Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dextran-g-lauric acid as IKK complex inhibitor carrier†
Chao-Ming Sua,
Ching Lina,
Chen-Yu Huang b,
Jih-Chao Yeha,
Tien-Yao Tsaia,
Tzong Rong Ger*a,
Ming-Chen Wang*a and
Shyh-Liang Lou*a
b,
Jih-Chao Yeha,
Tien-Yao Tsaia,
Tzong Rong Ger*a,
Ming-Chen Wang*a and
Shyh-Liang Lou*a
aDepartment of Biomedical Engineering, College of Engineering, Chung Yuan Christian University, Chungli 32023, Taiwan, Republic of China
bDepartment of Physics and Astronomy, Johns Hopkins University, MD 21218, USA
First published on 12th December 2017
Abstract
Copolymeric micelles made from dextran-g-lauric acid were optimized to carry IκB kinase-β inhibitor, IMD-0354, to form a drug delivery system and to verify the effects on drug loading and encapsulation efficiency. Dextran with different molecular weights was modified by lauric acid via an esterification process between the hydroxyl group of dextran and the carboxyl group of lauric acid. The hydrophilic dextran acts as a framework and lauric acid was grafted to its branches, forming an amphiphilic polymer. Fourier transform infrared spectrometry and 1H nuclear magnetic resonance (NMR) were used to confirm and calculate the grafting percentage of dextran-g-lauric acid. The critical micelle concentration (CMC) values were obtained using ultraviolet-visible spectrophotometry, and particle size was measured using dynamic light scattering and transmission electron microscopy (TEM). The effect of drug-loaded micelles was verified using cell viability and immunofluorescence staining. TEM images indicated that the self-assembled micelles are spherical in shape. The drug loading percentage of micelles was 0.76–4.67%, and the encapsulation efficiency was 15.4–93.75%. Regulating the hydrophilic and hydrophobic fragments of dextran-g-lauric acid changed the particle size, grafting rate, CMC, drug loading, and encapsulation efficiency of the polymer. IMD-0354-loaded micelles showed higher apoptosis expression in melanoma than free IMD-0354. This shows that controlled interactions of hydrophilic and hydrophobic regions on dextran-g-lauric acid may have profound effects on drug delivery for cancer treatment.
Introduction
Advances in nanotechnology have led to the development of many types of drug delivery vehicle, such as magnetic nanoparticles,1,2 gold nanoparticles,3,4 and functionalized quantum dots,5,6 for cancer diagnosis and clinical treatment. The accumulation of 10–100 nm scale materials on tumor tissue, which is described by the enhanced permeability and retention effect, dramatically improves tumor-selective targeting.7,8 The first application of polymeric micelles for drug delivery was reported in 1984.9 Polymeric micelles, whose size is in the range of 20–100 nm, have emerged as a potential colloidal carrier for poorly water-soluble drugs. However, some colloidal carrier systems can be captured easily by the reticuloendothelial system (RES) due to their large particle size which are then excreted out of the bloodstream. Therefore, a specific size of carrier systems must be used to achieve longevity in blood circulation and to avoid RES recognition.9,10 Understanding the characteristics of these polymeric nanoparticles, such as their hydrophobic and hydrophilic interaction, molecular weight, and particle size, are important for extending the circulation time of drug vehicles for tumor targeting and reducing the side effects of drugs on normal tissues.11Natural polymers have a wide range of applications due to their biocompatibility, biodegradability, and stability.12–14 Natural polymers have been used as a framework to modify specific components to achieve a specific function. Micelles, which are fabricated through the self-assembly of amphiphiles, are widely used as a drug-loading carrier. Drugs are physically encapsulated in the micelles and can be triggered through hydrolysis15,16 or environmental response such as pH17,18 and temperature.19–21 The natural polymer dextran is manufactured from Leuconostoc mesenteroides NRRL B-512(F). It consists of (1,6)-linked glucan with side chains attached to the 3-positions of the backbone glucose units. Onishi used dextran with various molecular weights in its graft copolymerization with methyl methacrylate and showed that the molecular weight of dextran affects the graft percentage of the copolymer.22 In another study, Francis applied dextran-grafted-polyethyleneglycol alkyl ether (DEX-g-PEG-Cn) to self-assembled micelles and carried cyclosporin A (CsA) to form a drug delivery system. The percentage of CsA loading to micelles was determined by varying the molecular weights of dextran and fragment number of PEG-Cn grafted per dextran chain. The results showed that increasing the number of hydrophobic fragments on the dextran main chain and decreasing the dextran molecular weight had a profound effect on the encapsulation of poorly water-soluble drugs such as CsA.23 Liu et al. synthesized dextran–agmatine bioconjugate and modified it with lauric acid for DNA delivery system. To graft dextran with lauric acid can promote the complexation with DNA due to hydrophobically cooperative binding effect, which is beneficial for hydrophobic drug loading.24 Ji et al. developed dextran-graft-(2-diazo-1,2-naphthoquinone) micelles for intracellular delivery of doxorubicin by NIR irradiation. They investigated the effects of changing the ratio of hydrophobic DNQ molecule on the micelle property such as critical micelle concentration or size, which results suggested hydrophobic molecule could enhance the micelle stability.25 Wen et al. synthesized deoxycholic acid-grafted dextran to encapsulate doxorubicin for drug delivery, which demonstrated a potential for cancer therapy due to the redox-responsive of micelles.26 Besides, study also show dextran modified with bile acids as hydrophobic fluorescent probe for investigating the fluorescence quantum yield and fluorescence emission maximum.27 Tiera et al. examined the adjustment of molar weight of modified dextran to the micelles, which show the drug efficiency related to the degrees of substitution and the increased interchain interactions of chemical structure are responsible for the formation of hydrophobic aggregates.28
A novel IκB kinase-β inhibitor, IMD-0354 inhibits the phosphorylation of IκB which in turn inhibits NF-κB activation into the nucleus. This suppresses the growth of human breast cancer cells, human mast cells, breast stem cancer cells, T-cell leukemia cells, and chronic lymphocytic leukemia by blocking the cell cycle and inducing apoptosis.29–34
In the previous literature, fatty acids such as palmitic acid (C16) or stearic acid (C18) was used to conjugate 8-amino-3,6-dioxaoctanoic acid and RGD to form amphiphilic structure.35 The CMC values were found to be lower for micelles with longer carbon chain than those with shorter carbon chain, while the size distributions were in the positive correlation with the carbon chain. The change of carbon chain length contributed by the conjugation of fatty acids would affect the micelles structure due to the change of hydrophilic or hydrophobic property. Lauric acid we used is a natural fatty acid with 12 carbon atom chain, which is of great anti-bacterial property. Lauric acid were previously prepared as micelles36,37 for drug delivery38 However, previous literatures do not investigate the features of micelles with lauric acid as hydrophobic fragments, which require optimization due to the short carbon chain make it difficult to form micelles with high hydrophilic framework.
