Open Access Article
Open Access ArticleSelective capture and rapid identification of E. coli O157:H7 by carbon nanotube multilayer biosensors and microfluidic chip-based LAMP†
Tianchan Lia,
Fanjiao Zhua,
Wei Guoa,
Hongxi Gua,
Jing Zhaoa,
Mei Yan*ab and
Shaoqin Liu *ab
*ab
aSchool of Life Science and Technology, State Key Laboratory of Urban Water Resource and Environment, Harbin Institute of Technology, Harbin 150080, China. E-mail: yanmei@hit.edu.cn; shaoqinliu@hit.edu.cn
bMicro- and Nanotechnology Research Center, Harbin Institute of Technology, Harbin 150080, China
First published on 13th June 2017
Abstract
We describe a sensitive approach for visual and point-of-care detection of E. coli O157:H7 and its toxic gene by combining carbon nanotube (CNT) multilayer biosensors and microfluidic chip-based loop-mediated isothermal amplification (LAMP). The anti-E. coli O157:H7 functionalized CNT multilayer biosensor can selectively capture the target bacterium E. coli O157:H7 in complex samples. After culturing, the captured bacteria can be released on demand by cleavage of the anti-E. coli O157:H7 antibody–bacteria interaction. The DNA concentration of the released bacteria was subsequently analyzed with microfluidic chip-based LAMP. After systematic optimization of capturing and detecting conditions, the proposed sensing platform was capable of detecting concentrations as low as 1 CFU mL−1 without complicated instrumentation, this is much more sensitive than previous reported methods. The distinct advantages of the proposed sensing platform, such as high specificity, low cost, good reproducibility and the ability of regenerating, make it a potential platform for detecting E. coli O157:H7 in related food safety and clinical diagnosis.
1. Introduction
Pathogenic strains of bacteria are a key concern for hospitals, environmental biology, the food industry and water supplies because bacterial infection can cause a wide variety of illnesses. For example, in the developing world, enterohemorrhagic Escherichia coli of serotype O157:H7 is one of the most abundant and dangerous pathogens,1–4 and often leads to hemorrhagic colitis, hemolytic uremic syndrome (HUS) and death.5–7Conventional methods of bacterial detection and identification rely on the submission of samples to laboratories for culturing in selective media.8 The cell culture-based assays are reliable, but are lengthy and generally take up several days to obtain a confirmed result, resulting in the loss of precious time between sample collection and treatment.9 Currently, a variety of detection methods have been developed to measure bacteria in samples, including enzyme-linked immunosorbent assay (ELISA),10,11 polymerase chain reaction (PCR),12–15 protein microassay,16 quartz crystal microbalance (QCM) system,17,18 surface plasmon resonance (SPR),19–21 flow cytometry (FC),22 chemiluminescence (CL),23–26 and surface enhanced Raman spectroscopy (SERS).27 Although these techniques are less time-consuming, they are associated with some limitations that hinder their wide applications. For example, PCR assays are expensive, complex and require a skilled technician. Therefore, a fast, reliable, portable and user-friendly technique able to detect specific bacteria and determine their concentrations is urgently needed.
Detection of bacteria involves two distinct processes: (1) selectively capturing of bacteria from samples and (2) identification of the captured bacteria. However, selective capture and recognition of bacteria is complicated by their low abundance (the infectious dose of pathogenic bacteria is about 10–100 cells).28 In response to the above pressing challenges, in this study, we proposed a simple strategy that combined loop-mediated isothermal amplification (LAMP)-based microfluidic chip with carbon nanotube multilayer structure to develop a portable sensing platform for the selective capture and recognition of the target bacterium. The antibody functionalized carbon nanotube multilayer is capable of selective capture, culture and release of bacteria. The captured bacteria could then be lysed for rapid and sensitive nucleic acid amplification with LAMP-based microfluidic chips. Using Escherichia coli O157:H7 as the targeted bacterium, we demonstrated the ability of the proposed sensing platform to selectively capture, culture, release and precisely detect and quantify bacteria. As compared to other biorecognition entities, the proposed sensing platform provides several advantages including low cost, facile and easy-to-use.29–32 Therefore it is foreseeable that the combination of carbon nanotube multilayer structure-based chip and LAMP microfluidic chip can be a fast, reliable and portable sensing platform for detection of the targeted bacterium.
