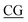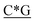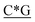Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Immunological effects of different types of synthetic CpG oligodeoxynucleotides on porcine cells
Ruiqiao Li†
a,
Lilin Zhang†a,
Peidian Shia,
Hui Denga,
Yi Li a,
Jie Rena,
Xubin Fub,
Lei Zhang*a and
Jinhai Huang
a,
Jie Rena,
Xubin Fub,
Lei Zhang*a and
Jinhai Huang *a
*a
aSchool of Life Sciences, Tianjin University, No. 92, Weijin Road, Nankai District, Tianjin, 300072, China. E-mail: jinhaih@tju.edu.cn; Fax: +86-22-27403902; Tel: +86-22-27403902
bTianjin Ruipu Biotechnology Limited Co., Tianjin, 300012, China
First published on 7th September 2017
Abstract
The agonists of toll-like receptor 9, synthetic oligodeoxynucleotides (ODNs) containing CpG sequences, stimulate innate and adaptive immune responses in humans and a variety of animal species. The aim of this study was to evaluate the immunomodulatory effects of four types of CpG ODNs as potential porcine vaccine adjuvants. In this study, four classes of swine CpG ODNs with distinct structural and biological properties were designed. Their abilities to trigger the expression of TLRs, antigen-presenting molecules and cytokines and to induce antiviral effects were determined in three different types of porcine cells (IPEC-J2, peripheral blood mononuclear cells (PBMCs), and 3D4/21). Our results showed that the four CpG ODNs triggered various types of molecular expression changed at the level of transcription. They activated the cytosolic antivirus TLRs at different levels, altered the mRNA expression of antigen-presenting molecules and intracellular signal transduction molecules, and triggered mRNA of cytokine production in a CpG type-specific and cell-specific manner at CpG ODNs, with the exception of type D, significantly increased IFN-α, IFN-β and TNF-α production and induced efficient anti-viral effects in IPEC-J2 cells but had little effect on MHC I expression. The designed CpG ODNs are useful for promoting early innate immunity and mucosal immunity and may also be promising adjuvant candidates for the treatment of porcine infectious diseases.
1. Introduction
Immune adjuvants are molecules, compounds or macromolecular complexes that can enhance the antigen response.1 Most adjuvants can assistant the antigen response through initiating the innate immune system.2 Active protection is mediated by the innate and adaptive immune systems.3 The innate immune system senses pathogen-associated molecular patterns (PAMPs) that are expressed by a wide variety of infectious microorganisms through pattern recognition receptors (PRRs), which trigger the activation of antipathogenic defenses and stimulate the adaptive immune response.4,5 Several classes of PRRs have been identified and characterized. These PRR classes include toll-like receptors (TLRs), nucleotide-binding oligomerization domain (Nod)-like receptor (TLR), leucine-rich repeat-containing receptors (NLRs), RIG-I-like receptors (RLRs), C-type lectin receptors (CLRs) and AIM-2 like receptor and intracellular sensors of nucleic acids such as OAS proteins and cGAS.6–8 Live-attenuated vaccines and some inactivated whole-pathogen vaccines contain a heterogeneous mixture of diverse pathogen components with the exception of antigens, and they successfully activate the immune system, including the innate immune system.9,10 However, due to a lack of PAMPs, highly purified vaccines, such as vaccines containing recombinant protein or DNA, do not effectively activate the initial innate immune response, which results in an ineffective activation of the downstream adaptive response.9,11,12 Accordingly, adjuvants are added to restore or improve immunogenicity to antigens with the objective of not altering tolerability.1The agonists of toll-like receptor 9, synthetic oligodeoxynucleotides (ODNs) containing CpG sequences, are recognized as a “danger” signal by the mammalian immune system.13,14 As a consequence, CpG ODNs stimulate both innate and adaptive immune responses in humans and a variety of animal species.15 To date, four classes of synthetic CpG ODNs have been described, and each class has distinct structural and biological properties.16–20 D-Type ODNs (also referred to as A type), which is constructed of a mixed phosphodiester/phosphorothioate backbone, contain a single CpG motif flanked by palindromic sequences. Another distinctive characteristic is the polyG tails at the 3′- and 5′-ends.17 The D-type ODNs stimulate peripheral blood mononuclear cells (PBMCs), which secrete IFN-α and IFN-β and promote maturation and IFN-α secretion of pDCs18,21 but have no influence on B cells.22 K-Type ODNs, also referred to as B type, contain multiple CpG motifs and compared with D-Type ODNs, all the phosphodiester backbones are replaced with phosphorothioate backbones to enhance resistance to nuclease digestion.23,24 K-Type ODNs trigger pDCs to differentiate and produce TNF-α and trigger B cells to proliferate and secrete IgM.25 C-Type ODNs, which contain the beneficial features of both D and K classes, are composed entirely of phosphororthioate nucleotides similar to K-type ODNs. C-Type ODNs contains palindromic CpG motifs, which allow this ODN class to stimulate B cells to secrete IL-6 and pDCs to produce IFN-α.18,19 P-Type ODNs contain two palindromic sequences and can form multimeric units to create higher order structures. The type I interferon-inducing potency and efficacy of the double-palindromic P-type ODNs are substantially higher than those of C-type ODNs. P-Type ODNs also stimulate superior cytokine production upon in vivo application.16 The different mechanisms of the classes of CpG ODNs have not been well characterized. Some researchers have declared that the nature of the pDC response is determined by the higher order structure and endosomal location of the CpG oligonucleotides. The distinct activities of K-type versus D-type ODNs have been traced to the retention time of CpG TLR-9 complexes in the endosomes of pDCs. Multimeric D-type ODNs together with the MyD88–IRF7 complex are retained for longer periods in the transferrin receptor (TfR)-positive endosome vesicles than the K-type ODNs and trigger a signaling cascade that supports IFN-α production. Whereas monomeric K-type ODNs are rapidly transported through (TfR)-positive endosomes into lysosome-associated membrane protein (LAMP)-1-positive endosomes, which promotes the maturation of PDCs. C-Type sequences are localized to both type of endosomes, which consequently induce both IFN-α and maturation of PDCs.26,27 CpG ODNs are recognized by both protomers in the dimer, in particular by the amino-terminal fragment (LRRNT-LRR10) from one protomer and the carboxy-terminal fragment (LRR20–LRR22) from the other.28
Species- and sequence-specific recognition differences have been demonstrated for TLR9 receptors.29 However, the majority of publications regarding the four types of CpG ODNs and the use of CpG-motifs are based on rodent models. Almost no information is available for the relationship between the effects of CpG ODNs on the native immune system and the different ODN structures in livestock or domestic animals such as cattle, pigs, horses and poultry.30 CpG ODNs possess the capacity to enhance innate and adaptive immune responses; thus, we regard CpG ODNs as a promising component in adjuvants for veterinary vaccines in animal husbandry. On the basis of previous research, we know that the ATCGAT hexamer at the core of ODNs is optimal for stimulating porcine PBMCs, and the palindrome created by this hexamer is critical to ODN activity.31 Therefore, we designed four synthetic oligodeoxynucleotide sequences in accordance with each characteristic of the abovementioned four classes of ODNs, and all of them contained the ATCGAT hexamer. The aim of this study is to evaluate the immunomodulatory effects of four CpG as potential porcine vaccine adjuvants. We simulated the 3D structure of the four classes of ODNs using Chem 3D to investigate the relationship between the structure and performance of the four classes of ODNs.