In this study, self-assembled amphiphile dextran-g-lauric acid was synthesized by varying the molecular weight of dextran to control the hydrophilic fragment of the micelle and by using different graft ratios of dextran to lauric acid to control the hydrophobic region. The micelle was loaded with IκB kinase B inhibitor, IMD-0354, to create a drug delivery system. The effects of inhibitor loading and the encapsulation efficiency of the drug were studied in terms of the interaction between the hydrophilic and hydrophobic regions.
Materials and methods
Materials
Dimethyl sulfoxide (DMSO) (>99.9%) was received from J.T. baker (N.J., USA). Dextran from Leuconostoc spp. (MW = ∼40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 6000), dodecanoic acid (lauric acid) (98%), N,N′-dicyclohexylcarbodiimide (DCC) (99%), 4-(dimethylamino)pyridine (DMAP) (>99%), calcium hydride (CaH2) (99.99%), sodium hydrogen carbonate, Triton X-100, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), and fluorescein isothiocyanate isomer I were purchased from Sigma-Aldrich (MO, USA). Potassium bromide (KBr) (>99%) was purchased from Scharlau (Spain). All chemicals, reagents, and solvents were used without further purification. Penicillin–streptomycin, fetal bovine serum (FBS), and Dulbecco's Modified Eagle Medium (DMEM) were purchased from Gibco (N.Y., USA). Goat anti-mouse IgG was purchased from Millipore (Darmstadt, Germany). Anti-NFκB, p65 subunit (monoclonal antibody) and goat anti-mouse IgG were purchased from Millipore.
000 and 6000), dodecanoic acid (lauric acid) (98%), N,N′-dicyclohexylcarbodiimide (DCC) (99%), 4-(dimethylamino)pyridine (DMAP) (>99%), calcium hydride (CaH2) (99.99%), sodium hydrogen carbonate, Triton X-100, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), and fluorescein isothiocyanate isomer I were purchased from Sigma-Aldrich (MO, USA). Potassium bromide (KBr) (>99%) was purchased from Scharlau (Spain). All chemicals, reagents, and solvents were used without further purification. Penicillin–streptomycin, fetal bovine serum (FBS), and Dulbecco's Modified Eagle Medium (DMEM) were purchased from Gibco (N.Y., USA). Goat anti-mouse IgG was purchased from Millipore (Darmstadt, Germany). Anti-NFκB, p65 subunit (monoclonal antibody) and goat anti-mouse IgG were purchased from Millipore.
Synthesis of dextran-g-lauric acid
DMSO and CaH2 were mixed under vacuum concentration to obtain water-free DMSO. Lauric acid and dextran were dissolved in 5 mL of DMSO, and DCC and DMAP were added to the solution with a molarity ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (lauric acid
3 (lauric acid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DCC
DCC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMAP). The synthesis parameters are shown in Table 1. The solution was heated to 60 °C for 48 h under a nitrogen atmosphere. Then, it was placed in a dialysis membrane with a molecular weight cut-off of 1200 Da and dialyzed for 48 h. The dialyzed solution was allowed to stand in a separatory funnel for 48 h. The aqueous layer was removed and dialyzed for 24 h. The polymer lauric acid-g-dextran was obtained after freeze drying.
DMAP). The synthesis parameters are shown in Table 1. The solution was heated to 60 °C for 48 h under a nitrogen atmosphere. Then, it was placed in a dialysis membrane with a molecular weight cut-off of 1200 Da and dialyzed for 48 h. The dialyzed solution was allowed to stand in a separatory funnel for 48 h. The aqueous layer was removed and dialyzed for 24 h. The polymer lauric acid-g-dextran was obtained after freeze drying.
| Compound | Dextran (Da) | Dextran![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lauric acid lauric acid |
Reaction time (h) | GR% |
|---|---|---|---|---|
| Dex40LA0.75 | 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75 0.75 |
48 | 11 |
| Dex40LA1.5 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 1.5 |
12 | ||
| Dex6LA0.75 | 6000 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75 0.75 |
14.7 | |
| Dex6LA1.5 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 1.5 |
18.7 |
Determination of critical micelle concentrations on dextran-g-lauric acid micelles
Dextran-g-lauric acid with various concentrations (0.001–1 wt%) was dissolved in 4 μM DPH solution and sonicated for 30 min. The solution was allowed to stand for 4 h in the dark. The CMC of dextran-g-lauric micelles was measured at 356 nm using an ultraviolet/visible (UV/vis) spectrophotometer.Preparation of dextran-g-lauric acid micelles loaded with IMD-0354
5 mg mL−1 dextran-g-lauric acid was dissolved in DMSO and stirred for 30 min. The solvent was mixed with an equal volume of deionized water. The solution was dialyzed using a dialysis membrane for 48 h. The molecular weight cut off (MWCO) of dialysis membrane used is less than 1200 Da. IMD-0354 DMSO solution was added to 5 mg mL−1 dextran-g-lauric acid in DMSO solvent and stirred for 30 min. Dextran-g-lauric acid and IMD-0354 were prepared in a ratio of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The solution was mixed into an equal volume of deionized water and dialyzed for 48 h using a dialysis membrane (MWCO < 1200 Da). Inhibitor-loaded micelles were obtained after freeze drying.
1. The solution was mixed into an equal volume of deionized water and dialyzed for 48 h using a dialysis membrane (MWCO < 1200 Da). Inhibitor-loaded micelles were obtained after freeze drying.
