E. Riley, J. E. Šponer and P. Hobza, J. Chem. Theory Comput., 2009, 5, 1166–1179 CrossRef PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- P. Nissen, J. A. Ippolito, N. Ban, P. B. Moore and T. A. Steitz, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 4899–4903 CrossRef CAS PubMed.
- K. Y. Sanbonmatsu, Biochimie, 2006, 88, 1075–1089 CrossRef CAS PubMed.
- P. L. Adams, M. R. Stahley, A. B. Kosek, J. Wang and S. A. Strobel, Nature, 2004, 430, 45–50 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra05284g |
| This journal is © The Royal Society of Chemistry 2017 |