2. Materials and methods
2.1 Oligodeoxynucleotide synthesis
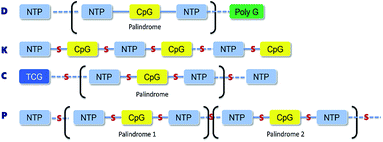
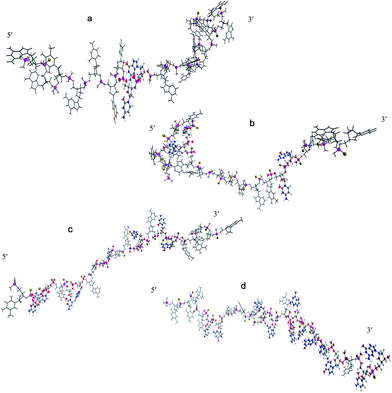
The D-type ODNs have been shown to have the earliest immunization function.31 The remaining ODNs were designed in accordance with that feature. All four of the oligodeoxynucleotide sequences contain at least one unmethylated CpG dinucleotide of the optimal sequence ATCGAT (Fig. 1). The K-type contains two ATCGAT motifs with a phosphorothioate backbone, and the C-type contains four CpG units with the exception of the TCG unit at the 5′-terminal with only one ATCGAT motif and a single palindrome embedded in the sequence. The P-type contains six CpG units with only one ATCGAT motif and two palindromes embedded in the sequence (Table 1). All ODNs were synthesized by GENEWIZ®. The 3D structure of all CpG ODN were simulated by ChemDraw 15.0 software (CambridgeSoft, V15.0.0.106), and all structures were displayed in minimum energy state (Table 2).| Types | Stretch | Bend | Stretch–bend | Torsion | Non-1,4 VDW | 1,4 VDW | Dipole/dipole | Total energy (kcal mol−1) |
|---|---|---|---|---|---|---|---|---|
| CpG D | 30.362 | 462.9365 | −0.7032 | 613.0337 | −255.9804 | 259.0518 | −302.5947 | 806.1057 |
| CpG K | 37.1046 | 384.6596 | −2.0501 | 296.2335 | −225.9389 | 292.0475 | −227.5121 | 554.54401 |
| CpG C | 45.9453 | 468.5275 | −6.4705 | 315.9276 | −224.8503 | 323.3768 | −273.5019 | 648.9545 |
| CpG P | 47.8179 | 506.8061 | −5.6828 | 353.4175 | −265.7135 | 354.865 | −290.3626 | 701.1475 |
2.2 Preparation of cells
Large white pigs were maintained at the Tianjin Institute of Experimental Animal Center in China. The approval number from the Tianjin government authority for the use of animals in experiments is SYXK-Jin 2011-0008. All animal experiments were performed in 6 week-old pigs in compliance with the Tianjin University Institutional Animal Care and Use Committee regulations (TJIACUC), state, and national guidelines. PBMCs were collected with heparin-containing anticoagulation tubes from healthy large white pigs, and the PBMCs were separated as previously described.32 PAM cells that were used in one independent experiment were obtained by bronchoalveolar lavage after necropsy.33 The cells were washed three times in RPMI 1640 supplemented with 2% FBS and were then incubated in RPMI 1640 with 10% FBS at 37 °C in a 5% CO2 in an incubator. IPEC-J2 cells (porcine intestinal epithelial cell) and porcine alveolar macrophage cells (3D4/21) were incubated in high glucose DMEM with 10% FBS at 37 °C in a 5% CO2 in an incubator.2.3 Stimulation of cells and statistical analysis
For the analysis of TLRs, cytokines and interferon mRNA levels, 0.5 × 106 cells were stimulated with ODNs and poly(I:C) to a final concentration 10 μg ml−1 for 24 h in 6-well cell culture plates. The cells were harvested from cultures and immediately homogenized in 1 ml of Bizol reagent (Biomiga, America). RNA was purified from the pellet and transcribed into cDNA using a TransGen Biotech TransScript First-Strand cDNA Synthesis Super Mix and oligodT18 as recommended by the manufacturer (Transgen Biotech, China). The reaction mix was incubated at 42 °C for 30 min followed by heating for 5 min at 85 °C to activate the reverse transcriptase. This cDNA (final volume 20 μl) was diluted 50 times and was amplified and quantified via real-time quantitative PCR using an EvaGreen 2× qPCR MasterMix kit produced by abm® (ABM, Canada). The reaction mixture consists of a master Mix-R containing Taq polymerase, dNTPs, MgCl2, fluorescent dye (detection), reference dye and proprietary buffer components, 0.4 μM of each primer and 2 μl of template cDNA in total volume of 20 μl. The subsequent steps included an initial denaturation for 10 min at 95 °C, followed by 45 cycles of denaturation for 15 s at 94 °C, annealing for 30 s at 56 °C and extension for 30 s at 68 °C. Confirmation of the specificity of the PCR-products was performed by subjecting these products to a melting curve analysis. The primers were designed from published nucleic acid sequences available from the GenBank databases using the program oligo7, as showed in Table 3. In all cases, the primer pairs that were selected spanned introns. All primers were synthesized at GENEWIZ®. Q-PCR was performed on an Applied Biosystems ABI 7500 as recommended by the manufacturer. For each sample, the β-actin gene was amplified and used as an internal control. Specific amplification was confirmed by sequencing the PCR products. The threshold cycle for the target genes and the difference between their Ct values (ΔCt) were determined. The relative transcript levels of the target gene are equal to the 2−ΔΔCt threshold method. The data represent the fold increase in the mRNA levels. The data are expressed as the mean ± SEM of three independent experiments. Calculations were performed with GraphPad Prism 6 (GraphPad Software Inc.) analysis software. Differences between the CpG and poly(I:C) stimulated groups in mRNA expression and the non-CG stimulated group were analyzed for statistical significance with unpaired two-tailed Student's t test. P < 0.05 was considered statistically significant.| Genes | Genbank | Primers (5′–3′) | Products/bp | Tm/°C |