Property measurement of drug-loaded micelles
Inhibitor-loaded micelles in phosphate-buffered saline (PBS) were dialyzed (MWCO < 1200) for 72 h. The concentration of IMD-0354 was measured at 299 nm using a UV-vis spectrophotometer. Encapsulation efficiency (EE%) was calculated as:| EE% = (weightloaded drug)/(weighttotal drug) × 100% | (1) |
The inhibitor-loaded micelles were added into DMSO and sonicated for 15 min to release the drug from the micelles. The solution was filtered using a 0.45 μm filter and mixed into a co-solvent (PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO = 1
DMSO = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (wt%)). The drug loading percentage was calculated as:
1 (wt%)). The drug loading percentage was calculated as:
| DL% = (weightloaded drug)/(weightpolymer) × 100% | (2) |
The experiment began with 10 mg of IMD-Dex6LA1.5 being dissolved in 10 mL of PBS. The solution was placed in a MWCO 1 kDa dialysis membrane. Then, it was placed in a beaker containing 30 mL of PBS maintained at 37 °C and stirred at 60 rpm. For the measurement, 0.7 mL of the outer dialysate was extracted to mix with 0.7 mL of DMSO. Note that PBS is mixed with equal volume of the dialysate. The mixed solution was measured at 299 nm using a UV-vis spectrophotometer and then the calibration curve was calculated for IMD-0354 dissolved in PBS/DMSO solution (PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO = 1
DMSO = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).
Analysis of micelle properties
The dextran-g-lauric acid was mixed with KBr and pressed into disks for analysis using an FT-IR spectrometer (FT/IR-4200, Jasco, Japan). The grafting rate was measured using a nuclear magnetic resonance (NMR) spectrometer (Ascend™ 600, Bruker, Germany). The dextran-g-lauric acid was dissolved in D2O for grafting rate analysis. The particle size of micelles was measured using dynamic light scattering (DLS) (LS series, Beckman Coulter, USA). The solution was filtered using a 0.45 μm filter when the concentration of the polymer solution was higher than the CMC. A drop of the solution was placed on a copper grid to measure the particle size using transmission electron microscopy (TEM; JEM-200CX, JEOL, Japan).Cell culture
In order to determine the effect of the synthesized micelles on tumour cells, we obtained the B16F10 melanoma cell line from National Cheng Kung University Hospital, Tainan, Taiwan. Cells were maintained in DMEM, supplemented with 10% FBS, 3.7 g of sodium bicarbonate, and 1% penicillin–streptomycin.Cell survival
The following preparation was conducted for the MTT study to examine whether the carrier was more effective with IMD-0354 loading. B16F10 melanoma cells were seeded in a 96-well plate with 1 × 103 cells per well. After 24 h, the medium was replaced with 0.1–10 μg mL−1 IMD-Dex6LA1.5 or free IMD-0354 containing medium. After 72 h culture period, the medium was removed and washed with PBS two times. MTT reagent (0.5 mg mL−1) was added to the medium and incubated for 4 h at 37 °C. Afterwards, the medium was replaced with 200 μL of DMSO to solubilize the formazan. The optical density (OD) was measured at 570 nm by enzyme-linked immunosorbent assay reader (1500-490, Thermo, USA) to determine the cell survival.Micelles uptake and cellular apoptosis
Dex6LA1.5 and FITC was dissolved in DMSO for 30 min, then 5 mL deionized water was added and reacted 24 h. The solution was dialyzed (MWCO < 1200) for 48 h and filtered using a 0.22 μm filter, then FITC-Dex6LA1.5 micelles was freeze dried. B16F10 cells were seeded in a 8-well culture slide with cell concentration of 4 × 103 cells per well, and then cultured with medium contain FITC-Dex6LA1.5 micelles (0.1 mg mL−1) after 24 h. After washed with PBS two times, the cells were fixed in 3.7% formaldehyde for 30 min. Confocal microscopy (LSM 510 Meta, Zeiss, Germany) was used to observe cell uptake micelles.Cellular apoptosis induced by IMD-Dex6LA1.5 or free IMD-0354 was determined by the following procedures. B16F10 melanoma cells were seeded in a 24-well plate with cell concentration of 7 × 103 cells per well. After 24 h, the medium was replaced by 0.5 μg mL−1 IMD-Dex6LA1.5 or free IMD-0354 in medium and then cultured for 4–72 h. The medium was removed and the cells were washed with PBS two times. The cells were then fixed in 3.7% formaldehyde for 30 min and washed again with PBS two times. 0.1% Triton™ X-100 surfactant in PBS were added into the cells for 10 min. The solution was removed and then 5% bovine serum albumin was added to the cells and allowed to react for 30 min. The anti-NFκB antibody p65 (200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was reacted with the cells at 4 °C for at least 12 h. The secondary goat anti-mouse IgG FITC antibody was used and reacted at room temperature for 1 h. The cells were washed twice with PBS, and then goat anti-mouse IgG antibodies were added in the dark for 1 h. The cells were washed and 10 μg mL−1 Hoechst 33258 was added for 20 min. The cells were again washed to remove the reacted solution. Finally, the cells were observed under a fluorescence microscope.
1) was reacted with the cells at 4 °C for at least 12 h. The secondary goat anti-mouse IgG FITC antibody was used and reacted at room temperature for 1 h. The cells were washed twice with PBS, and then goat anti-mouse IgG antibodies were added in the dark for 1 h. The cells were washed and 10 μg mL−1 Hoechst 33258 was added for 20 min. The cells were again washed to remove the reacted solution. Finally, the cells were observed under a fluorescence microscope.
To observe the nuclear morphology changes, cells were seeded, treated with 0.5 μg mL−1 of sample (Dex6LA1.5, IMD-Dex6LA1.5, free IMD-0354) for 4–72 hours. Cells were then fixed in 3.7% formaldehyde and incubated with 10 μg mL−1 of Hoechst 33258 to stain for 30 min.
