Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biofouling formation and structure on original and modified PVDF membranes: role of microbial species and membrane properties
Guili Zhao abc and
Wei Ning Chen*c
abc and
Wei Ning Chen*c
aNanyang Environment and Water Research Institute, Interdisciplinary Graduate School, Nanyang Technological University, 1 Cleantech Loop, Singapore 637141, Singapore
bAdvanced Environmental Biotechnology Centre, Nanyang Environment and Water Research Institute, Nanyang Technological University, 1 Cleantech Loop, Singapore 637141, Singapore
cSchool of Chemical and Biomedical Engineering, Nanyang Technological University, 62 Nanyang Drive, Singapore 637459, Singapore. E-mail: WNChen@ntu.edu.sg; Tel: +65 6316 2870
First published on 2nd August 2017
Abstract
In our previous study, three types of modified PVDF membranes, PVDF-g-PHEMA, PVDF-g-PNIPAAm, and PVDF-g-P(HEMA-co-NIPAAm), were successfully prepared by grafting the functional polymers PHEMA and PNIPAAm. In this study, bacterial adhesion tests on the original and modified PVDF membranes were performed using two model microbial species, i.e. Gram-negative Escherichia coli and Gram-positive Staphylococcus epidermidis, to investigate the effect of microbial species and membrane properties on the formation and structure of biofouling. The results indicated that less biofouling was found on the modified membranes as compared to that on the original membrane in both mono-species tests and mixed-species tests; this finding confirmed that the hydrophilicity of the membrane significantly affected the formation of biofouling such that the antifouling properties of the PVDF membranes were remarkably enhanced after grafting of the hydrophilic polymers on them. Less biofouling was observed on the modified hydrophilic membranes, with scattered biofouling spots for mono-species, whereas a thin layer structure of biofouling was observed for mixed-species; however, heavy biofouling covered the original membrane with a thick cake layer for both mono-species and mixed-species. This demonstrated that the microbial species significantly affected the formation and structure of biofilms since the species influenced the bacterial adhesion and biofouling behavior of each other based on their type and characteristics; moreover, the hydrophilicity of membrane also displayed an essential effect on the biofouling structure. The novelty of this study is as follows: it demonstrates that the microbial species and membrane properties play an essential role in the biofouling formation and structure as well as how to influence biofouling on different membranes; this study can provide useful insight and indications on how to effectively control and inhibit biofouling in the future.
Introduction
Membrane bioreactors (MBRs) are considered as a promising technology in wastewater treatment applications due to their several advantages such as a small footprint, less sludge production, and higher effluent quality as compared to the conventional-activated sludge systems.1–3 However, there still exist several challenges for the wider application and faster development of the MBR technology. One of the major obstacles that limits the potential of this technology is membrane fouling, which significantly reduces the filtration flux, shortens the lifespan of the membrane, and increases energy consumption and overall costs.4,5 Membrane fouling is mainly caused by the deposition of inorganic and organic substances on the membrane surface and its pores, wherein microorganisms and microbial foulants contribute to the formation of biofouling/biofilms. In a previous study, it was demonstrated that biofilms formed on a microfiltration membrane under observation via confocal laser scanning microscopy (CLSM), which resulted in a decline of system performance.6It is known that the formation of biofilms starts with the early attachment of bacteria and their subsequent proliferation on the substrate surface,7 wherein the microorganisms usually produce extracellular polymeric substances (EPSs), which are responsible for the growth and adhesion of microorganisms on the surface.8,9 It has been reported that the species and characteristics of microorganisms, for example, their different cell walls and composition of the EPSs produced, and the microbial community affect the formation of biofilms by affecting the interactions with the membrane material.10,11 In a study reported by Dreszer et al.,12 the development of the formation, structure, and detachment of biofilms was investigated via optical coherence tomography (OCT) to discover their relation with the transmembrane pressure and membrane performance. The results indicated that an increase in biofilm thickness played an important role in decline of membrane performance, whereas biofilm thickness decreased with an increase in permeate flux, leading to a denser structure. These phenomena demonstrated that both the thickness and the structure of biofilms affected the membrane performance. Moreover, different types of membranes had a significant effect on the biofouling behavior owing to different properties of the membranes. A previous study13 indicated that the hydrophilicity of a membrane significantly influenced the maturation and structure of biofouling, wherein uniform and thin biofilms were rapidly deposited on a hydrophobic polyvinyl difluoride (PVDF) membrane, whereas bacterial attachment was resistant to occur on the hydrophilic surface of the polyvinyl alcohol (PVA) membrane. Therefore, it is necessary to investigate the effect of microbial species and the properties of the supporting membrane on the formation and structure of biofouling for the control and removal of unwanted biofouling.