|---|---|---|---|---|
| TLR3 | AB258451 | F-CTTTTCCTTTCAATGGCTAA | 183 | 49.1 |
| R-AGAGGAGAATCAGCGAGTG | ||||
| TLR7 | AB188301 | F-GACGCCTTTGTTATCTACT | 242 | 52.5 |
| R-TGGGCAATCTCATACTCA | ||||
| TLR8 | AB092975 | F-GCTGCCGTTGTTAGAAGT | 406 | 52 |
| R-CGGAAACTGCTGGAGTAATG | ||||
| TLR9 | NM213985 | F-ACAATGACATCCATAGCCGAGT | 103 | 57.9 |
| R-TCTCCCTCAGCCCACATCCG | ||||
| MyD88 | AB292176.1 | F-GATGGTAGCGGTTGTCTCTGAT | 148 | 56.5 |
| R-GATGCTGGGGAACTCTTTCTTC | ||||
| NF-κB | EU399817.1 | F-GTGTGTAAAGAAGCGGGACCT | 139 | 56.5 |
| R-CACTGTCACCTGGAAGCAGAG | ||||
| p38 | XM001929490.1 | F-CTTACGGATGACCCACGTTCAGT | 127 | 56.5 |
| R-GCTCACAGTCTTCATTCACAGC | ||||
| IRF3 | NM_213770 | F-AGACGCTCACCACGCTACACC | 136 | 61.3 |
| R-CGTCCCGCGCCATGTCTACCAG | ||||
| IL-1β | M86725 | F-TGTTCTGCATGAGCTTTGTG | 358 | 55 |
| R-TCTGGGTATGGCTTTCCTTAG | ||||
| IL-2 | FJ5431091 | F-GCTGCTGGATTTACAGTTGC | 194 | 53 |
| R-TTGCTGAGTCAGAGTTTTTG | ||||
| IL-4 | HQ236500 | F-CCAACCCTGGTCTGCTTACT | 175 | 56 |
| R-CTTCTCCGTCGTGTTCTCTG | ||||
| IL-6 | NM_214399 | F-ACTGGCAGAAAACAACCTGA | 173 | 54.6 |
| R-CCTCGACATTTCCCTTATTGCT | ||||
| IL-10 | NM_214041 | F-CCCCTTTGACAGCTAATATTCCG | 146 | 52.8 |
| R-TGCTTCTAGTTAAAACCGAGT | ||||
| IL-12 p35 | NM_213993 | F-TCATGCCTTCAGAATTCGT | 189 | 52.8 |
| R-TATACCAGCTTAGATAGGACCA | ||||
| IL-12 p40 | NM_214013 | F-CACTGGTACCTCTAGGCAAG | 222 | 52.2 |
| R-AGCAACCAGCAATTCTGACA | ||||
| IL-17 | KF646141 | F-CACTCGGGCTGTATCAATGCT | 178 | 56.6 |
| R-AAATATGGCGGACGATGG | ||||
| TNF-α | NM_214022 | F-GAGATCAACCTGCCCGACT | 158 | 56 |
| R-CTTTCTAAACCAGAAGGACGTG | ||||
| IFN-α | NM_214393 | F-CACCTCAGCCAGGACAGAAG | 225 | 59.9 |
| R-TGAGGGGATCCAAAGTCCCT | ||||
| IFN-β | NM_001003923 | F-GCAGTATTGATTATCCACGAGA | 121 | 52.8 |
| R-TCTGCCCATCAAGTTCCAC | ||||
| IFN-γ | NM_213948 | F-AAGCTGATTAAAATTCCGGTA | 112 | 50.6 |
| R-TCTTCCGCTTTCTTAGGTT | ||||
| MHC I | KJ922760 | F-CGGGTACAGACAGGACGCCTAC | 183 | 60.1 |
| R-AGGTATCTGCGGAGCGACTC | ||||
| MHC II | KJ922761 | F-GCCCACAGTGACGGTGTATCCT | 238 | 59.7 |
| R-CACTCGGCAGCTGTAGACCTCT | ||||
| CD86 | AK231046 | F-AAGGACAAGGGCTCATATC | 291 | 51.3 |
| R-TACATTGTAGAGTTCCGTGATG | ||||
| β-Actin | XM_003124280 | F-CAAATGCTTCTAGGCGGACT | 201 | 52 |
| R-TGCTGTCACCTTCACCGTTC |
2.4 Immunoblot analysis
Cells treated with different CpG ODN were lysed in RIPA buffer (Cell Signaling Technology, Danvers MA, USA) in the presence of protease inhibitor cocktail (Roche, Basel, Switzerland). Lysed material was clarified by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at 4 °C. Clarified extracts representing 2 × 106 cells per lane were fractionated on a 12% SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane using a transfer apparatus according to the manufacturer's protocol (Bio-Rad, Hercules, CA, USA). Membranes were blocked for 1 h with 5% nonfat milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl abd 0.5% Tween 20). Membranes were washed 1× in TBST, and incubated with anti-β-actin (1
000g for 10 min at 4 °C. Clarified extracts representing 2 × 106 cells per lane were fractionated on a 12% SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane using a transfer apparatus according to the manufacturer's protocol (Bio-Rad, Hercules, CA, USA). Membranes were blocked for 1 h with 5% nonfat milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl abd 0.5% Tween 20). Membranes were washed 1× in TBST, and incubated with anti-β-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, TRANSGEN Co., China), anti-MyD88-rabbit polyclonal antibody (1
1000 dilution, TRANSGEN Co., China), anti-MyD88-rabbit polyclonal antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution, IMMUNOWAY, USA), anti-NF-κB-rabbit polyclonal antibody (1
2000 dilution, IMMUNOWAY, USA), anti-NF-κB-rabbit polyclonal antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution, Santa Biotechnology), and anti-IRF3-rabbit polyclonal antibody (1
2000 dilution, Santa Biotechnology), and anti-IRF3-rabbit polyclonal antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution, IMMUNOWAY) for 18 h at 4 °C. Membranes were washed three times with TBST and were incubated with a 1
2000 dilution, IMMUNOWAY) for 18 h at 4 °C. Membranes were washed three times with TBST and were incubated with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution of horseradish peroxidase-conjugated polyclonal goat antibody to rabbit (TIANGEN Co., China) or the goat anti-mouse IgG(H + L) HRP conjugate (TIANGEN Co. China) for 1 h at RT. Membranes were washed three times with TBST, developed with the ECL system (Amersham Biosciences) and visualized on a ChemiDoc imaging system (Bio-Rad, Hercules, USA) per the manufacturer's protocols.
2000 dilution of horseradish peroxidase-conjugated polyclonal goat antibody to rabbit (TIANGEN Co., China) or the goat anti-mouse IgG(H + L) HRP conjugate (TIANGEN Co. China) for 1 h at RT. Membranes were washed three times with TBST, developed with the ECL system (Amersham Biosciences) and visualized on a ChemiDoc imaging system (Bio-Rad, Hercules, USA) per the manufacturer's protocols.