Results and discussion
Functional group analysis
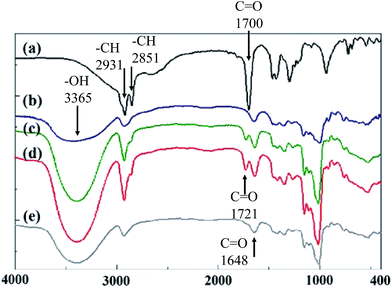
The grafting of the –OH functional group on dextran and the –COOH functional group on lauric acid resulted in a dehydration reaction. The hydrophilic dextran was acted as a framework of dextran-g-lauric acid and lauric acid was grafted to its branches, forming an amphiphilic polymer. FT-IR spectra was used to analyze the functional groups of the polymer and to evaluate their relationship. As shown in Fig. 1a, lauric acid has the symmetrical stretching vibration of the –CH group at 2851 cm−1. The absorption peaks at 1291 and 1700 cm−1 of lauric acid indicate the presence of –OH and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively.39 In the FT-IR spectrum of dextran (Fig. 1b), the peak at 1648 cm−1 confirms the presence of C
O, respectively.39 In the FT-IR spectrum of dextran (Fig. 1b), the peak at 1648 cm−1 confirms the presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration.40,41 In the lauric acid grafted dextran (DexLA), the C
O stretching vibration.40,41 In the lauric acid grafted dextran (DexLA), the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O functional groups in 1700 cm−1 t was shift to 1727 cm−1 during esterification reaction process, which result suggests the grafting of lauric acid to dextran. The peak at 1291 cm−1, indicating –OH stretching vibration, on lauric acid is not noticeable. The grafting of lauric acid to dextran consumed the hydrogen from the hydroxyl group, thereby weakening the functional group's absorption in the spectra. However, in Fig. 1e, the characteristic signal of lauric acid grafted to higher-molecular-weight dextran is lower compared to that obtained with other experimental parameters, indicating poor efficiency of the reaction.
O functional groups in 1700 cm−1 t was shift to 1727 cm−1 during esterification reaction process, which result suggests the grafting of lauric acid to dextran. The peak at 1291 cm−1, indicating –OH stretching vibration, on lauric acid is not noticeable. The grafting of lauric acid to dextran consumed the hydrogen from the hydroxyl group, thereby weakening the functional group's absorption in the spectra. However, in Fig. 1e, the characteristic signal of lauric acid grafted to higher-molecular-weight dextran is lower compared to that obtained with other experimental parameters, indicating poor efficiency of the reaction.
Nuclear magnetic resonance analysis
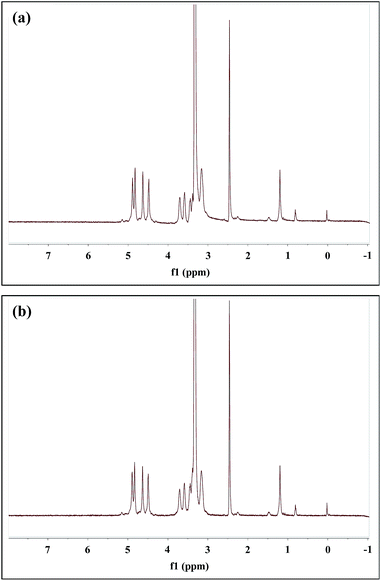
The peaks of dextran of DexLA has 1H NMR spectral resonances appears in the range of 3.1–3.8 ppm which show hydrogen protons of glucopyranosyl ring. The peaks belong to lauric acid of DexLA has 1H NMR spectral resonances at δ = 0.85, the hydrogen protons of methyl, and δ = 1.24, 1.51, 2.30, and 11.9 were hydrogen protons of carbon chain.42–44 The response at δ = 11.9 represents the hydroxyl group of lauric acid, where the region of grafting to dextran occurs. After the graft reaction of dextran and lauric acid, the δ = 11.9 peak disappeared (Fig. 2s†).The grafting rate (GR%) (eqn (3)) of lauric acid in DexLA at Fig. 2 and 3 are shown in Table 1. The percentage of the C1 position (δ = 4.9) of hydroxyl in the dextran graft to the C12 position (δ = 0.85) of carboxyl in lauric acid represents the quantity of dextran-g-lauric acid.
| GR% = (A0.8 ppm ÷ 3)/A4.9 ppm × 100% | (3) |
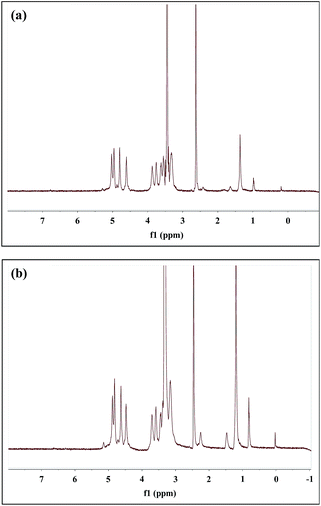 | ||
Fig. 3 1H NMR spectrum of dextran-g-lauric acid in D2O. Molecular weight of dextran is 6k. Molecular ratio of dextran to lauric acid is (a) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75 (Dex6LA0.75) and (b) 1 0.75 (Dex6LA0.75) and (b) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 (Dex6LA1.5). 1.5 (Dex6LA1.5). | ||
Grafting rate percentages of 11% and 12% were obtained for Dex40LA0.75 and Dex40LA1.5, respectively. In the framework of dextran (MW = 6000), Dex6LA0.75 and Dex6LA1.5 have GR% of 14.7% and 18.7%, respectively. The results showed that the higher GR% could be retrieved by using low molecular weight of dextran (MW = 6000) compared with high molecular weight dextran (MW = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). For Dex40LA micelles, the hydroxyl groups of higher molecular weight dextran might be hidden due to the long carbon chain form random coils which caused inefficient esterification during reaction. The GR% increased with increasing number of lauric acid chains and decreasing dextran molecular weight.
000). For Dex40LA micelles, the hydroxyl groups of higher molecular weight dextran might be hidden due to the long carbon chain form random coils which caused inefficient esterification during reaction. The GR% increased with increasing number of lauric acid chains and decreasing dextran molecular weight.