2. Experimental
2.1 Reagents and equipment
Multi-walled carbon nanotubes (MWCNTs), sodium polystyrenesulfonate (PSS, MW 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), polyethyleneimine (PEI, MW 25
000), polyethyleneimine (PEI, MW 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), n-hydroxysuccinimide (NHS), propidium iodide (PI) and (3-aminopropyl)triethoxysilane were purchased from Sigma-Aldrich. The E. coli O157:H7 strain (ATCC 43889) and anti-E. coli O157:H7 polyclonal antibody were purchased from Prajna Biology (Shanghai, China). Other chemicals of analytical grade were obtained from commercial sources and used as received. All aqueous solutions were prepared with ultrapure water from a Milli-Q water purification system (18.2 MΩ, Milli-Q, Millipore). The ITO (about 15 Ω sq−1) coated glass was obtained from Leaguer Film Technology (Shenzhen). Poly(dimethylsiloxane) (PDMS) was purchased from Dow Corning Co. Ltd. LAMP amplification reagents were purchased from Summus Co. Ltd., bacterial lysis reagent was from Huafeng Co. Ltd. (Guangzhou, China). LAMP primers used in this work were synthesized by Sangon Biotech Co. Ltd. (Shanghai, China).
000), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), n-hydroxysuccinimide (NHS), propidium iodide (PI) and (3-aminopropyl)triethoxysilane were purchased from Sigma-Aldrich. The E. coli O157:H7 strain (ATCC 43889) and anti-E. coli O157:H7 polyclonal antibody were purchased from Prajna Biology (Shanghai, China). Other chemicals of analytical grade were obtained from commercial sources and used as received. All aqueous solutions were prepared with ultrapure water from a Milli-Q water purification system (18.2 MΩ, Milli-Q, Millipore). The ITO (about 15 Ω sq−1) coated glass was obtained from Leaguer Film Technology (Shenzhen). Poly(dimethylsiloxane) (PDMS) was purchased from Dow Corning Co. Ltd. LAMP amplification reagents were purchased from Summus Co. Ltd., bacterial lysis reagent was from Huafeng Co. Ltd. (Guangzhou, China). LAMP primers used in this work were synthesized by Sangon Biotech Co. Ltd. (Shanghai, China).
UV-Vis absorption spectra were measured by using a U-4100 spectrophotometer (Hitachi, Japan). Cyclic voltammetry (CV) measurements were carried out with a CHI 860D electrochemistry work station (CHI, USA), and electrochemical impedance spectroscopy (EIS) was performed using a Solartron 1260A Impedance/Gain-phase Analyzer (Ametek, UK). The impedance spectra were recorded within the frequency range of 0.1 Hz–100 kHz at a 0 V bias potential. The amplitude of the applied sine wave potential in each case was 10 mV. Electrochemical measurements were performed with a conventional three electrode system comprised of a platinum wire as auxiliary electrode, a standard Ag/AgCl reference electrode and the modified ITO electrode as the working electrode. The PDMS was bonded with the glass slide by plasma cleaner (Harrick, USA). The quantitative detection of E. coli O157:H7 was monitored by digital fiber optic sensor (FS-N18N, Keyence Corporation, Osaka) and the optical fibers (FU-75F, Keyence Corporation, Osaka).
2.2 Preparation of carbon nanotube (CNT) multilayer-based biosensor
The ITO-coated glass substrates were cut into a definite size (30 mm × 10 mm), cleaned with Piraha solution,33 and deposited into toluene solution containing (3-aminopropyl)trimethoxysilane to yield an amine-functionalized surface. At the same time, the MWCNTs was acidized by ultrasonication in a mixture of concentrated HNO3 and H2SO4 (v/v, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) for 24 h, followed by extensive washing with deionized water until the filtrate was neutral. Then it was dried in a vacuum. Generally, van der Waals forces among CNTs are large enough to make them stick together and form large bundles. Thus the MWCNT multilayer prepared from pure MWCNT solution is not uniform. To improve the dispersion in solution and meanwhile remain their high conductivity, the resulting MWCNTs are in 1 mg mL−1 PSS solution to form the final concentration at 1 mg mL−1. The π-like stacking of the benzene rings benefits the binding between PSS and CNTs, leading to a high fraction of individualized CNTs after dispersion. It is found that MWCNTs dispersed in PSS solution is stable for several months at room temperature. As shown in Scheme 1, CNT multilayer-based biosensor was prepared by alternatively immersing amine-functionalized ITO into MWCNTs solution and 1 mg mL−1 PEI, using an immersion time of 5 min, and rinsed with pure water and dried under N2 flow after each layer deposited. The CNT multilayers-coated ITO electrode was then immersed into the activation solution (45 mM EDC and 15 mM NHS) for 2 h to activate the carboxyl groups. After rinsing with water, 10 μg mL−1 of anti-E. coli O157:H7 was immediately dropped on the surface and then incubated at 4 °C for 24 h. Then the surface was incubated with BSA for 1 h at room temperature to close the bound sites. After washed with PBS and water, the modified electrodes were stored at 4 °C for use.