2.5 VSV-GFP bioassay for IFN production
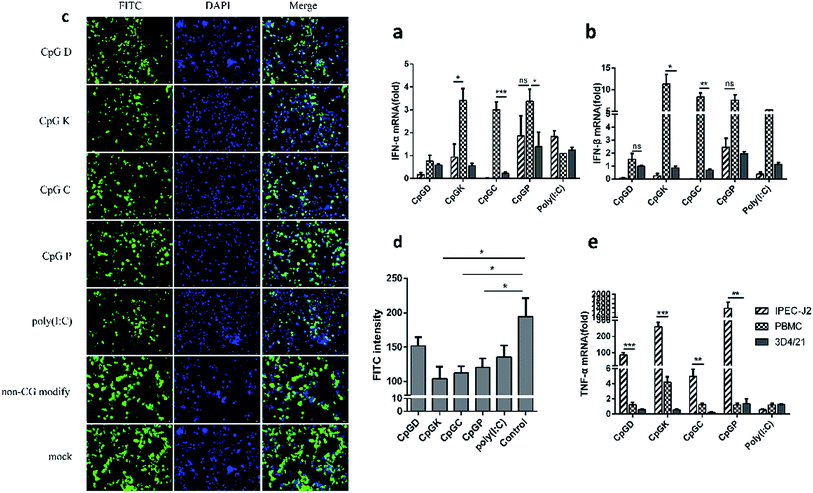
For the analysis of the production of IFN, 0.5 × 106 IPEC-J2 cells were stimulated in vitro with ODNs, poly(I:C) and a non-CG sequence (10 μg ml−1) for 6 h in 12-well cell culture plates, and the IPEC-J2 cells were then infected with VSV-GFP virus at 0.1 MOI for 18 h.34 GFP expression was assessed by fluorescence microscopy. We recorded the quantity of green fluorescence in different views from the same experimental group. By comparing the amount of green fluorescence in the different experiment groups, we assayed the ability of the four classes of CpG ODNs to alter the interferon production in the IPEC-J2 cells.3. Results
3.1 3D structure simulation of synthetic CpG ODNs
The 3D structures of four synthetic CpG ODNs were simulated using Chem 3D software, and the minimum energy state was calculated by Chem 3D. As shown in Fig. 2, D-type CpG ODNs, which is constructed with a mixed phosphodiester/phosphorothioate backbone, has a polyG tail in the phosphorothioate backbone. CpG D contains just a single CpG motif flanked by palindromic sequences. Compared with the other three oligodeoxynucleotides, CpG D is at the maximum energy level when the other ODN are at the minimum energy level. CpG D, K and P contain more than one CpG motif; meanwhile, they have a phosphorothioate backbone without phosphodiesters. CpG K contains a distortion structure at the 5′-end, and there is a CpG unit encircled by other nucleotides. CpG K is at a lower minimum energy level than the other ODN. The structure of CpG C and CpG P are displayed as a line, and they do not have a polyG tail like CpG D or a distortion structure like CpG K. CpG C and CpG P contain more than three CpG units, and most CpG units are arranged continuously on the P-type oligodeoxynucleotides near the 3′ end.3.2 Effect of CpG ODN treatment on the toll-like receptor mRNA expression
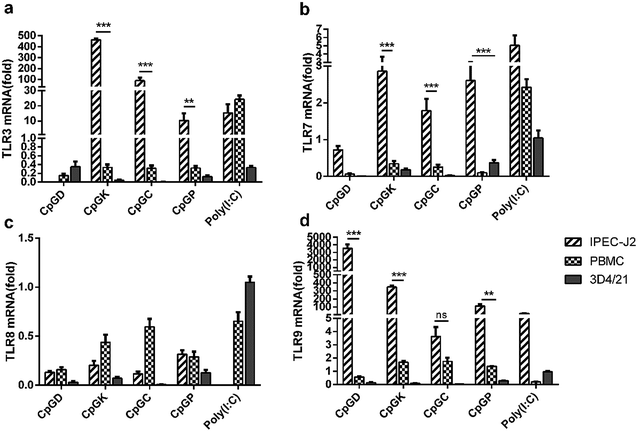
The TLR mRNA expression levels after 24 h of ODN stimulation showed a high variation among the three species of cells (Fig. 3). The TLR9 mRNA expression in the IPEC-J2 cells was significantly higher compared with the other groups, especially in the D-type treatment group, and the TLR8 mRNA expression level decreased in all cells (Fig. 3c and d). Beyond our expectations, the K-type, C-type and P-type ODNs promoted mRNA expression levels of TLR3 and TLR7 in the IPEC-J2 cells (Fig. 3a and b). However, the four types of ODNs did not stimulate TLR9 mRNA expression in the PBMCs and 3D4/21 cells as efficiently as in the IPEC-J2 cells.3.3 Effect of CpG ODN on the TLR signaling molecule mRNA expression
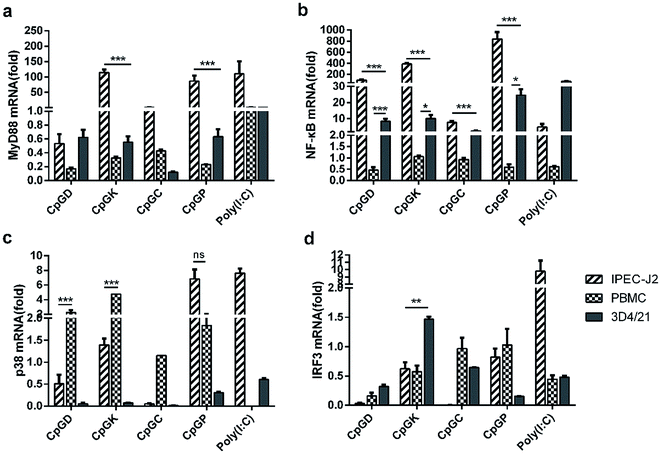
Toll-like receptor (TLR) agonists can promote the signaling cascades for the activation of genes that mediate the inflammatory response. This signal involves a MyD88-dependent pathway and MyD88-independent pathway.35,36 Most of the TLR family members, with the exception of TLR3, recruit the adaptor protein MyD88 and thus initiate a MyD88-dependent pathway, which involves the signal transduction intermediates IRAKs, TRAF6, TAK1, TAB1 and TAB2. Ultimately activating the two signaling pathways, one of the signaling pathways contains the MAPK family members p38, JNK, and ERK, and the other pathway involves the NF-κB transcription factor, which subsequently induces production of inflammatory cytokines.6,37 The TLR3 signaling pathway is completely dependent on the protein TIRF or TRAF3 and then recruits TRAF and TRAF6 and ultimately induces NF-κB activation and IRF3 phosphorylation.15,35,38 Therefore, we chose the key molecules, including MyD88, NF-κB, p38 and IRF3, to explore the change of mRNA expression that are triggered by the different types of ODNs. The mRNA expression of MyD88 and NF-κB in the IPEC-J2 cells was more efficient than in the PBMCs and 3D4/21 cells (Fig. 4). Specifically, all the CpG ODNs more strongly activated the MyD88 and NF-κB mRNA expression than the p38 mRNA expression, especially in the K-type and P-type ODN groups. We detected an elevation in the p38 mRNA expression level triggered by D-type ODNs and K-type ODNs in the PBMCs (Fig. 4c). The D-type and K-type CpG ODNs more strongly activated the p38 mRNA expression in the PBMCs than in the IPEC-J2. We did not detect elevated IRF3 mRNA expression levels among any of the three cell types, with the exception of a lower expression in 3D4/21 cells treated by the K-type ODNs (Fig. 4d). Consequently, CpG ODNs had the potency to influence the TLRs signal pathway. The MyD88 and NF-κB mRNA expression was strongly elevated in the IPEC-J2 cells. Three signal transduction molecules (MyD88, NFκB-p65, and IRF3) were detected to understand the signal transduction pathway in different CpG ODN treated IPEC-J2 cells. No IRF3 was detected in any CpG treated IPEC-J2 cells. Compare to non-CpG treatment cells, a rising MyD88 protein expression ratio in CpGD and CpGK groups, and NFκB-p65 protein in CpGC and CpGP groups were detected after 24 h CpG ODN treatment (Fig. 5), which revealed a cross-signal transduction molecules maybe produced and activated in different CpG treatment cells. To some degree, the mRNA expression of p38 from MAPK family can be elevated by D, K, and P-type ODNs in the PBMCs and IPEC-J2 cells. The mRNA expression of molecules related on MyD88-independent pathway cannot be efficiently triggered in any of the CpG ODNs treatment cells.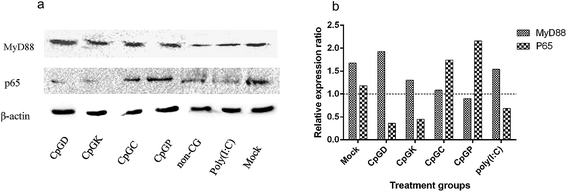 | ||