Measurement of Dex–LA micelle CMC
Table 2 and Fig. 1s† shows the measured CMC of amphiphiles obtained with various grafting parameters. The CMC values of Dex40LA0.75, Dex40LA1.5, Dex6LA0.75, and Dex6LA1.5 are 0.788, 0.785, 1.274, and 0.705 mg mL−1, respectively. This shows that almost the same CMC values were obtained as using higher molecular weight dextran (=40k). Instead, for micelles with lower molecular weight (=6k), as the graft ratio of lauric acid increase, the CMC value decrease, which indicates hydrophobicity of the polymer increased. Therefore, the amphiphilic micelles have to reach a specific concentration and balance between hydrophilic and hydrophobic forces. The similar GR% values of Dex40LA0.75 and Dex40LA1.5 indicates that the micelles have identical numbers of hydrophilic and hydrophobic fragments, which is important in the formation of micelles.| Micelle type | CMC (mg mL−1) | Particle size (nm) | PDIa | |
|---|---|---|---|---|
| TEM | DLS | |||
| a PDI is according to DLS results. | ||||
| Dex40LA0.75 | 0.788 | 82.2 ± 16.97 | 83.62 ± 1.35 | 0.69 ± 0.12 |
| Dex40LA1.5 | 0.785 | 57.44 ± 16.08 | 63.44 ± 33.67 | 0.50 ± 0.28 |
| Dex6LA0.75 | 1.274 | 35.35 ± 5.39 | 47.18 ± 1.64 | 0.52 ± 0.03 |
| Dex6LA1.5 | 0.705 | 17.57 ± 2.51 | 20.41 ± 11.9 | 0.63 ± 0.43 |
Micelle morphology and particle size
The particle size and morphology of micelles were observed using TEM and DLS. TEM images revealed spherical Dex–LA micelles. The particle sizes of Dex40LA0.75, Dex40LA1.5, Dex6LA0.75, and Dex6LA1.5 are 82.20, 57.44, 35.35, and 17.57 nm, respectively. The images are shown in Fig. 4 and the results are shown in Table 2. From DLS data (Table 2), the particle sizes of Dex40LA0.75, Dex40LA1.5, Dex6LA0.75, and Dex6LA1.5 are 83.62, 63.44, 47.18, and 20.41 nm, respectively. The size distribution of Dex–LA micelles has two different ranges depending on the molecular weight (40k or 6k) of the polymeric framework. The particle size of Dex40LA is higher than that of Dex6LA, which signifies that hydration is augmented in Dex40LA, promoting micelle swelling.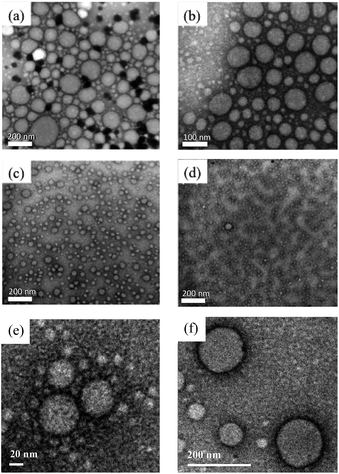 | ||
| Fig. 4 TEM image of micelles formed with dextran graft lauric acid. (a) Dex40LA0.75, (b) Dex40LA1.5, (c) Dex6LA0.75, (d) and (e) Dex6LA1.5, (f) Dex6LA1.5 loaded with IMD-0354. | ||
It is believed that the size of micelles depends on the interaction between its hydrophilic and hydrophobic regions. In Dex6LA micelles, the hydrophobic force is relatively stronger, making the micelles shrink. In this study, the hydrophilic fragments were controlled by using different molar weights of dextran in the Dex–LA micelles. The hydrophilic region of the polymer can be modified by adding dextran with a larger molecular weight or modifying the hydrophobic region by increasing the ratio of lauric acid to dextran. Through the reaction of hydrophobic and hydrophilic fragments, many of the physical properties of micelles were changed, such as CMC and particle size.
The DLS test show that micelles present high polydispersity index (PDI) values (0.50–0.69), showing a rather polydispersed distribution. The dextran with a higher molecular weight as the polymer framework may cause partial folding of the dextran, which in term cause instability of the micelle structure, and then the hydroxyl groups are hidden and limits the esterification reaction. The PDI value of Dex40LA is higher than that of Dex6LA, showing that the size of micelle was not uniform at higher molecular weight dextran. An appropriate MWCO of dialysis membrane and extension of the dialysis duration can minimize the nonuniformity problem.
Encapsulation efficiency and drug loading
UV-vis spectrophotometer was used to determine the presence of IMD-0354. For free Dex6LA1.5 micelles, there is no obvious absorbed peaks, but or IMD-Dex6LA1.5 and IMD-0354 both show absorption at 250–300 nm (Fig. 3s†). The result show that drug IMD-0354 has been embedded. The loading capacity of drug to micelles was evaluated in terms of encapsulation efficiency and drug loading percentage. By increasing the graft ratio of lauric acid to dextran has led to a slight increase in the drug loading of micelles. Moreover, increasing the IMD-0354 drug concentration ratio on micelles can effectively improve the drug loading of micelles.The micelle formation is contributed by both hydrophilic and hydrophobic blocks and our study suggest that the balance between hydrophilic (dextran) and hydrophobic (lauric acid) component allow the completion of micelles structure (Fig. 4s†). The lauric acid which composed of carbon chain would become the hydrophobic core. From the result of Table 3, the encapsulation efficiency (EE%) of IMD-Dex40LA0.75 and IMD-Dex40LA1.5 were 15.4 and 39.46%, respectively, which suggests that the content of lauric acid in micelles could affect encapsulating content of hydrophobic drug IMD-0354. The encapsulation of Dex40LA efficiency was low may be caused by the inadequate hydrophobic compartments integrated to the micelles. IMD-Dex6LA1.5 show a high encapsulation efficiency which is near 94%. The ratio (w/w) of DexLA to drug were adjusted to be 10 or 20, which show drug loading of 93.75 and 91.58%, respectively. This results implied that the Dex6LA1.5 was a for drug loading in the ratio of DexLA to drug was 20 (Table 1s†).
| Compound | Micelles![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) drug (wt%) drug (wt%) |
EE% | DL% |
|---|---|---|---|
| IMD-Dex40LA0.75 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
15.4 | 0.76 |
| IMD-Dex40LA1.5 | 39.47 | 1.96 | |
| IMD-Dex6LA0.75 | 38.1 | 1.9 | |
| IMD-Dex6LA1.5 | 93.75 | 4.67 |
In addition, from both EE% and DL% data, the increase of lauric acid ratio show increasing the drug-loading in micelles, which suggests that hydrophobicity can control the relationship between micelles and the drug. Moreover, the particle size of micelles increase from 17.57 ± 2.51 nm to 60.8 ± 9.28 nm after drug loading (Fig. 4e).