3) for 24 h, followed by extensive washing with deionized water until the filtrate was neutral. Then it was dried in a vacuum. Generally, van der Waals forces among CNTs are large enough to make them stick together and form large bundles. Thus the MWCNT multilayer prepared from pure MWCNT solution is not uniform. To improve the dispersion in solution and meanwhile remain their high conductivity, the resulting MWCNTs are in 1 mg mL−1 PSS solution to form the final concentration at 1 mg mL−1. The π-like stacking of the benzene rings benefits the binding between PSS and CNTs, leading to a high fraction of individualized CNTs after dispersion. It is found that MWCNTs dispersed in PSS solution is stable for several months at room temperature. As shown in Scheme 1, CNT multilayer-based biosensor was prepared by alternatively immersing amine-functionalized ITO into MWCNTs solution and 1 mg mL−1 PEI, using an immersion time of 5 min, and rinsed with pure water and dried under N2 flow after each layer deposited. The CNT multilayers-coated ITO electrode was then immersed into the activation solution (45 mM EDC and 15 mM NHS) for 2 h to activate the carboxyl groups. After rinsing with water, 10 μg mL−1 of anti-E. coli O157:H7 was immediately dropped on the surface and then incubated at 4 °C for 24 h. Then the surface was incubated with BSA for 1 h at room temperature to close the bound sites. After washed with PBS and water, the modified electrodes were stored at 4 °C for use.
2.3 LAMP reaction in microfluidic chip
A PDMS microfluidic chip used in this work had a simple layout with brief channels geometry (8 parallel lines, 10 mm × 0.6 mm × 0.8 mm, volume of about 5 μL). Microfluidic channels were formed in PDMS by a photolithography technique.34,35 The LAMP reaction was carried out with a 5 μL reaction mixture containing 1 × ThermoPol buffer, 8 mM MgSO4, 0.8 M Betaine, 1 mM deoxynucleoside triphosphates (dNTPs), 0.32 U Bst Polymerase, 0.2 mM (each) F3 (primer sequence: AACTACTGTAAGTAATGGAACG) and B3 (GTGATTTTTTGTTCTATGTCACT), 1.6 mM (each) FIP (TGTTGGAACAATAACTTCATCTCCTGTTGCTCTTCATTTAGCTTTG) and BIP (AATGCTATAAAATACACAGGAGCCACAGACATTTGCCAAGTTTCA).Amplification was performed in a water bath at 63 °C for 1 h. The detection result was determined by a fiber optic sensor according to the turbidity of the solution during LAMP amplification, which was then confirmed by agarose gel electrophoresis.
2.4 Sample preparation
The E. coli O157:H7 were spiked into apple juice and milk to form different concentrations, 1 CFU mL−1, 5 CFU mL−1, 10 CFU mL−1, 100 CFU mL−1 respectively. All samples were stored at 4 °C and analyzed with the proposed assays.2.5 Fluorescence microscopy analysis
The sample containing E. coli O157:H7 strains were incubated with the biosensors for 45 min, after the bacteria were captured, the biosensor was immersed into 4% paraformaldehyde solution for 30 min, and then washed with PBS for two times. The biosensor was then immersed into PI fluorescent dye for 15 min, washed by PBS for two times and dried in air. The bacteria were viewed using a fluorescence microscopy (Olympus BX53, Tokyo, Japan). Fluorescence images were obtained using a 100 × oil lens.2.6 Detecting E. coli O157:H7 with CNT multilayer-based LAMP assay
The CNT multilayers-based biosensor was incubated with 1 mL E. coli O157:H7 sample solution for 45 min, rinsed with PBS solution, cultured in LB nutrient broth medium at 37 °C, and immersed into the mixture of 5 mM [Fe(CN)6]3−/4− and 5 mM PBS for EIS measurements. Next, DNA templates of the captured bacteria were extracted with DNA extraction kit for bacteria. 0.4 μL nucleic acid extracted from the E. coli O157:H7 was then introduced via the inlet. Further, 4.6 μL LAMP reaction mixture was drawn slowly into the microchannel by capillary force. The inlet and outlet were tightly sealed by uncured PDMS to form an integral microchannel for LAMP reaction. The results were analyzed with the optical detection unit including optical fibers and digital fiber optic sensor. The fiber optic sensor employs a high-intensity red light-emitting diode (LED) light at 640 nm and a phototransistor.The launching and collecting optical fibers were inserted carefully into the fiber channels that oppose each other. The detection length is 12 mm. The reduction of optical density was used to indicate the turbidity generation of the LAMP reaction. The turbidity increased with the amplification of DNA, so optical density = ln(I0/I1) = turbidity, where I0 is the intensity of incident light and I1 is the intensity of transmitted light.
3. Results and discussion
3.1 Principle of microfluidic chip-based LAMP coupled with CNT multilayer biosensor
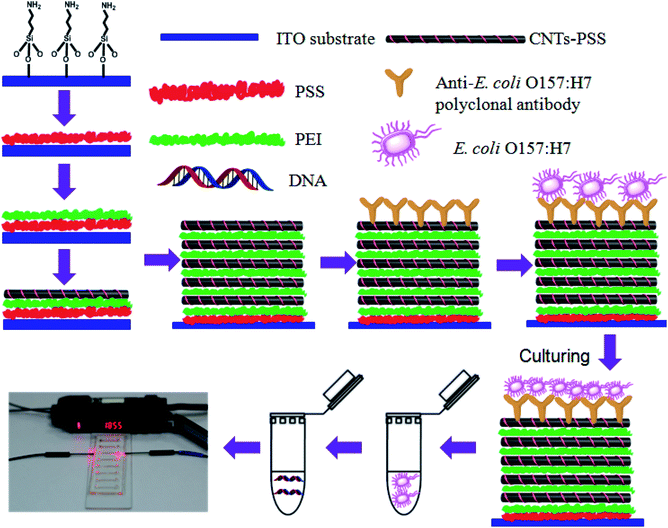
The protocol in this study combines the amplification features of LAMP with the ability of anti-E. coli O157:H7 polyclonal antibody-modified CNT multilayer electrode to selectively capturing E. coli O157:H7. The principle of the sensing platform is shown in Scheme 1. In this multilayer biosensor, MWCNTs was used to increase the surface area and electrical conductivity of sensors. The large surface area of MWCNTs allows more anti-E. coli O157:H7 polyclonal antibodies linked to MWCNTs in the multilayers via a carbodiimide-mediated wet-chemistry approach. The resulting anti-E. coli O157:H7 polyclonal antibody-modified CNT multilayer electrode are then used as the nanoscale anchorage substrates to effectively capture E. coli O157:H7 on the electrode surface through antigen–antibody interaction. When E. coli O157:H7 is present in the sample, it will be captured by the anti-E. coli O157:H7 polyclonal antibody-modified CNT multilayers through the antigen–antibody interactions, resulting in the changes in the charge transfer resistance of the electrode. The captured bacteria are then cultured in modified E. coli culture broth to increase the number of E. coli O157:H7 and improve the detection sensitivity. This culture enrichment process is inevitable because the contamination level often falls below the detection limit. DNA templates of the captured bacteria are extracted and analyzed using a microfluidic chip-based LAMP coupled with an optical turbidity sensor.3.2 Preparation, characterization and bacterial capture capability of the CNT multilayers
As shown in Scheme 1, the anti-E. coli O157:H7 polyclonal antibody-modified CNT multilayer electrode is prepared by alternatively immersing amine-functionalized ITO into negatively charged MWCNTs solution and positively charged PEI, followed by functionalization with anti-E. coli O157:H7 polyclonal antibody. UV-Vis spectroscopy, cyclic voltammetry and electrochemical impedance spectroscopy (EIS) confirmed the successful preparation of CNT multilayer electrode (see details in Part S1 of ESI and Fig. S1 and S2†).We first tested the capability of the anti-E. coli O157:H7 polyclonal antibody functionalized CNT multilayers for selectively capturing E. coli O157:H7. The antibody functionalized CNT multilayers were exposed to various concentrations of E. coli O157:H7. Since the capture efficiency depends on the incubation time and antibody concentration on the surface of electrode, we investigated the influence of incubation time and antibody concentration during the functionalization process on E. coli O157:H7 capture efficiency (Fig. S3†). A suitable concentration of anti-E. coli O157:H7 polyclonal antibody was found to be 10 μg mL−1. The optimal incubation time for the electrode was 45 min for the target capture.
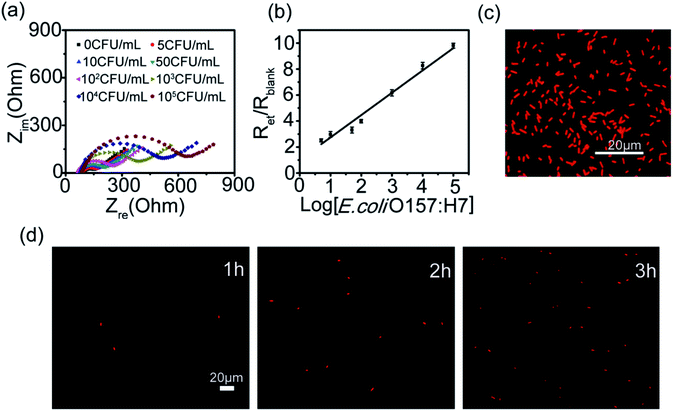
Based on the above optimized capture conditions, the capability of the electrode for capturing E. coli O157:H7 at different concentrations were analyzed with EIS and fluorescence microscopy. Fig. 1a illustrates the Nyquist plot showing a mixed charge transfer and diffusion resistance process that varies gradually with increasing the concentration of E. coli O157:H7. As shown in Fig. 1a, the charge transfer resistance (Ret) of the antibody functionalized CNT multilayers is ca. 75.71 ± 5.23 Ω. In the presence of bacteria, a greater increase in Ret is observed due to the binding of bacteria through antigen–antibody interaction. Furthermore, the Ret increases with an increase in target E. coli O157:H7 in the solution. A linear relationship between the ratio of Ret/Rblank and the logarithm of the E. coli O157:H7 concentration was found in the range of 5–105 CFU mL−1 (Fig. 1b). The fluorescence microscopy also confirmed the successful capture of E. coli O157:H7 by the antibody functionalized CNT multilayers. As shown in Fig. 1c, the E. coli O157:H7 stained by PI fluorescent dye are found to adhere to the antibody functionalized CNT multilayers and maintain good bacteria morphology.
The cell viability of the captured bacteria was next evaluated by immersing the electrodes in culture broth for 1 h, 2 h and 3 h, respectively. The captured bacteria was then stained with PI and observed with fluorescence microscope. As shown in Fig. 1d and S4,† after culturing with culture broth for 1 h, 2 h and 3 h, the number of captured bacteria on the electrode increases, indicating that the CNT multilayers has good biocompatibility and does not influence the reproduction of the target bacteria. By using staining method and microscopic counting method, we found that about 60% of target bacteria were captured in the range of 1–100 CFU mL−1 and nearly 95% cells were still viable. These results verify the anti-E. coli O157:H7 polyclonal antibody functionalized CNT multilayers possesses higher capture efficiency and good compatibility.
3.3 Quantification of the captured bacterial gene by microfluidic chip-based LAMP
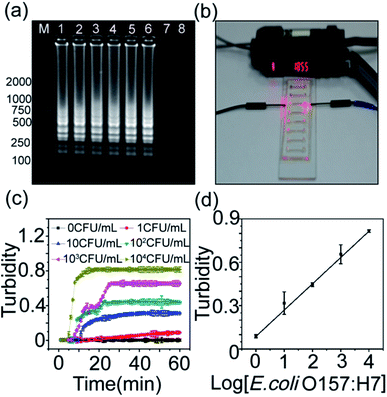
DNA templates of the captured bacteria were extracted with DNA extraction kit for bacteria and quantified further by microfluidic chip-based LAMP. The microfluidic chip-based LAMP relies on an auto-cycling strand displacement of DNA synthesis performed by the Bst polymerase and is able to synthesize large amounts of DNA under isothermal conditions in less than an hour.36 Moreover, the product of microfluidic chip-based LAMP can be detected by monitoring the turbidity of the white precipitate magnesium pyrophosphate using the optical sensors.37 Therefore, microfluidic chip-based LAMP assays are sensitive, faster, easier to use, and suitable for point-of-care detection.We first investigated the specificity of the microfluidic chip-based LAMP reaction. DNA templates extracted from the E. coli O157:H7, Bst polymerase, and a set of four primers including two inner primers and two outer primers were injected into the microchannels and reacted for 1 h at 63 °C. The product of LAMP was analyzed by agarose gel electrophoresis. As shown in Fig. 2a, in the presence of DNA templates, the LAMP gave positive results, but no positive result was observed in the absence of any DNA templates, indicating that the microfluidic chip-based LAMP reaction is specific.
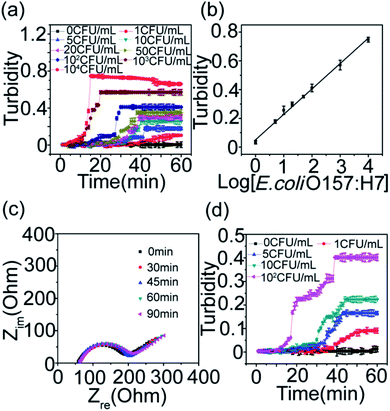
The visual detection of bacterial gene was then performed on the microfluidic chip-based LAMP reaction coupled with the turbidity sensor (Fig. 2b). Fig. 2c shows the dynamic curves of the turbidity during the microfluidic chip-based LAMP reaction. It can be found that the turbidity increases quickly with the reaction process. Moreover, the turbidity increases linearly with the bacterial concentration (Fig. 2d). This means that the bacterial concentration could be correlated to the gene quantities detected by the microfluidic chip-based LAMP reaction.
3.4 Analytic performance of the combination microfluidic chip-based LAMP coupled with CNT multilayer biosensor
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[E. coli O157:H7] + 0.083 with a correlation coefficient of 0.9929 (n = 6). The detection limit (LOD) was calculated to be 1 CFU mL−1 at 3σ. The detection limit of the proposed sensing platform was lower than those of other electrochemical methods (LOD: 8.0 × 102 cells mL−1), chemiluminescence (LOD: 103 CFU mL−1), PCR (LOD: 102 CFU mL−1) etc. (Table S1†).38–40
log[E. coli O157:H7] + 0.083 with a correlation coefficient of 0.9929 (n = 6). The detection limit (LOD) was calculated to be 1 CFU mL−1 at 3σ. The detection limit of the proposed sensing platform was lower than those of other electrochemical methods (LOD: 8.0 × 102 cells mL−1), chemiluminescence (LOD: 103 CFU mL−1), PCR (LOD: 102 CFU mL−1) etc. (Table S1†).38–40
Regeneration of the CNT multilayer biosensor was another advantage of the proposed sensing platform. After electrochemical detection, the CNT multilayer biosensor was immersed in pH 2.2 glycine–HCl buffer solutions to break the captured bacteria-antibodies on CNT multilayers interaction and release the captured bacteria. EIS and fluorescence staining method were performed to demonstrate the detachment and attachment of E. coli O157:H7. As shown in Fig. S5,† the regenerating biosensor still remained 95% of its original capturing efficiency for two times. After 6 times, the bacterial-capture efficiency decreased to 85% of its original capturing efficiency. These results confirmed that the CNT multilayer biosensor could be regenerated without significant loss of the capture efficiency.
| Sample | Added (CFU mL−1) | Proposed method | Plate count method | ||
|---|---|---|---|---|---|
| Measured (CFU mL−1) | Recovery (%) | Measured (CFU mL−1) | Recovery (%) | ||
| Apple juice | 0 | 0.76 ± 0.16 | 0.33 ± 0.58 | ||
| Apple juice | 1 | 1.01 ± 0.16 | 101.0% | 1.12 ± 0.35 | 112.0% |
| Apple juice | 5 | 5.23 ± 0.02 | 104.6% | 5.12 ± 0.63 | 102.4% |
| Apple juice | 10 | 10.89 ± 0.03 | 108.9% | 10.45 ± 0.14 | 104.5% |
| Apple juice | 100 | 102.81 ± 0.26 | 102.8% | 100.67 ± 0.21 | 100.7% |
| Milk | 0 | 0.72 ± 0.12 | 0.33 ± 0.58 | ||
| Milk | 1 | 1.21 ± 0.16 | 121.0% | 1.06 ± 0.06 | 106.0% |
| Milk | 5 | 5.50 ± 0.01 | 110.0% | 5.26 ± 0.03 | 105.2% |
| Milk | 10 | 10.13 ± 0.03 | 101.3% | 10.35 ± 0.12 | 103.5% |
| Milk | 100 | 112.12 ± 0.04 | 112.1% | 102.17 ± 0.42 | 102.2% |
4. Conclusion
In conclusion, we have demonstrated the ability of the CNT multilayer biosensor and the microfluidic chip-based LAMP combination to capture, culture, lyse, detect and quantify E. coli O157:H7. The detection sensing platform displayed high specificity and detect E. coli O157:H7 for concentrations as low as 1 CFU mL−1 without complicated instrumentation. Compared with other solid phase-based separation techniques such as magnetic beads, the capture and culture process of bacteria can be monitored in situ by electrochemical methods. In addition, the proposed sensing platform has many advantages such as low cost and simplified operations. These merits make it a potential platform for point-of-care detection of E. coli O157:H7 in hospitals, environmental biology, food industry and water supplies.Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant no. 51402069 and 51372054) and State Key Laboratory of Urban Water Resource and Environment (Harbin Institute of Technology) (Grant No. HC201527).Notes and references
- Centers for Disease Control and Prevention, Surveillance for Foodborne Disease Outbreaks, 2009–2010: Morbidity and Mortality Weekly Report. MMWR, United States, 2013, vol. 62, p. 41 Search PubMed.
- Centers for Disease Control and Prevention, in Surveillance for Foodborne Disease Outbreaks, United States, 2011, Annual Report, US Department of Health and Human Services, Atlanta, Georgia, 2014 Search PubMed.
- Centers for Disease Control and Prevention, in Surveillance for Foodborne Disease Outbreaks, United States, 2012, Annual Report, US Department of Health and Human Services, Atlanta, Georgia, 2014 Search PubMed.
- Centers for Disease Control and Prevention, in Surveillance for Foodborne Disease Outbreaks, United States, 2013, Annual Report, US Department of Health and Human Services, Atlanta, Georgia, 2015 Search PubMed.
- L. J. Yang, Y. B. Li and G. F. Erf, Anal. Chem., 2004, 76, 1107–1113 CrossRef CAS PubMed.
- K. Rijal and R. Mutharasan, Analyst, 2013, 138, 2943–2950 RSC.
- Y. X. Wang, J. F. Ping, Z. Z. Ye, J. Wu and Y. B. Ying, Biosens. Bioelectron., 2013, 49, 492–498 CrossRef CAS PubMed.
- H. Aycicek, H. Aydogan, A. Kucukkaraaslan, M. Baysallar and A. C. Basustaoglu, Food Control, 2004, 15, 253–259 CrossRef.
- C. Fournier-Wirth, M. Deschaseaux, C. Defer, S. Godreuil, C. Carriere, X. Bertrand, V. Tunez, T. Schneider, J. Coste and P. Morel, Transfusion, 2006, 46, 220–224 CrossRef PubMed.
- E. Nazemi, S. Aithal, W. M. Hassen, E. H. Frost and J. J. Dubowski, Sens. Actuators, B, 2015, 207, 556–562 CrossRef CAS.
- N. Wang, M. He and H. C. Shi, Anal. Chim. Acta, 2007, 590, 224–231 CrossRef CAS PubMed.
- U. Dharmasiri, M. A. Witek, A. A. Adams, J. K. Osiri, M. L. Hupert, T. S. Bianchi, D. L. Roelke and S. A. Soper, Anal. Chem., 2010, 82, 2844–2849 CrossRef CAS PubMed.
- P. Patel, J. A. Garson, K. I. Tettmar, S. Ancliff, C. McDonald, T. Pitt, J. Coelho and R. S. Tedder, Transfusion, 2012, 52, 1423–1432 CrossRef CAS PubMed.
- K. Sen, J. L. Sinclair, L. Boczek and E. W. Rice, Environ. Sci. Technol., 2011, 45, 2250–2256 CrossRef CAS PubMed.
- P. Elizaquível, G. Sánchez and R. Aznar, Food Control, 2012, 25, 704–708 CrossRef.
- G. Maddalo, F. Stenberg-Bruzell, H. r. Götzke, S. Toddo, P. Björkholm, H. Eriksson, P. Chovanec, P. Genevaux, J. Lehtiö and L. L. Ilag, J. Proteome Res., 2011, 10, 1848–1859 CrossRef CAS PubMed.
- X. Jiang, R. Wang, Y. Wang, X. Su, Y. Ying, J. Wang and Y. Li, Biosens. Bioelectron., 2011, 29, 23–28 CrossRef CAS PubMed.
- X. Mao, L. Yang, X. L. Su and Y. Li, Biosens. Bioelectron., 2006, 21, 1178–1185 CrossRef CAS PubMed.
- A. Subramanian, J. Irudayaraj and T. Ryan, Biosens. Bioelectron., 2006, 21, 998–1006 CrossRef CAS PubMed.
- N. Tawil, E. Sacher, R. Mandeville and M. Meunier, Biosens. Bioelectron., 2012, 37, 24–29 CrossRef CAS PubMed.
- I. Yazgan, N. M. Noah, O. Toure, S. Zhang and O. A. Sadik, Biosens. Bioelectron., 2014, 61, 266–273 CrossRef CAS PubMed.
- C. Piñero-Lambea, G. Bodelón, R. Fernández-Periáñez, A. M. Cuesta, L. Álvarez-Vallina and L. A. n. Fernández, ACS Synth. Biol., 2015, 4, 463–473 CrossRef PubMed.
- S. C. Donhauser, R. Niessner and M. Seidel, Anal. Chem., 2011, 83, 3153–3160 CrossRef CAS PubMed.
- M. Magliulo, P. Simoni, M. Guardigli, E. Michelini, M. Luciani, R. Lelli and A. Roda, J. Agric. Food Chem., 2007, 55, 4933–4939 CrossRef CAS PubMed.
- A. Wolter, R. Niessner and M. Seidel, Anal. Chem., 2008, 80, 5854–5863 CrossRef CAS PubMed.
- Y. Zhang, C. Tan, R. Fei, X. Liu, Y. Zhou, J. Chen, H. Chen, R. Zhou and Y. Hu, Anal. Chem., 2014, 86, 1115–1122 CrossRef CAS PubMed.
- W. Premasiri, D. Moir, M. Klempner, N. Krieger, G. Jones and L. Ziegler, J. Phys. Chem. B, 2005, 109, 312–320 CrossRef CAS PubMed.
- Food and Drug Administration (FDA), Bacteriological Analytical Manual (BAM), U.S., 8th edn, 2009 Search PubMed.
- J. T. Connelly, J. P. Rolland and G. M. Whitesides, Anal. Chem., 2015, 87, 7595–7601 CrossRef CAS PubMed.
- Y. Higa, R. Uemura, W. Yamazaki, S. Goto, Y. Goto and M. Sueyoshi, J. Vet. Med. Sci., 2016, 78, 1343–1346 CrossRef PubMed.
- X. Jiang, W. Jing, X. Sun, Q. Liu, C. Yang, S. Liu, K. Qin and G. Sui, ACS Sens., 2016, 1, 958–962 CrossRef CAS.
- D. Lee, Y. T. Kim, J. W. Lee and T. S. Seo, Biosens. Bioelectron., 2016, 79, 273–279 CrossRef CAS PubMed.
- Z. Z. Zheng, Y. L. Zhou, X. Y. Li, S. Q. Liu and Z. Y. Tang, Biosens. Bioelectron., 2011, 26, 3081–3085 CrossRef CAS PubMed.
- D. C. Duffy, J. C. McDonald, O. J. A. Schueller and G. M. Whitesides, Anal. Chem., 1998, 70, 4974–4984 CrossRef CAS PubMed.
- J. C. McDonald and G. M. Whitesides, Acc. Chem. Res., 2002, 35, 491–499 CrossRef CAS PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, 7 CrossRef.
- X. E. Fang, Y. Y. Liu, J. L. Kong and X. Y. Jiang, Anal. Chem., 2010, 82, 3002–3006 CrossRef CAS PubMed.
- L. Xu, J. Du, Y. Deng and N. He, J. Biomed. Nanotechnol., 2012, 8, 1006–1011 CrossRef CAS PubMed.
- Y. Liu and A. Mustapha, Int. J. Food. Microbiol., 2014, 170, 48–54 CrossRef CAS PubMed.
- J. Bai, X. Shi and T. Nagaraja, J. Microbiol. Methods, 2010, 82, 85–89 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra04583b |
| This journal is © The Royal Society of Chemistry 2017 |