| Fig. 5 The western-blot results of MyD88 and p65 molecule expression in CpG ODN treated IPEC-J2 cells. (a) MyD88 and p65 molecules expression; (b) relative expression ratio. | ||
3.4 Effect of ODNs on immune related molecules mRNA expression
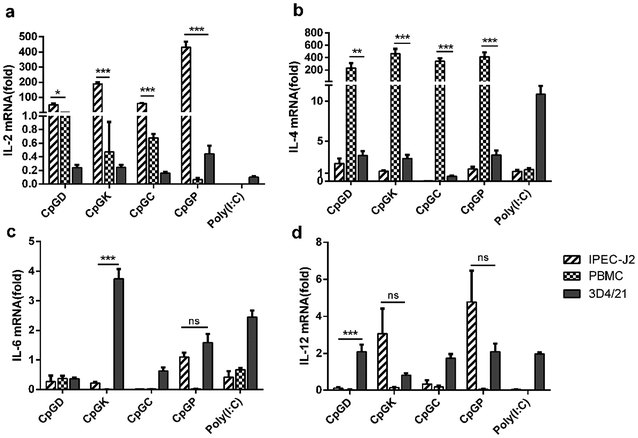
The sensing of agonists by PRRs expressed on antigen-presenting cells leads to the activation of adaptive immune responses.39 Toll-like receptor agonists can promote a shift in the immune response from the allergy-promoting Th2 response towards a Th1 and/or regulatory response.40 The signal evoked by diverse pathogenic infections along with T cell receptor stimulation results in the differentiation of T helper lineages. Naive T cells differentiate into Th1 subgroups that are regulated by signals from interferons and IL-12. Meanwhile, Th1 cells mainly secrete IFN-γ to further promote the Th1 cell differentiation. IL-4 is the master regulator for naïve T cell differentiation into Th2 subgroups with the essential helper IL-2 signaling pathway, and Th2 cells mainly secrete IL-4 and promote their own differentiation. Cytokines such as IL-6 and IL-23 promote Th17 cell differentiation.41 All T-helper cell groups can inhibit the activation of the other groups using their own cytokines and interleukins.42,43 In this study, we attempted to explore the different influences of the adaptive immune responses triggered by different classes of CpG ODNs. We detected the mRNA expression levels of IL2, IL-4, IL-6 and IL-12 in three types of cells treated with four classes of CpG ODNs after 24 h. All CpG ODNs effectively up-regulated IL-2 expression in the IPEC-J2 cells and significantly promoted IL-4 expression in PBMCs. The four classes of CpG ODNs induced IL-2 expression in the IPEC-J2 cells, and the P-type ODNs especially evoked IL-2 mRNA expression but not in the rest of the lineages (Fig. 6a). In contrast, IL-4 mRNA was weakly expressed in the IPEC-J2 cells but strongly expressed in the PBMCs. Moreover, type D, K and P ODNs also triggered a slight elevation in the IL-4 mRNA expression levels (Fig. 6b). Consequently, the regulator of Th2 cells was up-regulated after treatment with all of the ODNs. IL-6 and IL-12 mRNAs were weakly expressed in most situations, but IL-6 mRNA levels were slightly elevated in the 3D4/21 cells treated with the K-type ODNs, and IL-12 mRNA levels were slightly elevated in the 3D4/21 cells treated with the type D, C and P ODNs (Fig. 6c and d).3.5 Anti-viral effects of CpG ODN treatment on cells
To understand how the different classes of ODNs contribute to the innate immune system, the mRNA expression levels of interferons and tumor necrosis factor-α (TNFα) were also detected. We found that IFN-β was more upregulated than IFN-α (Fig. 7a and b). Meanwhile, interferons were more intensely up-regulated in the PBMCs treated with the K, C and P-type ODNs than with the D-type ODNs. The IPEC-J2 cells hardly expressed IFN-α and IFN-β. However, the P-type ODNs stimulated the expression levels of interferons in all cells, even in the 3D4/21 and IPEC-J2 cells (Fig. 7a and b). In contrast, TNF-α was highly up-regulated in the IPEC-J2 cells compared with the other cell types (Fig. 7e). The IPEC-J2 cells, treated with the four types of ODNs, were inoculated with the VSV-GFP virus to evaluate the type I IFN production. Then, the positive green fluorescent cells were counted and compared with the poly(I:C) group and the non-CpG ODN group. As shown in Fig. 6c and d, the K, C and P-type CpG ODNs inhibited the VSV-GFP virus proliferation well as the poly(I:C) treatment; however, the VSV-GFP proliferation rates in the cells in the non-CpG and negative groups were unaffected.3.6 Influence of ODNs on the antigen presentation molecules
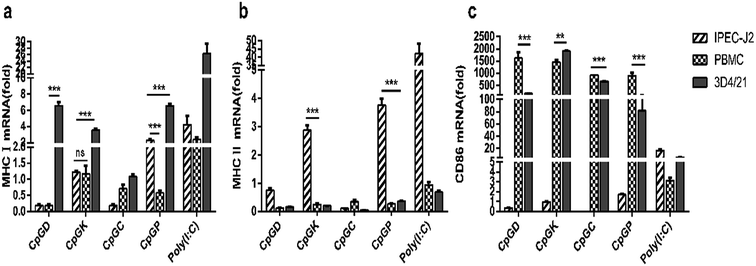
The impact of the different classes of ODNs on antigen presentation molecules were investigated. With the exception of the C-type ODNs, the other CpGs enhanced the MHC I mRNA expression in the 3D4/21 cells. The P-type ODNs elevated the MHC I mRNA expression levels in both the 3D4/21 cells and PBMCs (Fig. 8a). MHC I mRNA expression was only detected in the IPEC-J2 cells treated with the K- and P-type ODNs (Fig. 8b). Meanwhile, all four classes of the ODNs increased the CD86 mRNA expression levels in the 3D4/21 cells and PBMCs (Fig. 8c).4. Discussion
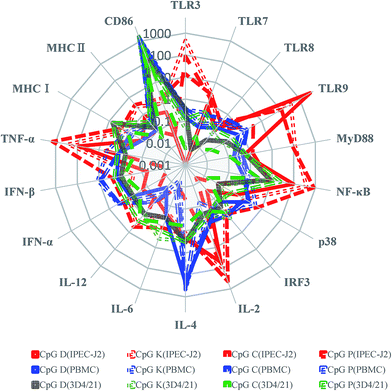