The ability of micelles to carry drugs depends on the interaction of their hydrophilic and hydrophobic regions. In a study of cyclosporin A (CsA) loaded DEX-g-PEG-Cn micelles by Leroux,23 the molecular weight of dextran used were 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 40
000 and 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da. The drug loading property of CsA-loaded DEX-g-PEG-Cn micelles changed with dextran molecular weight. The micelles formed by dextran with a higher molecular weight provided higher hydrophilicity, whereas dextran with a lower molecular weight formed micelles with lower hydrophilicity. Therefore, micelles with higher-molecular-weight dextran will have decreased hydrophobic drug CsA loading content. In our study, the drug loading percentages of IMD-Dex40LA0.75 and IMD-Dex6LA0.75 micelles are 0.76% and 1.9%, respectively. IMD-0354 encapsulated in micelles is a hydrophobic drug. Dex–LA micelles with higher-molecular-weight dextran have poor drug loading content. IMD-Dex6LA1.5 has higher drug loading percentage (4.76%) compared to IMD-Dex40LA1.5 due to increased hydrophobic segments. This drug loading content result is similar to that obtained in a study of Leroux.23
000 Da. The drug loading property of CsA-loaded DEX-g-PEG-Cn micelles changed with dextran molecular weight. The micelles formed by dextran with a higher molecular weight provided higher hydrophilicity, whereas dextran with a lower molecular weight formed micelles with lower hydrophilicity. Therefore, micelles with higher-molecular-weight dextran will have decreased hydrophobic drug CsA loading content. In our study, the drug loading percentages of IMD-Dex40LA0.75 and IMD-Dex6LA0.75 micelles are 0.76% and 1.9%, respectively. IMD-0354 encapsulated in micelles is a hydrophobic drug. Dex–LA micelles with higher-molecular-weight dextran have poor drug loading content. IMD-Dex6LA1.5 has higher drug loading percentage (4.76%) compared to IMD-Dex40LA1.5 due to increased hydrophobic segments. This drug loading content result is similar to that obtained in a study of Leroux.23
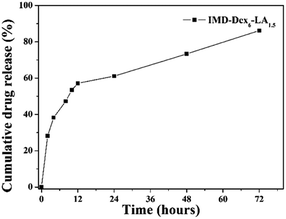
Fig. 5 shows the drug release curve, which can be divided into two slopes, one in the 0–12 h period and the other in the 12–72 h period. In the 0–12 h period, water diffused into the micelles and caused the gradient of drug concentration to change. This drove the drugs out from the micelles via diffusion. In 12 h, the cumulative drug release reached 53.37%. These results are similar to those reported for doxorubicin-loaded stearate-g-dextran micelles (50% drug release was obtained in 10 h).32 In the 12–72 h period, the hydrolysis of micelle ester linkages caused the structure of micelles to loose inducing an increase in drug release. The hydrolysis of micelles improved water diffusing into micelles and the increase of the ester linkage degradation lead to drug release. The cumulative drug release of IMD-Dex6LA1.5 reached 86.15% in 72 h.
As discussed above, our carriers are based on the mechanism of hydrolysis ester linkage. Utilizing polymers with ester linkages to synthesize micelles has some advantages. The ester linkages of micelles is regarded as short hydrophobic fragments. It is preferable to employ short hydrophobic molecules for carriers and to link them with an ester group, for good human metabolism. By turning lengthy hydrophobic molecules this way the lysis of bond between the ester group and hydrophobic molecules occur which further adds on for metabolism to take place in an effective manner. In another study, Holmberg compared the hydrolysis rates of micelles formed by ester and carbonate surfactants. It was shown that micelles with ester surfactant has slower rates of hydrolysis than the carbonate surfactant implying that an ester surfactant prolongs micelle circulation in the body.45
Nakano provided another example that favors hydrolysis of ester linkage mechanism.46 They showed that the catalytic enzyme β-glucosidase cleaves the glucosyl ester linkage much more weakly than the glucosidic linkage. Even so, the glucosyl ester linkage requires 24 h to activate its cleaved effect. For micelles that must be retained in the human body for a period of time, the hydrolysis of ester linkage compounds should be a favorable choice. Our micelle design requires drugs to remain for 12 h in the body, which is consistent with Nakano's study.46
The developed IMD-0354 loaded-micelles were applied for anti-tumor applications. IMD-0354 (N-(3,5-bis-trifluoromethyl-phenyl)-5-chloro-2-hydroxy-benzamide) is a novel inhibitor of the IKK-induced phosphorylation of IkBβ and hence protects it from undergoing degradation, which in turn results in the inhibition of NF-κB activation.47 NF-βB is overexpressed in tumors, and thus IMD-0354 as a novel NF-βB inhibitor has attracted the attention of researchers.29,47 Due to the enhanced permeability and retention (EPR) effect, the time of drug circulation of micelles loaded with IMD-0354 in the human body is extended.
Cell viability and mechanism of IMD-0354-loaded micelles
Fig. 6 shows that melanoma co-cultured with IMD-Dex6LA1.5 or free IMD-0354 could induce cell death at a low dosage. IMD-0354 with a molecular weight lower than 1000 g mol−1 is diffused into the cell.8 Comparing with free IMD-0354, IMD-Dex6LA1.5 has a more efficient cellular uptake through endocytosis of melanoma, which increases the cumulative IMD-0354 dosage. Therefore, IMD-Dex6LA1.5 is more efficient in inducing apoptosis than the free IMD-0354. These results show that IMD-Dex6LA1.5 could enhance the drug effect by reducing the dosage to achieve better treatment. Though, free IMD-0354 and IMD-0354 encapsulated in Dex6LA1.5 both induce apoptosis in melanoma, the more effective treatment result of drug-loaded micelles could be achieved even in low dosage than the free IMD-0354 treatment.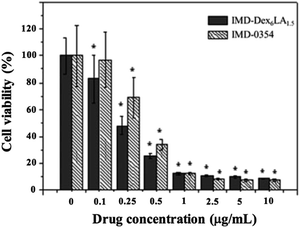 | ||
| Fig. 6 Cell survival rate of B16F10 melanoma co-culture with various concentrations of IMD-Dex6LA1.5 or free IMD-0354 at 72 h. | ||
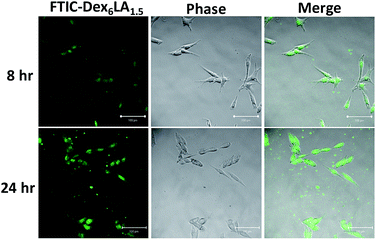
To evaluate the micelles accumulative phenomenon, we cultured melanoma with FITC-Dex6LA1.5 micelles to observe the uptake of micelles by fluorescent images. From the results of Fig. 7, the green fluorescent was accumulated in cytoplasm, which confirmed that the micelles were uptake and accumulated as incubation time increased.