Currently, the development of antifouling membranes is considered as one of effective strategies to reduce membrane fouling. In our previous study,14 the functional polymers poly(hydroxyethyl methacrylate) (PHEMA) and poly(N-isopropylacrylamide) (PNIPAAm) were utilized to modify a PVDF membrane to improve its antifouling properties owing to their high resistance to protein adsorption and bacterial adhesion, which led to the formation of PVDF-g-PHEMA, PVDF-g-PNIPAAm, and PVDF-g-P(HEMA-co-NIPAAm) membranes. In particular, PNIPAAm, which is a typical thermosensitive polymer, exhibited biofouling release properties in response to changes in temperature; this was due to change in the hydrophilicity at the lower critical solution temperature (LCST) of around 32 °C. The preliminary results indicated that the hydrophilicity of the PVDF membrane was significantly improved after the functional polymers were grafted, and the antifouling properties of the modified membranes were remarkably enhanced in comparison with those of the pure PVDF membrane. In particular, PVDF-g-P(HEMA-co-NIPAAm) exhibited better resistance ability to protein adsorption and bacterial adhesion among the modified membranes. It was reported that the high complexity of microbial communities in activated sludge systems had a significant effect on the antifouling efficiency and it increased the difficulty of the study.15 Therefore, it is very important to investigate the effect of microbial species and the properties of the supporting membrane on the formation of biofilms and their structure. This study was aimed at investigating biofouling on the original PVDF membrane and three types of modified membrane prepared in our laboratory and revealing the effect of microbial cells on the formation and structure of biofouling using two types of well-studied model organisms. Hence, it can provide indications on how to effectively control and inhibit biofouling in the future, which is one of the novelties of this study. Furthermore, dynamic filtration experiments were also conducted using a microbial suspension and synthetic wastewater-containing proteins and humic acid to evaluate the antifouling properties of the membranes and confirm the great potential of the modified membranes for practical wastewater treatment.
Experimental
Materials
A commercial PVDF membrane (diameter: 50 mm; average pore size: 0.45 μm) was provided by ACME Research Support Pte Ltd., Singapore. The commercial PVDF membrane was used as a base matrix for surface modification, and three types of modified membrane, i.e. PVDF-g-PHEMA, PVDF-g-PNIPAAm, and PVDF-g-P(HEMA-co-NIPAAm), were prepared according to our previous study.14 Bovine serum albumin (BSA, >99%, Mn = 67 kDa) and humic acid were supplied by Chemicals Testing & Calibration Laboratory Pte Ltd., Singapore. The dye SYTO 63 was purchased from Thermo Fisher Scientific Life Technologies Holdings Pte Ltd., Singapore. LIVE BacLight™ Bacterial Gram Stain Kit L7005, which contained two dyes SYTO 9 and hexidium iodide, was also purchased from Thermo Fisher Scientific Life Technologies Holdings Pte Ltd., Singapore.Antibacterial activity and biofouling release test
To evaluate the fouling resistance of the modified membranes to different bacterial strains, their antimicrobial activity was determined via bacterial adhesion on the membrane surface using the typical Gram-negative bacterium Escherichia coli16 and the Gram-positive bacterial strain Staphylococcus epidermidis.17 The protocol was adapted from our previous study.14 In detail, E. coli was cultured in a flask with an LB medium, which consisted of 10 g L−1 tryptone, 5 g L−1 yeast extract, and 10 g L−1 sodium chloride. The bacteria were cultured in an incubator for 5 h at 37 °C at a shaking speed of 200 rpm, and finally, the cell concentration was approximately 109 cells per mL. Membrane samples were placed in a 5 mL bacterial suspension in a 6-well plate for 5 h at 25 °C and 37 °C. After this, the membranes were washed using a 0.01 M phosphate buffered saline (PBS) solution, and bacterial cells deposited on the membrane surface were observed via SEM (JSM-6701F, JEOL Asia Pte. Ltd.) according to the previous literature.18,19 The amount of bacterial adhesion on the membrane surface was determined by counting the bacterial cells from the SEM images within a certain area (at least 8 SEM images). In the Gram-positive bacteria test, S. epidermidis was cultured in a YPD (yeast extract-peptone-dextrose) medium containing 10 g L−1 yeast extract, 20 g L−1 peptone and 20 g L−1 dextrose at a shaking speed of 200 rpm at 37 °C. After incubation for 5 h, the cell concentration reached 109 cells per mL. The next step of the bacterial adhesion experiment was similar to that used for the E. coli strain.The bacterial release properties of the membranes with respect to different bacterial species were also assessed via a colony forming unit (CFU) test using E. coli and S. epidermidis.20,21 Specifically, as abovementioned in the antibacterial activity test, the membrane samples were immersed in a 5 mL E. coli suspension for incubation for 5 h at 37 °C. After this, the samples were taken out and slightly rinsed with a sterile PBS solution. Then, parallel samples of the same membrane were placed in a 5 mL sterile PBS solution for bacterial detachment for further 5 h at 25 °C and 37 °C. After this, the remaining bacterial cells adhering to the membranes were thoroughly obtained by rinsing, and the cleansed solution was diluted 100-fold. Then, 0.1 mL of the diluent was inoculated onto an LB agar plate for culture for 15 h at 37 °C. The CFU number of bacteria obtained from the membrane after detachment for 5 h at different temperatures was used to evaluate the biofouling release properties. The CFU tests were performed using 3 independent samples for each measurement, and each measurement was carried out on 3 replicate agar plates. For the Gram-positive bacteria test, note that S. epidermidis was cultured in a YPD medium, and the cleansed solution was diluted 1000-fold in the biofouling release test.
Biofilm analysis using confocal laser scanning microscopy
To study the formation and construction of biofilms caused by the Gram-negative bacteria or Gram-positive bacteria, the biofilms of E. coli or S. epidermidis on the membrane surface were also visualized and analyzed by confocal laser scanning microscopy (CLSM). The biofilms were formed after the deposition of bacteria on the membranes for 12 h at 25 °C and 37 °C, and this process was the same as abovementioned in the bacterial adhesion experiment. Thereafter, the E. coli or S. epidermidis biofilms on different membrane samples were stained with SYTO 63 for 10 min and then washed with a 0.9% NaCl solution. The samples were observed by CLSM, and a laser was set at 638 nm to detect fluorescence. For the analysis of the biofilms of E. coli and S. epidermidis mixture, the membranes were immersed in a 5 mL of bacterial suspension mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for contact for 12 h at 25 °C and 37 °C, respectively. After this, the membranes were taken out and rinsed with the 0.9% NaCl solution. Then, the samples were dyed using the Gram Stain Kit L7005 (volume of solutions A and B = 1
1) for contact for 12 h at 25 °C and 37 °C, respectively. After this, the membranes were taken out and rinsed with the 0.9% NaCl solution. Then, the samples were dyed using the Gram Stain Kit L7005 (volume of solutions A and B = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 10 min and then washed with 0.9% NaCl solution. The samples were observed by CLSM using an excitation laser at 488 nm to measure green fluorescence at 500 nm emission for Gram-negative bacteria and red fluorescence at 625 nm emission for the Gram-positive bacteria. The volume and structure of the biofilm were analyzed using the Imaris 8.1.2 software (software for 3D and 4D real-time interactive data visualization and management).
1) for 10 min and then washed with 0.9% NaCl solution. The samples were observed by CLSM using an excitation laser at 488 nm to measure green fluorescence at 500 nm emission for Gram-negative bacteria and red fluorescence at 625 nm emission for the Gram-positive bacteria. The volume and structure of the biofilm were analyzed using the Imaris 8.1.2 software (software for 3D and 4D real-time interactive data visualization and management).
Dynamic membrane fouling analysis via microfiltration
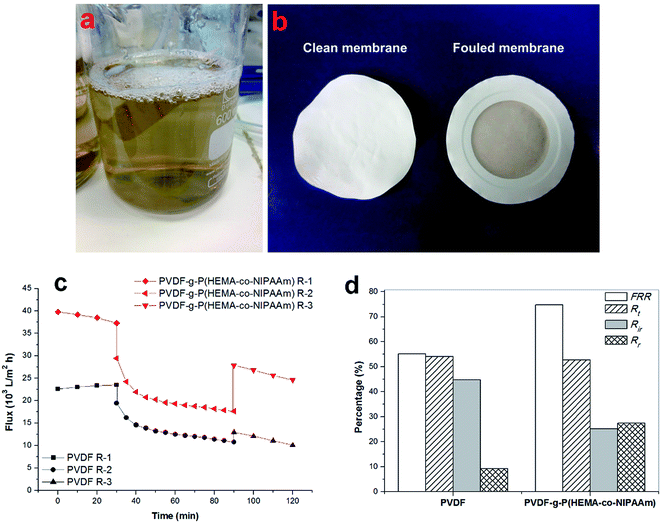
Cross-flow microfiltration was carried out with an E. coli suspension (107 cells per mL) via several cycles of fouling-washing to study the dynamic formation of biofouling. The effective area of the filtration module was 3.8 cm2. First, the system was filtered with distilled water for 1 h under a pressure of 1 bar to ensure a steady state. The flux was determined by the following equation:
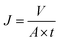 | (1) |
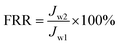 | (2) |
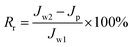 | (3) |
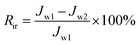 | (4) |
| Rt = Rr + Rir | (5) |
In addition, the fouling test was performed by dynamic filtration using synthetic wastewater at 1 bar and 25 °C. First, the stable water flux was determined and obtained for filtration with pure water for 0.5 h after pre-filtration for 1 h. Second, the feed solution was replaced with synthetic wastewater, and the flux was obtained as Jp during filtration for 1 h. Third, the membrane filtration system was washed using pure water for 1 h, and then, filtration was conducted with pure water for 0.5 h again, and one cycle was completed. The synthetic wastewater consisted of 0.3 g L−1 BSA, 9 mM L−1 NaCl, 2 mM L−1 NaHCO3, 1 mM L−1 CaCl2, and 1 mg L−1 humic acid (HA),25–27 and a suspension of this mixture was used as the filtration feed after natural sedimentation for 2 h. Dynamic fouling analysis was also carried out via several cycles of the fouling-washing process at different temperatures using synthetic wastewater filtration. The fouling-washing process was similar to that in the filtration experiments of the bacterial suspension, whereas the fouling process was conducted at 37 °C followed by cold water washing process at 15 °C. As results, the flux recovery ratio (FRR) and fouling resistance ratios (Rr, Rir, and Rt) were calculated via eqn (2), (3), (4), and (5), respectively.
Results and discussion
Study of anti-biofouling properties of membranes using Gram-negative and Gram-positive bacteria
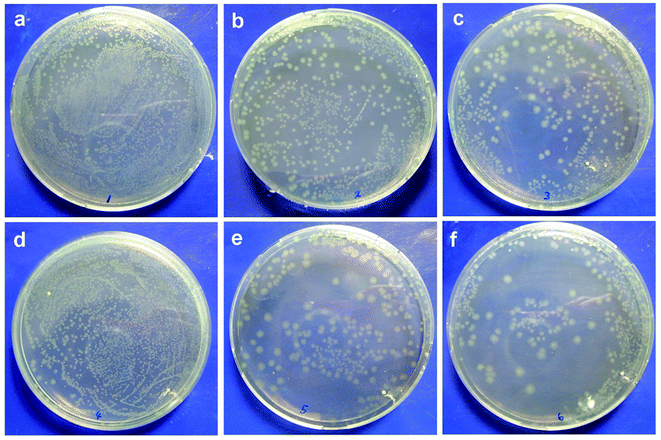
The antimicrobial properties of the membranes were determined via bacterial adhesion of Gram-negative E. coli and Gram-positive S. epidermidis in this study. For the E. coli test, Fig. 1 showed that significantly fewer bacteria were observed on the modified membranes than those on the pristine PVDF membrane. More specifically, lower values of bacterial adhesion were observed on the PVDF-g-PNIPAAm and PVDF-g-P(HEMA-co-NIPAAm) membranes at 25 °C, with the reduction of 67% and 71%, respectively, in comparison with that for the original PVDF membrane; this was a good indication of high resistance to bacterial adhesion on these modified membranes. It is known that bacteria readily adhere to hydrophobic surfaces via hydrophobic–hydrophobic interactions between their cell wall and the membrane surface. In general, water molecules were repulsed away from the original PVDF membrane surface owing to the absence of hydrogen bonding interactions in the boundary layer between the hydrophobic membrane interface and water; this enabled the bacteria to approach the boundary layer and adhere to the membrane. On the other hand, a hydration layer tends to appear on the hydrophilic-modified membranes through hydrogen bonding interactions between water and the membrane interface, and the hydration layer prevents bacteria from adhering to the membrane and further getting deposited on the surface.28 Hence, the modified membranes with high hydrophilicity exhibited antimicrobial properties. The E. coli release activity was also confirmed by the CFU tests, as shown in Fig. 2. The results indicated that the remaining amounts of bacteria adhering to the modified membranes after detachment at 25 °C were obviously less than those in the groups treated at 37 °C, indicating good biofouling release ability owing to thermosensitive properties. Moreover, after bacterial release at 25 °C, much fewer bacterial colonies were observed on the plate with the PVDF-g-P(HEMA-co-NIPAAm) membrane in comparison with those for the original and PNIPAAm membranes; this was mainly due to not only the increase in the fouling resistance of the copolymer-modified membrane at 37 °C upon grafting PHEMA (less bacterial attachment on the copolymer-modified membrane before detachment) but also the improvement in fouling release in response to changes in temperature upon grafting PNIPAAm.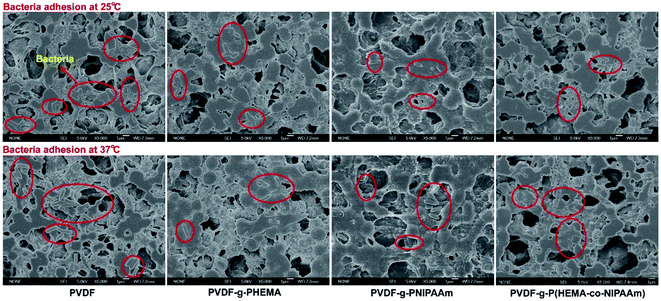 | ||
| Fig. 1 SEM images of adhesion of Escherichia coli on different membrane surfaces at 25 °C and 37 °C. | ||
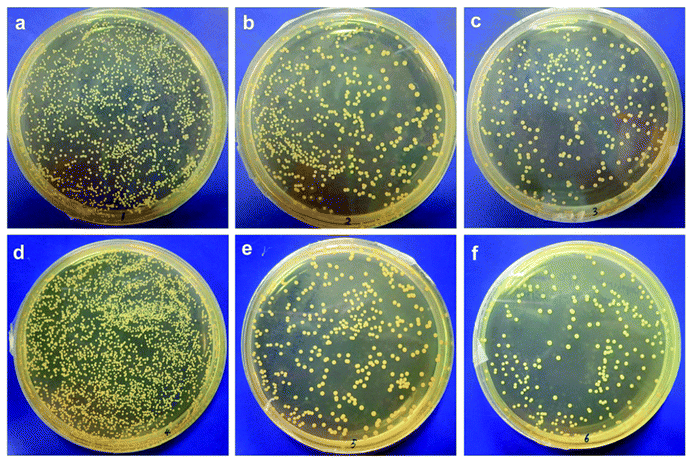
The adhesion of S. epidermidis to the membranes was also observed by SEM, and the results are shown in Fig. 3. It can clearly be seen that fewer bacteria were observed on the modified membranes in comparison with those on the original membrane at both 25 °C and 37 °C; this indicated better antifouling property of the modified membrane, and the results were similar to those of the E. coli attachment test (Fig. 1). However, the amounts of S. epidermidis attached to the PNIPAAm and copolymer-modified membranes at 25 °C were roughly the same as in the groups treated at 37 °C, and the results were different from those for the adhesion of E. coli at different temperatures. Through comparison of the original and modified membranes, it was observed that hydrophilicity had a significant effect on the fouling resistance; however, via comparison of the modified membranes at different temperatures in the S. epidermidis tests, it was observed that tunable hydrophilicity caused by thermosensitive properties slightly affected the fouling resistance. In Venault's study,18 it was reported that both E. coli and S. epidermidis favored pristine and hydrophobic PVDF surfaces more than those of the modified membranes blended with hydrophilic functional polymers, with over 99% reduction of bacterial attachment on the modified membranes. It can be seen from Fig. 4 that bacterial colonies on the plates of the modified membranes are visibly fewer than those on the plates of the original membrane after bacterial detachment for 5 h, whereas there is no obvious difference in the biofouling release with respect to the same modified membrane at different temperatures; this suggests that the thermosensitive properties slightly influence the release of S. epidermidis.
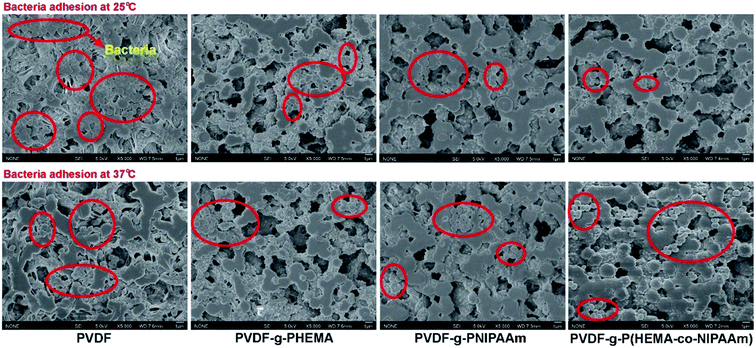 | ||
| Fig. 3 SEM images of adhesion of Staphylococcus epidermidis to different membrane surfaces at 25 °C and 37 °C. | ||
Moreover, other Gram-positive bacteria, except S. epidermidis, can be used to evaluate the antifouling properties of the modified thermosensitive membranes in future studies. In a previous study,20 Wang et al. designed a low-fouling RO membrane via surface modification by grafting a zwitterionic polymer, and two model microorganisms, namely, E. coli and Bacillus subtilis (Gram-positive bacterium), were utilized to examine the antimicrobial properties of the membrane. They reported that bacterial mortality for the modified membrane was nearly 100% for both E. coli and B. subtilis, which suggested strong antimicrobial properties of the modified membrane against Gram-negative and Gram-positive bacteria. Xu et al.19 prepared an antimicrobial polysulfone membrane blended with Ag/Cu2O hybrid nanowires, and the blended membrane exhibited excellent antibacterial performance against both Gram-negative E. coli and Gram-positive Staphylococcus aureus. SEM images also indicated that the adhesion of E. coli and S. aureus to the blended membrane was hindered, and their growth was also remarkably inhibited on the modified surface.
Study of the formation and structure of biofilms on the membranes with CLSM
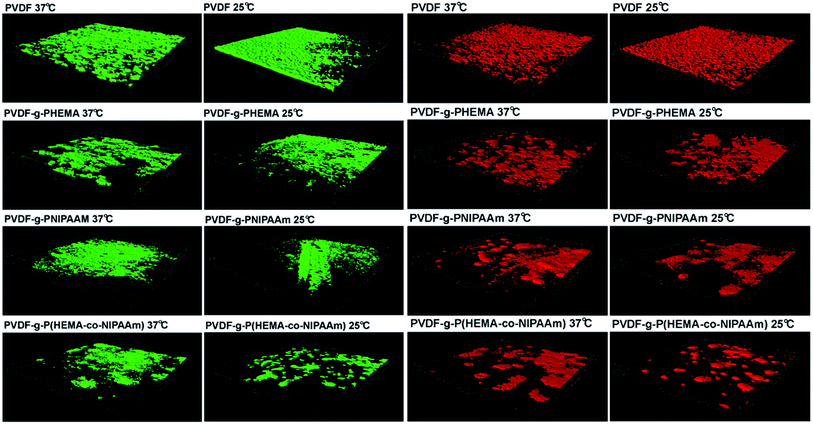 | ||
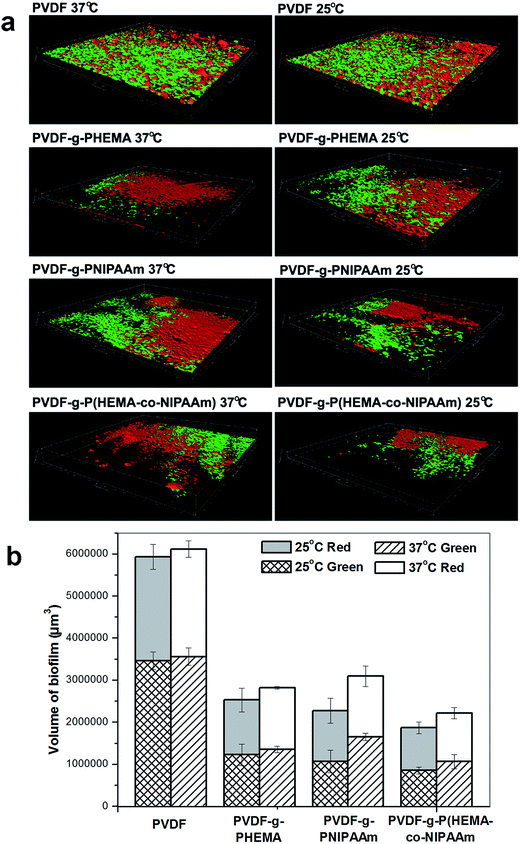
| Fig. 5 Three-dimensional reconstructions of biofilms formed by adhesion of E. coli (green) and S. epidermidis (red) based on confocal analysis. | ||
It can also be seen from Fig. 5 that the structure of the S. epidermidis biofilm was very similar to that formed by E. coli, such that an integrated and thick layer of biofouling formed on the surface of the original membrane, whereas biofouling spots scattered over the surface of the modified membranes. In addition, no obvious difference in the volumes of the biofilms on the modified membranes was found in the tests performed at different temperatures; this suggested that the effect of thermosensitive properties on the adhesion of S. epidermidis was not significant, which was consistent with previous results. It is worth noting that bacterial adhesion is also strongly dependent on the properties of the bacterial species. For example, different cell walls and compositions of the EPSs produced affect the interaction with the membrane materials. It was reported by Carretier et al.29 that different amounts of bacterial adhesion were found on the same membrane in the Gram-negative E. coli tests and Gram-positive Streptococcus mutans tests owing to the different nature of the bacterial species.
Dynamic membrane fouling analysis via microfiltration
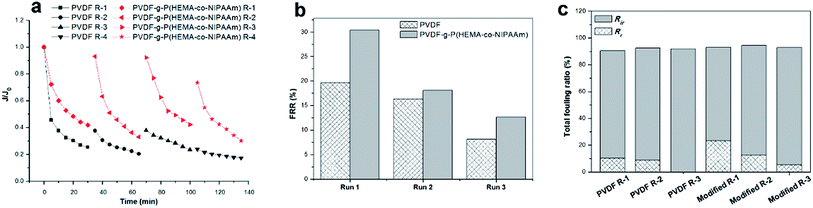
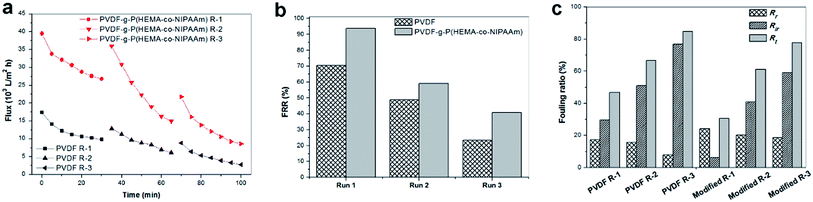
To investigate the fouling release ability exhibited by the thermosensitive membranes prepared in this study, three filtration cycles were conducted using synthetic wastewater, and one cycle process consisted of 0.5 h pollutant filtration at 37 °C, followed by cleaning with cold water for 1 h at 15 °C. Fig. 9(a) clearly shows that the modified membrane had higher permeability than the pure PVDF membrane. Fig. 9(b) shows that the modified membrane exhibits an FRR value as high as 94%, whereas the value is only 70% for the original membrane after the first cycle; this demonstrated that the flux of the modified membrane was very easily recovered by cold water cleaning. It was found from Fig. 9(c) that higher reversible fouling was deposited on the modified membrane and further cleaned by flushing with cold water in comparison with that on the pristine membrane; this proved that the fouling release ability of the modified membrane was higher owing to its tunable hydrophilicity in response to changes in temperature. However, the FRR value of the modified membrane decreased to 41% after three filtration cycles. This phenomenon was probably due to two reasons. First, progressive fouling was formed on the membrane surface during consecutive filtration runs. Second, as abovementioned in the discussion, a dense cake layer easily formed in the presence of Ca2+ and other multiple pollutants, which led to heavy membrane fouling, and internal fouling might even occur in this case. Thus, other multiple cleaning methods can be employed in combination with hydraulic cleaning below the LCST of PNIPAAm to promote the removal of fouling in practical filtration processes for long-term operation. In a literature review,35 Wang et al. stated that according to the fouling removal mechanisms, membrane cleaning strategies can be divided into four types: physical, chemical, physicochemical, and biological/biochemical cleaning. In addition, the time interval for in situ cleaning in MBRs is usually once every 10 min or several weeks or months depending on the fouling conditions and cleaning methods used, and its frequency is much higher than that of ex situ cleaning. Therefore, for thermosensitive membranes hydraulic cleaning with temperature control provides an auxiliary and feasible strategy for promoting fouling release and enhancing flux recovery in combination with classical cleaning methods for wastewater treatment in MBRs.
Conclusion
In this study, the formation and structure of biofilms were investigated via confocal observation using mono-species and mixed-species of Gram-negative E. coli and Gram-positive S. epidermidis. The results confirmed that the membrane properties (hydrophilicity and morphology/roughness) markedly affected the formation of biofouling such that the antifouling properties of the PVDF membrane were significantly improved after the hydrophilic polymers were grafted. Moreover, it was suggested that the microorganism species also played an essential role in the formation and structure of biofilms as the species influenced each other's bacterial adhesion and biofouling behavior. Less biofouling was observed on the modified hydrophilic membranes, with scattered biofouling spots for mono-species, but a thin layer structure of biofouling for the mixed-species was observed, whereas heavy biofouling covered the original membrane with a thick cake layer for both the mono-species and mixed-species. Moreover, the modified membranes exhibited higher fouling resistance during filtration of synthetic wastewater and higher flux recovery after hydraulic cleaning in comparison with the pure PVDF membrane. These results also revealed that the hydrophilicity of the membrane affected the structure of biofilms such that biofouling formed on the modified membranes was easily removed by flushing with water owing to the thin layer or cluster structure of the biofouling. Therefore, the contribution of this study lies in providing useful insights and indications on how to control and inhibit biofouling effectively in the future.Acknowledgements
The authors acknowledge the financial support received from the Singapore National Research Foundation under its Environmental & Water Technologies Strategic Research Program and administered by the Environment & Water Industry Program Office (EWI) of the PUB (grant 1102-IRIS-08-08). We also thank the support received from the Nanyang Environment and Water Research Institute, Nanyang Technological University.References
- J. A. Howell, T. C. Arnot and W. Liu, Ann. N. Y. Acad. Sci., 2003, 984, 411–419 CrossRef CAS PubMed.
- F. Fan, H. Zhou and H. Husain, Water Res., 2005, 40, 205–212 CrossRef PubMed.
- K. Kimura, H. Hara and Y. Watanabe, Desalination, 2005, 178, 135–140 CrossRef CAS.
- A. Y. Hu and D. C. Stuckey, J. Environ. Eng., 2006, 132, 190–198 CrossRef CAS.
- H. J. Lin, K. Xie, B. Mahendran, D. M. Bagley, K. T. Leung and S. N. Liss, et al., Water Res., 2009, 43, 3827–3837 CrossRef CAS PubMed.
- J. Luo, J. Zhang, X. Tan, D. McDougald, G. Zhuang, A. G. Fane, S. Kjelleberg, Y. Cohen and S. A. Rice, Biofouling, 2014, 30, 1093–1110 CrossRef CAS PubMed.
- J. W. Costerton, Z. Lewandowski, D. E. Caldwell, D. R. Korber and H. M. Lappin-Scott, Annu. Rev. Microbiol., 1995, 49, 711–745 CrossRef CAS PubMed.
- A. Drews, C. H. Lee and M. Kraume, Desalination, 2006, 200, 186–188 CrossRef CAS.
- H. Nagaoka, S. Ueda and A. Miya, Water Sci. Technol., 1996, 34, 165–172 CAS.
- L. Vanysacker, P. Declerck, M. R. Bilad and I. F. J. Vankelecom, J. Membr. Sci., 2014, 453, 394–401 CrossRef CAS.
- K. Fish, A. M. Osborn and J. B. Boxall, Sci. Total Environ., 2017, 593–594, 571–580 CrossRef CAS PubMed.
- C. Dreszer, A. D. Wexler, S. Drusova, T. Overdijk, A. Zwijnenburg, H.-C. Flemming, J. C. Kruithof and J. S. Vrouwenvelder, Water Res., 2014, 67, 243–254 CrossRef CAS PubMed.
- D. Saeki, Y. Nagashima, I. Sawada and H. Matsuyama, Colloids Surf., A, 2016, 506, 622–628 CrossRef CAS.
- G. Zhao and W. N. Chen, Appl. Surf. Sci., 2017, 398, 103–115 CrossRef CAS.
- H. Daims, M. W. Taylor and M. Wagner, Trends Biotechnol., 2006, 24, 483–489 CrossRef CAS PubMed.
- H. Ma, B. S. Hsiao and B. Chu, J. Membr. Sci., 2014, 452, 446–452 CrossRef CAS.
- L. K. Ista, S. Mendeza and G. P. Lopez, Biofouling, 2010, 26, 111–118 CrossRef CAS PubMed.
- A. Venault, Y. H. Liu, J. R. Wu, H. S. Yang, Y. Chang and J. Y. Lai, et al., J. Membr. Sci., 2014, 450, 340–350 CrossRef CAS.
- Z. Xu, S. Ye, G. Zhang, W. Li, C. Gao and C. Shen, et al., J. Membr. Sci., 2016, 509, 83–93 CrossRef CAS.
- J. Wang, Z. Wang, J. X. Wang and S. Wang, J. Membr. Sci., 2015, 493, 188–199 CrossRef CAS.
- J. Nikkola, X. Liu, Y. Li, M. Raulio, H. L. Alakomi and J. Wei, et al., J. Membr. Sci., 2013, 444, 192–200 CrossRef CAS.
- W. Chen, Y. Su, L. Zheng, L. Wang and Z. Jiang, J. Membr. Sci., 2009, 337, 98–105 CrossRef CAS.
- Y. Hao, A. Moriya, T. Maruyama, Y. Ohmukai and H. Matsuyama, J. Membr. Sci., 2011, 376, 247–253 CrossRef CAS.
- X. Chen, B. Zhao, P. Han, W. Fu and L. Chen, React. Funct. Polym., 2014, 84, 10–20 CrossRef CAS.
- Y. X. Shen, K. Xiao, P. Liang, J. Y. Sun, S. J. Sai and X. Huang, J. Membr. Sci., 2012, 415, 336–345 CrossRef.
- S. Liang, G. Qi, K. Xiao, J. Sun, E. P. Giannelis and X. Huang, et al., J. Membr. Sci., 2014, 463, 94–101 CrossRef CAS.
- X. Zhao and C. Liu, J. Membr. Sci., 2016, 515, 29–35 CrossRef CAS.
- F. Liu, N. Awanis Hashim, Y. Liu, M. R. Moghaeh Abed and K. Li, J. Membr. Sci., 2011, 375, 1–27 CrossRef CAS.
- S. Carretier, L. A. Chen, A. Venault, Z. R. Yang, P. Aimar and Y. Chang, J. Membr. Sci., 2016, 510, 355–369 CrossRef CAS.
- K. Kjærgaard, M. A. Schembri, H. Hasman and P. Klemm, J. Bacteriol., 2000, 182, 4789–4796 CrossRef.
- E. S. Tarleton and R. J. Wakeman, Chem. Eng. Res. Des., 1993, 71, 399–410 CAS.
- X. Zhao, Y. Su, Y. Li, R. Zhang, J. Zhao and Z. Jiang, J. Membr. Sci., 2014, 450, 111–123 CrossRef CAS.
- X. Zhao, H. Xuan, Y. Chen and C. He, J. Membr. Sci., 2015, 494, 48–56 CrossRef CAS.
- A. Tiraferri, Y. Kang, E. P. Giannelis and M. Elimelech, ACS Appl. Mater. Interfaces, 2012, 4, 5044–5053 CAS.
- Z. Wang, J. Ma, C. Y. Tang, K. Kimura, Q. Wang and X. Han, J. Membr. Sci., 2014, 468, 276–307 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2017 |