There is a large amount of evidence demonstrating that the synthetic oligodeoxynucleotides containing CpG sequences can trigger the innate immunoreaction, such as significantly increased intestinal mRNA expression of Th1 cytokines, CC chemokines and CXC chemokines, and co-inhibitory receptor programmed cell death-1 (PD-1),44–46 to protect the organism against microorganism invasions. CpG motifs activate mammalian lymphocytes and macrophages that produce cytokines, including IFN-γ, IL-12, TNF-α and polyclonal Ig.47–50 Some CpG ODNs can also enhance Th1 responses correlated with enhanced CD4+ T cells used as an adjuvant for the PRV vaccine and provided protection against TGEV and other viruses in pigs.51–56 Another study demonstrated that there was a distinction between class-A and class-B CpG ODNs admixed with or incorporated into vaccine delivery vehicles, and encapsulation of the class-A CpG-ODNs strongly reduced the CpG-ODN uptake and intracellular trafficking in the cytosol. In contrast, encapsulation of the class-B CpG-ODNs increased its uptake and did not consistently influence intracellular trafficking into the nucleus.57On the basis of the previous study in humans and other species, we concentrated on the biological functions of CpG ODNs with different structures used for pigs. In our study, we found four classes of ODNs with variable biological effects on different cells. When the same cell line was treated with different CpG ODNs, a similar cellular transcription spectrum tendency occurred, but the same molecule induced variable mRNA expression levels. As shown in Fig. 9, most CpG triggered higher expression of TLR9, MyD88, NF-κB, IL-2, TNF-α mRNA in the IPEC-J2 cells than in the other cell types. All CpG promoted stronger CD86 mRNA expression in the PBMCs and 3D4/21 cells. Most CpG ODNs significantly promoted the mRNA expression of CD86 and IL-4, which is beneficial for enhanced recognition between APC (3D4/21 cells) and T cells and trigger the regulation of the Th2 immune response in PBMCs. The rising IL-2 mRNA levels in the IPEC-J2 cells promoted the activation of lymphocytes. Meanwhile, all of the CpG ODNs stimulated mRNA of interferon production more effectively in the PBMCs than in the IPEC-J2 cells, and a good anti-viral infection effect was observed for type-K, C and P CpG ODNs. Our results indicate that the designed CpG ODNs may be useful to activate the inflammatory response and to promote early innate immunity and mucosal immunity; therefore, the CpG ODNs are a promising adjuvant candidate for porcine infectious diseases. The P-type ODNs, which contained the largest number of CpG motifs, induced the strongest activation of the immune responses in all three cell types. The P-type ODNs elevated the mRNA expression levels of NF-κB, p38, IL-2, and TNF-α more than the other CpG ODNs. Similar to the P-type ODNs, the K-type ODNs performed efficiently in the three porcine cell types, and it induced higher mRNA expression levels of MyD88, IL-4, IL-6 and interferons than the other ODN. In this study, we observed that the TLR9 pathway-related molecule mRNA expression level was strongly elevated in the IPEC-J2 cells, especially related to the MyD88-dependent pathway, and we found that CpG ODNs hardly triggered the mRNA expression of IRF3, a marker of the MyD88-independent pathway. Therefore, we speculated that CpG ODNs stimulated the innate immune response via interaction with TLR9 and triggered the signaling through the MyD88-dependent pathway. Owing the complex signal activation of TLR pathway, including the MyD88-dependent and MyD88-independent pathway, and the existence of multi-molecule cross-talk and cross-interference, the expression of TLR signaling and transcription factors does not equivalent to activation of special TLR is possible. Owing to the impacts of CpG sequence, intracellular intake, and differentiate signal transduction pathway, the expression of these signaling and transcription factors does not fully equivalent to their activation. Our result showed that variable signal transduction molecules maybe produced or activated in different CpG treatment cells, and a cross-talk and cross-interference exist in intracellular signal pathways. It is necessary to investigate the special mechanisms and related cross-adaptor molecule involve in signal transduction caused by different CpG ODNs in detail.
All the CpG ODNs effectively stimulated CD86 mRNA expression in macrophage cells. MHC I mRNA was highly elevated in the 3D4/21 cells, and this result indicated that the D-type ODNs containing the ATCGAT hexamer optimally stimulated swine antigen-presenting cells. The CD86 molecule is expressed on antigen-presenting cells that provided the costimulatory signals necessary for T cell activation and survival, and this result indicated that CpG ODNs efficiently promoted antigen-presenting cell interactions with lymphocytes. Furthermore, all the CpG ODNs significantly promoted the CD86 and IL-4 mRNA expression levels in the PBMCs. The regulator of Th2 cells was more strongly expressed than the regulators of Th1 and Th17 among all three cell types. The regulators of Th2 cells acted in two ways; one way was through elevated expression of IL-2 in the IPEC-J2 cells, and another way was through elevated expression of IL-4 in the PBMCs. This result indicated that CpG ODNs enhanced the activation of Th2 cells.