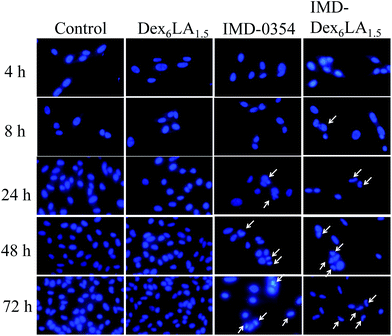
As shown in Fig. 8, melanoma cultured with IMD-Dex6LA1.5 or free IMD-0354 had no significant difference compared to the control or Dex6LA1.5 groups at 4 and 8 h. This means that the cumulative drug concentration of IMD-Dex6LA1.5 or free IMD-0354 uptaken by the cells was insufficient to inhibit NF-κB expression. In co-culture at 24 h, NF-κB expression was inhibited. NF-κB expression was obviously inhibited by IMD-Dex6LA1.5 or free IMD-0354 at 48 and 72 h. Of note, the fluorescence of the nucleus also decreased because IMD-0354 not only inhibited the NF-κB pathway on the cytoplasm but also inhibited upstream of the IKK complex to activate the NF-κB pathway, which suppresses NF-κB into the nucleus.40
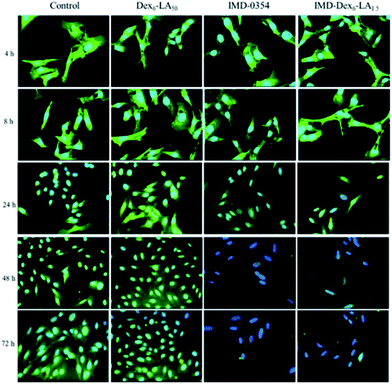 | ||
| Fig. 8 Immunofluorescence staining of B16F10 melanoma co-culture with Dex6LA1.5, IMD-Dex6LA1.5, or free IMD-0354. Green fluorescence represents NF-κB and blue fluorescence represents Hoechst 33258. | ||
IMD-0354 could inhibit phosphorylation of IKK complex that induced cancer cell apoptosis due to activation of IκB and NF-κB were block.29–34 In Fig. 9, the time-dependent changes in nuclear morphology of the cells treated with IMD-Dex6LA1.5, were compared with that of Dex6LA1.5, free IMD-0354 and non-treated (control). For free IMD-0354 and IMD-Dex6LA1.5 groups, the nucleus show atrophy and incomplete, the fluorescence intensity of nucleus also enhanced by chromatin condensation. The results shown that IMD-Dex6LA1.5 show excellent property to deliver IMD-0354 to cause cell apoptosis.
Conclusions
The goal of this study was to synthesize self-assembled amphiphiles dextran-g-lauric acid that carries the drug IMD-0354 for anti-tumor applications. By regulating the hydrophilic and hydrophobic segments of dextran-g-lauric acid micelles, the effects of particle size, CMC, and drug contents were studied. The micelles swelled to a larger size due to stronger hydrophilic interactions, but the number of hydrophobic segments did not affect CMC. IMD-0354 release from micelles was 86.15% in 72 h and increasing the amount of drug can improve DL% and EE%. The results of this study indicate that the IMD-loaded dextran-g-lauric acid micelles have potential for cancer treatment.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This manuscript is dedicated to the memory of Prof. Shyh-Liang Lou, who sadly passed away on 12 September 2016, whose wisdom and insight greatly contributed to benefit the original ideas for this paper. This work was supported partly by Ministry of Science and Technology, Taiwan, under Grant no. MOST 104-2112-M-033-008-MY2 and MOST 105-2221-E-033-006-.References
- H. Xu, Z. P. Aguilar, L. Yang, M. Kuang, H. Duan, Y. Xiong, H. Wei and A. Wang, Biomaterials, 2011, 32, 9758–9765 CrossRef CAS PubMed.
- T. Koyama, M. Shimura, Y. Minemoto, S. Nohara, S. Shibata, Y. Iida, S. Iwashita, M. Hasegawa, T. Kurabayashi and H. Hamada, J. Controlled Release, 2012, 159, 413–418 CrossRef CAS PubMed.
- G. Bisker, D. Yeheskely-Hayon, L. Minai and D. Yelin, J. Controlled Release, 2012, 162, 303–309 CrossRef CAS PubMed.
- G. Unak, F. Ozkaya, E. I. Medine, O. Kozgus, S. Sakarya, R. Bekis, P. Unak and S. Timur, Colloids Surf., B, 2012, 90, 217–226 CrossRef CAS PubMed.
- H. Hafian, A. Sukhanova, M. Turini, P. Chames, D. Baty, M. Pluot, J. H. Cohen, I. Nabiev and J.-M. Millot, Nanomedicine, 2014, 10, 1701–1709 CrossRef CAS PubMed.
- K. H. Lee, J. F. Galloway, J. Park, C. M. Dvoracek, M. Dallas, K. Konstantopoulos, A. Maitra and P. C. Searson, Nanomedicine, 2012, 8, 1043–1051 CrossRef CAS PubMed.
- S. D. Perrault, C. Walkey, T. Jennings, H. C. Fischer and W. C. Chan, Nano Lett., 2009, 9, 1909–1915 CrossRef CAS PubMed.
- T. M. Allen, Nat. Rev. Cancer, 2002, 2, 750–763 CrossRef CAS PubMed.
- H. Bader, H. Ringsdorf and B. Schmidt, Macromol. Mater. Eng., 1984, 123, 457–485 Search PubMed.
- K. Kataoka, A. Harada and Y. Nagasaki, Adv. Drug Delivery Rev., 2001, 47, 113–131 CrossRef CAS PubMed.
- K. F. Pirollo and E. H. Chang, Trends Biotechnol., 2008, 26, 552–558 CrossRef CAS PubMed.
- L. N. Hassani, F. Hendra and K. Bouchemal, Drug discovery today, 2012, 17, 608–614 CrossRef CAS PubMed.