TLR3 recognizes viral double-stranded RNA and its synthetic analog polyriboinosinic poly(I:C), and TLR9 recognizes synthetic oligodeoxynucleotides containing CpG sequences. However, in our results, with the exception of the D-type ODNs, the type K, C and P ODNs also triggered high TLR3 mRNA transcription in the IPEC-J2 cells, and the D-type ODNs promoted the highest levels of TLR9 mRNA. Different from the other ODNs, the D-type consists of a mixed phosphodiester/phosphorothioate backbone that contains only one CpG motif but has the maximum inner energy. Perhaps the structure of the D-type ODNs is most similar to the natural oligodeoxynucleotides of bacteria and other microorganisms; it induced the TLR9 mRNA expression levels more intensely than the other ODNs in the IPEC-J2 cells. The type K, C and P CpG ODNs contained more than one CpG motif (2, 5, and 6, respectively), which promoted the TLR3 mRNA level in the following order: type K > P > C > D. In CpG C, 5 discontinuous CpG motifs arranged on the oligodeoxynucleotides are ordered but a non-CG motif is present at the 3′-terminal. The CpG K contain a distortion structure at the 5′-end, 5 discontinuous CpG motifs encircled by other nucleotides and have the minimum inner energy compared to the others. In contrast, for CpG P, 6 CpG motifs containing two palindromic sequences arranged continuously at the 3′-terminal enable them to form multimeric units to form a higher ordered structure. Because of the deficiency of the phosphorothioate backbone, the TLR3 mRNA expression levels were only weakly induced by the D-type ODNs. Our results demonstrated that the CpG ODNs containing several discontinuous CpG motif and a lower inner energy at the 3′-end is important to induce the transcription of the TLR3 mRNA.
In conclusion, the CpG ODNs containing different CpG motifs regulate mRNA expression differently in vitro and perform at variable efficiencies in different cells. Meanwhile, the D-type ODNs triggered TLR9 most effectively, and the K-type and P-type ODNs have a strong capacity to initiate the early TLR3 immune response. Therefore, these CpG ODNs can be used as immune adjuvants together or individually to trigger appropriate vaccine responses.
Author contributions
Conceived and designed the experiments: Jinhai Huang. Performed the experiments: Ruiqiao Li, Peidian Shi, Hui Deng, Yi Li, Lilin Zhang, Jie Ren, and Xudong Fu. Analyzed the data: Hui Deng and Lei Zhang. Contributed reagents/materials/analysis tools: Jinhai Huang. Wrote the paper: Lei Zhang and Jinhai Huang.Conflicts of interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article. This manuscript does not contain any individual personal data, and individual consent to publish is not applicable.Acknowledgements
This work was supported by the National Natural Science Foundation of China (31272540) and the underprop project of Tianjin Science and Technology Committee in China (16YFZCNC00640 and 17JCZDJC33900).References
- O. Perez, A. Batista-Duharte, E. Gonzalez, C. Zayas, J. Balboa, M. Cuello, O. Cabrera, M. Lastre and V. E. Schijns, et al., Braz. J. Med. Biol. Res., 2012, 45, 681–692 CrossRef CAS PubMed.
- B. G. Gellin and D. M. Salisbury, Vaccine, 2015, 33, B44–B46 CrossRef CAS PubMed.
- T. J. Moyer, A. C. Zmolek and D. J. Irvine, J. Clin. Invest., 2016, 126, 799–808 CrossRef PubMed.
- R. Medzhitov, Nature, 2007, 449, 819–826 CrossRef CAS PubMed.
- C. R. Alving, K. K. Peachman, M. Rao and S. G. Reed, Curr. Opin. Immunol., 2012, 24, 310–315 CrossRef CAS PubMed.
- T. Kawai and S. Akira, Nat. Immunol., 2010, 11, 373–384 CrossRef CAS PubMed.
- J. Wu and Z. J. Chen, Annu. Rev. Immunol., 2014, 32, 461–488 CrossRef CAS PubMed.
- S. Akira, S. Uematsu and O. Takeuchi, Cell, 2006, 124, 783–801 CrossRef CAS PubMed.
- S. Awate, L. A. Babiuk and G. Mutwiri, Front. Immunol., 2013, 4, 114 Search PubMed.
- L. A. Brito and D. T. O'Hagan, J. Controlled Release, 2014, 190, 563–579 CrossRef CAS PubMed.
- A. D. Pasquale, S. Preiss, F. T. D. Silva and N. Garcon, Vaccines, 2015, 3, 320–343 CrossRef PubMed.
- D. Carter and S. G. Reed, Curr. Opin. HIV AIDS, 2010, 5, 409–413 CrossRef PubMed.
- F. Steinhagen, T. Kinjo, C. Bode and D. M. Klinman, Vaccine, 2011, 29, 3341–3355 CrossRef CAS PubMed.
- E. N. Miyaji, E. Carvalho, M. L. Oliveira, I. Raw and P. L. Ho, et al., Braz. J. Med. Biol. Res., 2011, 44, 500–513 CrossRef CAS PubMed.
- H. An, C. Qian and X. Cao, Sci. China: Life Sci., 2010, 53, 34–43 CrossRef CAS PubMed.
- U. Samulowitz, M. Weber, R. Weeratna, E. Uhlmann, B. Noll, A. M. Krieg and J. Vollmer, Oligonucleotides, 2010, 20, 93–101 CrossRef CAS PubMed.
- D. Verthelyi, K. J. Ishii, M. Gursel, F. Takeshita and D. M. Klinman, J. Immunol., 2001, 166, 2372–2377 CrossRef CAS.
- A. Krug, S. Rothenfusser, V. Hornung, B. Jahrsdorfer, S. Blackwell, Z. K. Ballas, S. Endres, A. M. Krieg and G. Hartmann, Eur. J. Immunol., 2001, 31, 2154–2163 CrossRef CAS PubMed.
- G. Hartmann, J. Battiany, H. Poeck, M. Wagner, M. Kerkmann, N. Lubenow, S. Rothenfusser and S. Endres, Eur. J. Immunol., 2003, 33, 1633–1641 CrossRef CAS PubMed.
- J. D. Marshall, K. Fearon, C. Abbate, S. Subramanian, P. Yee, J. Gregorio, R. L. Coffman and G. Van Nest, J. Leukocyte Biol., 2003, 73, 781–792 CrossRef CAS PubMed.
- Z. Waibler, M. Anzaghe, A. Konur, S. Akira, W. Muller and U. Kalinke, Eur. J. Immunol., 2008, 38, 3127–3137 CrossRef CAS PubMed.