- L. Lu, C. Zhang, L. Li and C. Zhou, Carbohydr. Polym., 2013, 94, 444–448 CrossRef CAS PubMed.
- Y. Ohya, S. Takeda, Y. Shibata, T. Ouchi, A. Kano, T. Iwata, S. Mochizuki, Y. Taniwaki and A. Maruyama, J. Controlled Release, 2011, 155, 104–110 CrossRef CAS PubMed.
- W. Xun, H.-Y. Wang, Z.-Y. Li, S.-X. Cheng, X.-Z. Zhang and R.-X. Zhuo, Colloids Surf., B, 2011, 85, 86–91 CrossRef CAS PubMed.
- T.-B. Ren, W.-J. Xia, H.-Q. Dong and Y.-Y. Li, Polymer, 2011, 52, 3580–3586 CrossRef CAS.
- A. Boudier, A. Aubert-Pouëssel, C. Gérardin, J.-M. Devoisselle and S. Bégu, Int. J. Pharm., 2009, 379, 212–217 CrossRef CAS PubMed.
- J. Niu, Z. Su, Y. Xiao, A. Huang, H. Li, X. Bao, S. Li, Y. Chen, M. Sun and Q. Ping, Eur. J. Pharm. Sci., 2012, 45, 216–226 CrossRef CAS PubMed.
- J.-C. Yeh, H.-H. Yang, Y.-T. Hsu, C.-M. Su, T.-H. Lee and S.-L. Lou, Colloids Surf., A, 2013, 421, 1–8 CrossRef CAS.
- V. San Miguel, A. Limer, D. M. Haddleton, F. Catalina and C. Peinado, Eur. Polym. J., 2008, 44, 3853–3863 CrossRef CAS.
- L. Wei, C. Cai, J. Lin and T. Chen, Biomaterials, 2009, 30, 2606–2613 CrossRef CAS PubMed.
- Y. Onishi, Polymer, 1980, 21, 819–824 CrossRef CAS.
- M. F. Francis, L. Lavoie, F. M. Winnik and J.-C. Leroux, Eur. J. Pharm. Biopharm., 2003, 56, 337–346 CrossRef CAS PubMed.
- J. Yang, Y. Liu, H. Wang, L. Liu, W. Wang, C. Wang, Q. Wang and W. Liu, Biomaterials, 2012, 33, 604–613 CrossRef CAS PubMed.
- G.-Y. Liu, C.-J. Chen, D.-D. Li, S.-S. Wang and J. Ji, J. Mater. Chem., 2012, 22, 16865–16871 RSC.
- Y. Song, B. Lou, J. Cheng, P. Zhao, C. Lin and X. Wen, J. Biomed. Nanotechnol., 2016, 12, 2083–2096 CrossRef CAS.
- M. Nichifor, A. Lopes, A. Carpov and E. Melo, Macromolecules, 1999, 32, 7078–7085 CrossRef CAS.
- N. A. B. Vieira, M. S. Moscardini, V. A. de Oliveira Tiera and M. J. Tiera, Carbohydr. Polym., 2003, 53, 137–143 CrossRef CAS.
- A. S. Baldwin, J. Clin. Invest., 2001, 107, 241 CrossRef CAS PubMed.
- S. Wang, Z. Liu, L. Wang and X. Zhang, Cell. Mol. Immunol., 2009, 6, 327–334 CrossRef CAS PubMed.
- M. Karin, Nature, 2006, 441, 431–436 CrossRef CAS PubMed.
- A. Tanaka, S. Muto, M. Konno, A. Itai and H. Matsuda, Cancer Res., 2006, 66, 419–426 CrossRef CAS PubMed.
- L. Mansouri, N. Papakonstantinou, S. Ntoufa, K. Stamatopoulos and R. Rosenquist, Semin. Cancer Biol., 2016, 39, 40–48 CrossRef CAS PubMed.
- A. Tanaka, M. Konno, S. Muto, N. Kambe, E. Morii, T. Nakahata, A. Itai and H. Matsuda, Blood, 2005, 105, 2324–2331 CrossRef CAS PubMed.
- N. M. Javali, A. Raj, P. Saraf, X. Li and B. Jasti, Pharm. Res., 2012, 29, 3347–3361 CrossRef CAS PubMed.
- K. Aramaki, S. Hoshida and S. Arima, Colloids Surf., A, 2012, 396, 278–282 CrossRef CAS.
- J. D. Hoefelmeyer, H. Liu, G. A. Somorjai and T. D. Tilley, J. Colloid Interface Sci., 2007, 309, 86–93 CrossRef CAS PubMed.
- J.-P. Nam, S.-C. Park, T.-H. Kim, J.-Y. Jang, C. Choi, M.-K. Jang and J.-W. Nah, Int. J. Pharm., 2013, 457, 124–135 CrossRef CAS PubMed.
- G. Fang, H. Li and X. Liu, Mater. Chem. Phys., 2010, 122, 533–536 CrossRef CAS.
- A. P. Rokhade, S. A. Patil and T. M. Aminabhavi, Carbohydr. Polym., 2007, 67, 605–613 CrossRef CAS.
- S. Kim, C. Won and C. Chu, Carbohydr. Polym., 1999, 40, 183–190 CrossRef CAS.
- R. K. Purama, P. Goswami, A. T. Khan and A. Goyal, Carbohydr. Polym., 2009, 76, 30–35 CrossRef CAS.
- M. H. P. B. Vettori, S. M. M. Franchetti and J. Contiero, Carbohydr. Polym., 2012, 88, 1440–1444 CrossRef CAS.
- K. Wojciechowski and J. Buffle, Biosens. Bioelectron., 2004, 20, 1051–1059 CrossRef CAS PubMed.
- A. Mahmud, X.-B. Xiong and A. Lavasanifar, Macromolecules, 2006, 39, 9419–9428 CrossRef CAS.
- Y.-Z. Du, Q. Weng, H. Yuan and F.-Q. Hu, ACS Nano, 2010, 4, 6894–6902 CrossRef CAS PubMed.
- A. R. Tehrani-Bagha, H. Oskarsson, C. Van Ginkel and K. Holmberg, J. Colloid Interface Sci., 2007, 312, 444–452 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra04544a |
| This journal is © The Royal Society of Chemistry 2017 |