- Z. K. Ballas, W. L. Rasmussen and A. M. Krieg, J. Immunol., 1996, 157, 1840–1845 CAS.
- M. Kremlitzka, B. Macsik-Valent and A. Erdei, Cell. Mol. Life Sci., 2015, 72, 2223–2236 CrossRef CAS PubMed.
- G. K. Mutwiri, A. K. Nichani, S. Babiuk and L. A. Babiuk, J. Controlled Release, 2004, 97, 1–17 CrossRef CAS PubMed.
- G. Hartmann and A. M. Krieg, J. Immunol., 2000, 164, 944–953 CrossRef CAS.
- K. Honda, Y. Ohba, H. Yanai, H. Negishi, T. Mizutani, A. Takaoka, C. Taya and T. Taniguchi, Nature, 2005, 434, 1035–1040 CrossRef CAS PubMed.
- C. Guiducci, G. Ott, J. H. Chan, E. Damon, C. Calacsan, T. Matray, K. D. Lee, R. L. Coffman and F. J. Barrat, J. Exp. Med., 2006, 203, 1999–2008 CrossRef CAS PubMed.
- U. Ohto, T. Shibata, H. Tanji, H. Ishida, E. Krayukhina, S. Uchiyama, K. Miyake and T. Shimizu, Nature, 2015, 520, 702–705 CrossRef CAS PubMed.
- J. Pohar, C. Yamamoto, R. Fukui, M. M. Cajnko, K. Miyake, R. Jerala and M. Bencina, J. Immunol., 2017, 198(5), 2093–2104 CrossRef CAS PubMed.
- K. Kapp, J. Schneider, L. Schneider, N. Gollinge, S. Jansch, M. Schroff, B. Wittig and C. Kleuss, Immun., Inflammation Dis., 2016, 4, 446–462 CrossRef CAS PubMed.
- S. Kamstrup, D. Verthelyi and D. M. Klinman, Vet. Microbiol., 2001, 78, 353–362 CrossRef CAS PubMed.
- A. J. Ulmer, W. Scholz, M. Ernst, E. Brandt and H. D. Flad, Immunobiology, 1984, 166, 238–250 CrossRef CAS PubMed.
- G. Zhang, S. Qiao, Q. Li, X. Wang, Y. Duan, L. Wang, Z. Xiao and C. Xia, Immunogenetics, 2006, 58, 845–849 CrossRef CAS PubMed.
- Z. Li, G. Liu, L. Sun, Y. Teng, X. Guo, J. Jia, J. Sha, X. Yang, D. Chen and Q. Sun, PLoS Pathog., 2015, 11, e1004783 Search PubMed.
- S. Ntoufa, M. G. Vilia, K. Stamatopoulos, P. Ghia and M. Muzio, Semin. Cancer Biol., 2016, 39, 15–25 CrossRef CAS PubMed.
- F. Leulier and B. Lemaitre, Nat. Rev. Genet., 2008, 9, 165–178 CrossRef CAS PubMed.
- T. Kawai and S. Akira, Ann. N. Y. Acad. Sci., 2008, 1143, 1–20 CrossRef CAS PubMed.
- S. Akira and K. Takeda, Nat. Rev. Immunol., 2004, 4, 499–511 CrossRef CAS PubMed.
- A. Iwasaki and R. Medzhitov, Nat. Immunol., 2015, 16, 343–353 CrossRef CAS PubMed.
- A. Ziegler, V. Gerber and E. Marti, Vet. J., 2017, 219, 6–11 CrossRef CAS PubMed.
- H. Hasegawa, I. Mizoguchi, Y. Chiba, M. Ohashi, M. Xu and T. Yoshimoto, Front. Immunol., 2016, 7, 479 Search PubMed.
- Y. Kanno, G. Vahedi, K. Hirahara, K. Singleton and J. J. O'Shea, Annu. Rev. Immunol., 2012, 30, 707–731 CrossRef CAS PubMed.
- A. M. Thornton and E. M. Shevach, J. Immunol., 2000, 164, 183–190 CrossRef CAS.
- A. Buermann, D. Romermann, W. Baars, J. Hundrieser, J. Klempnauer and R. Schwinzer, Xenotransplantation, 2016, 23, 347–356 CrossRef PubMed.
- J. Yang, M. Mao, S. Zhang, H. Li, Z. Jiang, G. Cao, D. Cao, X. Wang and L. Zhang, Int. Immunopharmacol., 2012, 12, 415–424 CrossRef CAS PubMed.
- X. Chen, Q. Zhang, Y. Luo, C. Gao, X. Zhuang, G. Xu and T. Qiao, OncoTargets Ther., 2016, 9, 6511–6518 CrossRef PubMed.
- Y. Yamamoto, S. Shigemori, S. Nigar, K. Oshiro, Y. Wang, T. Sato and T. Shimosato, Anim. Sci. J., 2016, 87, 710–717 CrossRef CAS PubMed.
- A. G. Dalgleish, Hum. Vaccines Immunother., 2014, 10, 3369–3374 CrossRef PubMed.
- H. Cai, Z. Kuang, K. Huang, J. Shi, X. Zhao, P. Chu, C. Huang, F. Ming, F. Xia, J. Yang and L. Zhang, Vet. Immunol. Immunopathol., 2014, 161, 66–76 CrossRef CAS PubMed.
- T. W. Dubensky Jr and S. G. Reed, Semin. Immunol., 2010, 22, 155–161 CrossRef PubMed.
- Y. L. Huang, V. F. Pang, M. C. Deng, C. Y. Chang and C. R. Jeng, Res. Vet. Sci., 2014, 96, 187–195 CrossRef CAS PubMed.
- H. J. Warshakoon, J. D. Hood, M. R. Kimbrell, S. Malladi, W. Y. Wu, N. M. Shukla, G. Agnihotri, D. Sil and S. A. David, Hum. Vaccines, 2009, 5, 381–394 CrossRef CAS.
- A. Manuja, B. K. Manuja, J. Kaushik, H. Singha and R. K. Singh, Immunopharmacol. Immunotoxicol., 2013, 35, 535–544 CrossRef CAS PubMed.
- J. Lin, C. Tu, C. Mou, X. Chen and Q. Yang, Vet. Immunol. Immunopathol., 2016, 172, 1–8 CrossRef CAS PubMed.
- F. Ming, J. Yang, P. Chu, M. Ma, J. Shi, H. Cai, C. Huang, H. Li, Z. Jiang, H. Wang, W. Wang, S. Zhang and L. Zhang, PLoS One, 2013, 8, e65536 CAS.
- W. Charerntantanakul, Vet. Immunol. Immunopathol., 2009, 129, 1–13 CrossRef CAS PubMed.
- T. Demoulins, P. Milona and K. C. McCullough, Nanomedicine, 2014, 10, 1739–1749 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |