Open Access Article
Open Access ArticleA 1,8-naphthalimide-based fluorescent probe for selective and sensitive detection of peroxynitrite and its applications in living cell imaging†
Xiulan Li‡
,
Jingli Hou‡,
Chao Peng,
Li Chen,
Wenbo Liu and
Yangping Liu *
*
Tianjin Key Laboratory on Technologies Enabling Development of Clinical Therapeutics and Diagnostics, School of Pharmacy, Tianjin Medical University, Tianjin 300070, P. R. China. E-mail: liuyangping@tmu.edu.cn
First published on 7th July 2017
Abstract
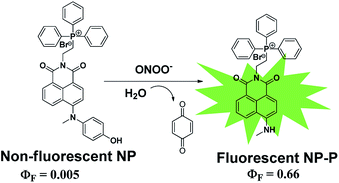
Peroxynitrite (ONOO−) is an extremely reactive oxygen species which can damage a wide array of biomolecules in cells and eventually result in mitochondrial dysfunction and cell death. Detection and imaging of this species is of significant importance to understand its role in biological processes. However, current methods are mostly limited by low sensitivity and/or specificity. In this work, we designed and synthesized a new 1,8-naphthalimide-based turn-on fluorescent probe NP for ONOO− in which the 1,8-naphthalimide fluorophore was directly conjugated with a methyl(4-hydroxyphenyl)amino group. The reaction of NP with ONOO− led to the oxidative N-dearylation of the methyl(4-hydroxyphenyl)amino group and produced a highly fluorescent product NP-P. The probe NP exhibited high sensitivity for ONOO− with a detection limit of 11 nM and high specificity over other reactive species in phosphate buffer. Moreover, NP was used to specifically detect ONOO− generated in RAW264.7 murine macrophages upon stimulation by LPS and IFN-γ. Our present study sheds light on the design of new probes for ONOO− and the resulting probe NP has great potential in enabling selective and specific detection and imaging of ONOO− in different biological and chemical systems.
1. Introduction
Peroxynitrite (ONOO−), a highly reactive oxidant and efficient nitrating agent in living systems, is endogenously generated through the diffusion-controlled reaction of nitric oxide (NO) and superoxide (O2˙−).1,2 This species can react with a wide array of biomolecules such as proteins, lipids, and nucleic acids, resulting in cell death. As a result, ONOO− is closely related to a variety of diseases, including Alzheimer's disease, arthritis, cancer, autoimmune and inflammatory diseases, and other disorders.3–6 On the other hand, accumulating evidence has demonstrated that ONOO− plays positive roles in either signal transduction via nitrating tyrosine residues7 or immunogenic response against invading pathogens.8–10 Therefore, development of specific methods for detection of cellular ONOO− is of considerable significance for both diagnosis of pertinent diseases and exploration of its diverse pathophysiology.Fluorescent probes have been recognized as the most efficient molecular tools for detection and imaging of trace amounts of biomolecules in biological systems due to their simplicity, sensitivity, visualization, real-time analysis and noninvasiveness.11–14 The early-stage fluorescent ONOO− probes were based on the reduced non-fluorescent rhodamine or fluorescein dyes, which could be converted into fluorescent products upon oxidization by ONOO−.15–19 However, the interference from other reactive oxygen species (ROS) such as ˙OH and ClO− was not negligible when using these probes. To overcome this issue, small-molecule fluorescent probes with the improved selectivity were developed on the base of the ONOO−-triggered specific reactions, including aromatic nitration,20 formation of dioxirane with activated ketones,21–23 oxidation of organoselenium/organotellurium,24–26 phenol27 or 4-hydroxyaniline groups,28,29 chemical transformation of boronic acid30,31 or boronate32–34 to phenol, and so on.35,36 Recently, fluorescent probes displaying the reversible reactions with ROS have accepted much attention.37,38
1,8-Naphthalimide has served as an attractive fluorophore owing to its intense fluorescence, high sensitivity, good photostability and insensitivity to pH in the physiological range.39–41 Herein, we present design and synthesis of a new 1,8-naphthalimide-based fluorescent probe by direct conjugation of the 1,8-naphthalimide fluorophore with a methyl(4-hydroxyphenyl)amino reaction group, which has high selectivity to ONOO−.29 It can be expected that the probe NP is non-fluorescent due to the PET process from the methyl(4-hydroxyphenyl) amino group with high electron density to the excited fluorophore. However, robust fluorescence can be observed due to the production of the green-emission product NP-P upon from oxidative N-dearylation of the methyl(4-hydroxyphenyl)amino group in NP by ONOO− (Scheme 1). We hypothesized that NP could be an ideal probe for detecting intracellular ONOO− due to the following virtues: high sensitivity and selectivity to ONOO−, insensitivity to pH and high photostability. Incorporation of the lipophilic triphenylphosphonium (TPP) cation group into 1,8-naphthalimide core is very helpful to facilitate its accumulation within cells since such lipophilic cations are shown to easily permeate lipid bilayers, driven by the membrane potential.42
2. Material and methods
2.1 Synthesis of NP
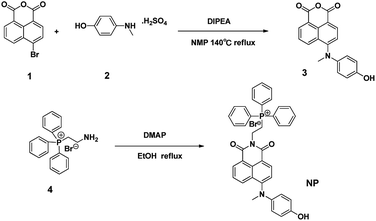
The probe NP was synthesized using the route as shown in Scheme 2. In brief, 4-bromo-1,8-naphthalic anhydride 1 reacted with 4-(methylamino)phenol 2 in NMP at 140 °C to afford the intermediate 3, which was further conjugated with (2-aminoethyl)triphenylphosphonium bromide 4 in the presence of DMAP at reflux in ethanol to give the desirable compound NP as a reddish-brown solid. The resulting fluorescent probe NP was well characterized by 1H-NMR, 13C-NMR and HRMS (Fig. S1–S4†).2.2 Preparation of the test solution
Deionized water and spectroscopic grade ethanol were used for spectroscopic studies. Peroxynitrite (ONOO−) solution was prepared as reported,42 and its concentration was estimated by using an extinction coefficient of 1670 M−1 cm−1 at 302 nm [CONOO− = Abs302 nm/1.67 (mM)]. Superoxide solution (O2˙−) was prepared by adding KO2 (1 mg) to dry dimethyl sulfoxide (1 mL) and the solution was stirred vigorously for 10 min. H2O2 solution was prepared by dilution of commercial H2O2 solution (30%) in deionized water. Hydroxyl radical (OH˙) was generated in situ by adding FeCl2 into 10 equiv. of H2O2 solution. ROO˙ was generated from 2,2′-azobis(2-amidinopropane)dihydrochloride, which was firstly dissolved in deionized water and then added into the probe testing solutions at 37 °C for 1 h. Nitric oxide (NO) was generated from NO donor of β-Gal-NONoate (10 μM) with β-galactosidase (2 unit) at 37 °C.2.3 Cell culture
RAW264.7 macrophage cells were kindly provided by Laboratory of Epigenetics in Development and Tumorigenesis Department of Cell Biology of Tianjin Medical University. Cells were grown in RPMI 1640 medium supplemented with 10% Fetal Bovine Serum (FBS) and 1% antibiotics at 37 °C in a 5% CO2/95% air incubator in a humidified atmosphere.2.4 Imaging the stimulation induced by ONOO− in RAW264.7 murine macrophages
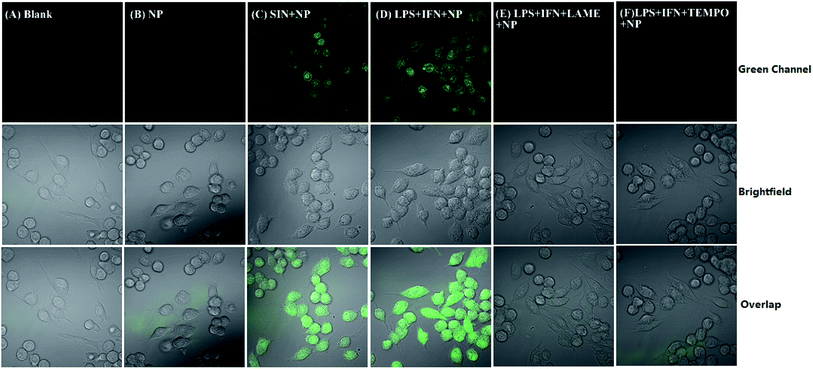
For imaging exogenously added ONOO−, the cells were pretreated with NP (5 μM) for 30 min, and then incubated with ONOO− (50 μM) donor SIN-1 for 30 min at 37 °C in DMEM medium. For imaging the endogenous ONOO−, the cells were pretreated with 1 μg mL−1 LPS and 50 ng mL−1 INF-γ for 4 h in DMEM medium at 37 °C, and then incubated with probe NP (5 μM) for 30 min at 37 °C in the same medium. For inhibition assays, the cells were activated with LPS (1 μg mL−1) and INF-γ (50 ng mL−1) in the presence of LAME (5 mM) or TEMPO (300 μM) for 4 h, and then loaded with probe NP (5 μM) for 30 min. After each treatment, the cells were washed with PBS for 3 times. Images were acquired through Olympus Fluo View™ FV1200 confocal microscope with band path of 500–600 nm upon excitation of NP at 488 nm.3. Results and discussion
3.1 Reactivity and selectivity of NP toward ONOO−
The reaction of NP with ONOO− was tested using HPLC and LC-MS analysis. NP (4 μM) was stable in phosphate buffer solution (100 mM, pH 7.4), and its retention time on the reversed C18 column was 18.5 min with the parent ion M+ at m/z 607.2 as a phosphonium cation (see details in ESI†). After addition of ONOO− (10 equiv.), a new peak with a retention time of 15.6 min appeared with the parent ion M+ at m/z 515.2 (Fig. 1), consistent with the expected product (NP-P) of the N-dearylation reaction. These results demonstrated that NP could be dearylated upon treatment with ONOO− to generate NP-P as showed in Fig. 1.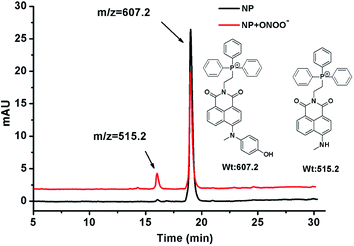 | ||
| Fig. 1 LC-MS analysis for the reaction of NP with ONOO−. See details in ESI.† | ||
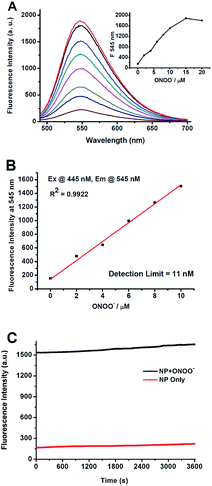
To verify whether the N-dearylation of NP induced by ONOO− is concentration-dependent, we examined the fluorescent response of NP towards ONOO−. As shown in Fig. 2A, NP (1 μM) in PBS (100 mM, pH 7.4) only showed weak fluorescence with an emission maximum at 545 nm (Φ = 0.005), indicative of highly efficient fluorescence quenching by the phenol group. Upon addition of ONOO−, the fluorescence intensity at 545 nm gradually increased due to the production of the highly fluorescent NP-P (Φ = 0.66, Fig. 2A). The fluorescent intensity reaches the maximum value when 15 equiv. of ONOO− was added, and a >12-fold enhancement in the fluorescence intensity was observed. Further addition of ONOO− resulted in a slight decrease of the fluorescent intensity (Fig. 2A). The fluorescence intensity at 545 nm was linearly related to the concentrations of ONOO− in the range of 0 to 10 μM (R2 = 0.99) (Fig. 2B). And the detection limit of 11 nM was calculated based on 3σ/κ. This value was comparable to those of other peroxynitrite probes such as HKGreen-4 (10 nM)43 and MITO-CC (11.3 nM).44 The low detection limit indicated that NP was highly sensitive to ONOO−.
While the sensitivity depends strongly on the fluorescence brightness of a probe before and after its reactivity with ONOO−, photo bleaching by a strong laser excitation may also lead to less sensitivity. Therefore, the stability of the product NP-P against light was investigated. The reaction mixture containing ONOO− (10 μM) and NP (1 μM) in PBS was exposed to a long-term irradiation with light (445 nm) (1 h). As shown in Fig. 2C, no significant decrease of its fluorescence was observed, indicating that NP-P was highly photostable. Similar photostability was also observed for NP except for the weaker fluorescence for NP. Therefore, NP can be applied for long-term measurement or imaging of ONOO−.
In addition to high reactivity, high selectivity is also required for NP as a good probe to detect ONOO− in biological systems. For this purpose, the reactivity of NP towards various biologically relevant ROS, such as H2O2, ˙OH, O2˙−, HOCl, ROO˙ and NO was also examined. Fig. 3 showed that almost no fluorescence enhancement was observed when excess (10 equiv.) of each species including NO, ˙OH, HOCl and ROO˙ was added into the NP solution (1 μM) in PBS. And only a 2-fold enhancement of fluorescence was observed after addition of O2˙− or H2O2 as compared to a 12-fold enhancement of fluorescence in the case of ONOO−. Meanwhile, the fluorescence response of NP towards reducing agents, amino acids and metal ions was also investigated (Fig. S6†). No significant fluorescence enhancements were observed upon treatment of NP with these agents. Collectively, NP has high selectivity and sensitivity toward ONOO− over other ROS, biologically related agents and metal ions, thus allowing for further investigation on imaging ONOO− in cells.
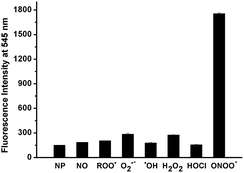 | ||
| Fig. 3 Comparison of the fluorescence responses of NP (1 μM) toward ONOO− and other ROS (10 μM for all ROS) in PBS (100 mM, pH 7.4) at 25 °C. | ||
3.2 Effect of pH on the fluorescence response of NP toward ONOO−
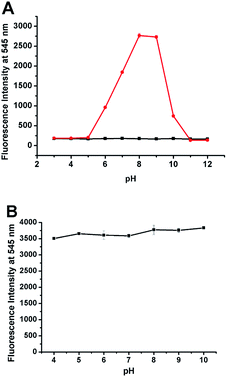
We also evaluated the reactivity of NP with ONOO− at different pHs. As shown in Fig. 4A, NP displayed pH-dependent reactivity to ONOO− with the significant fluorescence response in the pH range of 6–10 with an emission maximum at 545 nm after adding specific amount of ONOO−. And the enhancement was strongest in the pH range of 8–9, indicating that the probe NP can be used to detect ONOO− at physiological conditions. Furthermore, considering that pH within cells varies in a wide range, the effect of pH on the fluorescence of the product NP-P was also investigated. The pH values in the system were adjusted by adding high concentrations of PBS (100 mM) with designated pHs to the reaction mixture of ONOO− and NP. As shown in Fig. 4B, the intensity was almost the same across the pH values from 4 to 10 indicating that the fluorescence of NP-P was insensitive to pH. The above results indicated that the probe NP is well suitable to detect ONOO− in the wide range of pHs.3.3 Bioimaging of ONOO− in RAW264.7 cells
NP was used to detect ONOO− generated from RAW264.7 murine macrophages which are known to produce ONOO− upon stimulation with interferon-γ (IFN-γ) and lipopolysaccharide (LPS).45 As shown in Fig. 5B, when the cells were incubated with NP (5 μM), a very weak green fluorescence was observed due to endogenous ONOO− from macrophages on rest. Pretreatment of the cells with the ONOO− donor SIN-1 (3-morpholinosydnonimine)46 induced the marked fluorescence enhancement (Fig. 5C). Similarly, a strong fluorescence was observed when the cells were stimulated with IFN-γ/LPS (Fig. 5D). In order to confirm that the fluorescence originates from ONOO−, the cells were pretreated with either 2,2,6,6-tetramethylpiperidine-N-oxyl (TEMPO, the O2˙− scavenger)47 or labdanolic acid methyl ester (LAME, nitric oxide synthase inhibitor) before stimulation with IFN-γ/LPS. The almost complete inhibition of the fluorescence enhancements by TEMPO or LAME indicates that NP is specifically sensitive to ONOO− in RAW264.7 cells. To verify if NP could be accumulated in mitochondria due to the TPP cation group, the macrophages were pretreated with NP and MitoTracker Red. After extensive washing with PBS, the cells were incubated with SIN-1 (50 μM) for 30 min. The overlaid images (Fig. S7†) clearly showed that the NP probe was not site-specifically localized into the mitochondria but with additional distribution in the nuclear. The nonspecific distribution property of NP may be due to its lower hydrophobicity which has a significant role in controlling the targeting property of the TPP group.48,49 No significant cytotoxicity of NP below 5 μM (Fig. S8†) further ensures the use of NP in biological systems. These results consistently confirm that the probe NP with good biocompatibility can be used for bioimaging of ONOO− in cells.4. Conclusions
In the present study, we synthesized a novel fluorescent probe NP for ONOO− by the direct conjugation of a 1,8-naphthalimide dye with a sensitive methyl(4-hydroxyphenyl)amino reaction group. The reaction of the probe NP with ONOO− resulted in the oxidative N-dearylation to release the fluorescent product NP-P, as verified by LC-MS. NP exhibits high sensitivity to ONOO− with a detection limit of 11 nM. Moreover, this probe also shows high selectivity to ONOO− over various reactive species and strong fluorescent response to ONOO− at pHs from 7 to 9. High biocompatibility and photostability guarantee the wide use of NP in biological systems. Using a laser scanning confocal microscope, the specific imaging of the exogenous and stimulation-induced ONOO− in RAW264.7 murine macrophages was successfully achieved. Taken together, our new probe is well suitable for detection and imaging of ONOO− in biological systems which would deepen our understanding of various pathophysiological roles of ONOO− in cellular processes.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 81603064, 21572161 and 31500684), Science & Technology Projects of Tianjin (15JCZDJC32300 and 15JCYBJC23700) and Foundation of Tianjin Medical University (2110/2YY033).Notes and references
- R. Radi, J. Biol. Chem., 2013, 288, 26464–26472 CrossRef CAS PubMed.
- G. Ferrersueta and R. Radi, ACS Chem. Biol., 2009, 4, 161–177 CrossRef CAS PubMed.
- C. Szabo, H. Ischiropoulos and R. Radi, Nat. Rev. Drug Discovery, 2007, 6, 662–680 CrossRef CAS PubMed.
- P. Pacher, J. S. Beckman and L. Liaudet, Physiol. Rev., 2007, 87, 315–424 CrossRef CAS PubMed.
- T. Nagano, J. Clin. Biochem. Nutr., 2009, 45, 111–124 CrossRef CAS PubMed.
- G. Ferrer-Sueta and R. Radi, ACS Chem. Biol., 2009, 4, 161–177 CrossRef CAS PubMed.
- L. Liaudet, G. Vassalli and P. Pacher, Front. Biosci., Elite Ed., 2009, 14, 4809–4814 CrossRef.
- R. G. Allen, W. P. Lafuse, N. D. Powell, J. I. Webster Marketon, L. M. Stiner-Jones, J. F. Sheridan and M. T. Bailey, Infect. Immun., 2012, 80, 3429–3437 CrossRef CAS PubMed.
- P. A. Darrah, M. K. Hondalus, Q. Chen, H. Ischiropoulos and D. M. Mosser, Infect. Immun., 2000, 68, 3587–3593 CrossRef CAS PubMed.
- M. N. Alvarez, G. Peluffo, L. Piacenza and R. Radi, J. Biol. Chem., 2011, 286, 6627–6640 CrossRef CAS PubMed.
- J. Du, M. Hu, J. Fan and X. Peng, Chem. Soc. Rev., 2012, 41, 4511–4535 RSC.
- Y. Yang, Q. Zhao, W. Feng and F. Li, Chem. Rev., 2013, 113, 192–270 CrossRef CAS PubMed.
- F. Yu, X. Han and L. Chen, Chem. Commun., 2014, 50, 12234–12249 RSC.
- X. Jing, F. Yu and L. Chen, Chem. Commun., 2014, 50, 14253–14256 RSC.
- H. Possel, H. Noack, W. Augustin, G. Keilhoff and G. Wolf, FEBS Lett., 1997, 416, 175–178 CrossRef CAS PubMed.
- S. L. Hempel, G. R. Buettner, Y. Q. O'Malley, D. A. Wessels and D. M. Flaherty, Free Radical Biol. Med., 1999, 27, 146–159 CrossRef CAS PubMed.
- N. W. Kooy, J. A. Royall, H. Ischiropoulos and J. S. Beckman, Free Radical Biol. Med., 1994, 16, 149–156 CrossRef CAS PubMed.
- N. W. Kooy, J. A. Royall and H. Ischiropoulos, Free Radical Res., 1997, 27, 245–254 CrossRef CAS PubMed.
- G. Y. Wiederschain, Biochemistry, 2011, 76, 1276 CAS.
- T. Ueno, Y. Urano, H. Kojima and T. Nagano, J. Am. Chem. Soc., 2006, 128, 10640–10641 CrossRef CAS PubMed.
- D. Yang, H. L. Wang, Z. N. Sun, N. W. Chung and J. G. Shen, J. Am. Chem. Soc., 2006, 128, 6004–6005 CrossRef CAS PubMed.
- Z. N. Sun, H. L. Wang, F. Q. Liu, Y. Chen, P. K. H. Tam and D. Yang, Org. Lett., 2009, 11, 1887–1890 CrossRef CAS PubMed.
- T. Peng and D. Yang, Org. Lett., 2010, 12, 4932–4935 CrossRef CAS PubMed.
- F. Yu, P. Li, G. Li, G. Zhao, T. Chu and K. Han, J. Am. Chem. Soc., 2011, 133, 11030–11033 CrossRef CAS PubMed.
- N. Li, H. Wang, M. Xue, C. Chang, Z. Chen, L. Zhuo and B. Tang, Chem. Commun., 2012, 48, 2507–2509 RSC.
- F. Yu, P. Li, B. Wang and K. Han, J. Am. Chem. Soc., 2013, 135, 7674–7680 CrossRef CAS PubMed.
- Q. Zhang, Z. Zhu, Y. Zheng, J. Cheng, N. Zhang, Y. T. Long, J. Zheng, X. Qian and Y. Yang, J. Am. Chem. Soc., 2012, 134, 18479–18482 CrossRef CAS PubMed.
- T. Peng, N. K. Wong, X. Chen, Y. K. Chan, D. H. Ho, Z. Sun, J. J. Hu, J. Shen, H. El-Nezami and D. Yang, J. Am. Chem. Soc., 2014, 136, 11728–11734 CrossRef CAS PubMed.
- H. Zhang, J. Liu, Y. Q. Sun, Y. Huo, Y. Li, W. Liu, X. Wu, N. Zhu, Y. Shi and W. Guo, Chem. Commun., 2015, 51, 2721–2724 RSC.
- Z. J. Chen, W. Ren, Q. E. Wright and H. W. Ai, J. Am. Chem. Soc., 2013, 135, 14940–14943 CrossRef CAS PubMed.
- X. Sun, K. Lacina, E. C. Ramsamy, S. E. Flower, J. S. Fossey, X. Qian, E. V. Anslyn, S. D. Bull and T. D. James, Chem. Sci., 2015, 6, 2963–2967 RSC.
- F. Yu, P. Song, P. Li, B. Wang and K. Han, Analyst, 2012, 137, 3740–3749 RSC.
- J. Kim, J. Park, H. Lee, Y. Choi and Y. Kim, Chem. Commun., 2014, 50, 9353–9356 RSC.
- L. Chen, Z. Xu, J. Li, B. Zhou, M. Shan, Y. Li, L. Liu, B. Li and J. Niu, RSC Adv., 2014, 4, 1025–1031 RSC.
- K. K. Lin, S. C. Wu, K. M. Hsu, C. H. Hung, W. F. Liaw and Y. M. Wang, Org. Lett., 2013, 15, 4242–4245 CrossRef CAS PubMed.
- J. T. Hou, M. Y. Wu, K. Li, J. Yang, K. K. Yu, Y. M. Xie and X. Q. Yu, Chem. Commun., 2014, 50, 8640–8643 RSC.
- Z. Lou, P. Li and K. Han, Acc. Chem. Res., 2015, 48, 1358–1368 CrossRef CAS PubMed.
- B. Wang, P. Li, F. Yu, P. Song, X. Sun, Z. Lou and K. Han, Chem. Commun., 2013, 49, 1014–1016 RSC.
- T. S. Reddy and A. R. Reddy, Spectrochim. Acta, Part A, 2014, 128, 880–886 CrossRef PubMed.
- C. Wang, X. Zheng, R. Huang, S. Yan, X. Xie, T. Tian, S. Huang, X. Weng and X. Zhou, Asian J. Org. Chem., 2012, 1, 259–263 CrossRef CAS.
- Y. Kai, Y. Hu, K. Wang, W. Zhi, M. Liang and W. Yang, Spectrochim. Acta, Part A, 2014, 118, 239–243 CrossRef CAS PubMed.
- G. F. Kelso, C. M. Porteous, C. V. Coulter, G. Hughes, W. K. Porteous, E. C. Ledgerwood, R. A. Smithand and M. P. Murphy, J. Biol. Chem., 2001, 276, 4588–4596 CrossRef CAS PubMed.
- T. Peng, N. K. Wong, X. Chen, Y. K. Chan, D. H. Ho, Z. Sun, J. J. Hu, J. Shen, H. EI-Nezami and D. Yang, J. Am. Chem. Soc., 2014, 136, 11728–11734 CrossRef CAS PubMed.
- D. Cheng, Y. Pan, L. Wang, Z. Zeng, L. Yuan, X. Zhang and Y. T. Chang, J. Am. Chem. Soc., 2017, 139, 285–292 CrossRef CAS PubMed.
- R. B. Lorsbach, W. J. Murphy, C. J. Lowenstein, S. H. Snyder and S. W. Russell, J. Biol. Chem., 1993, 268, 1908–1913 CAS.
- N. Ashki, K. C. Hayes and F. Bao, Neuroscience, 2008, 156, 107–117 CrossRef CAS PubMed.
- R. B. R. Muijsers, E. V. D. Worm, G. Folkerts, C. J. Beukelman, A. S. Koster, D. S. Postma and F. P. Nijkamp, Br. J. Pharmacol., 2000, 130, 932–936 CrossRef CAS PubMed.
- M. F. Ross, T. A. Prime, I. Abakumova, A. M. James, C. M. Porteous, R. A. Smith and M. P. Murphy, Biochem. J., 2008, 411, 633–645 CrossRef CAS PubMed.
- M. F. Ross, G. F. Kelso, F. H. Blaikie, A. M. James, H. M. Cochemé, A. Filipovska, T. Da Ros, R. A. Smith and M. P. Murphy, Biochemistry, 2005, 70, 222–230 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures, characterization data and additional spectra. See DOI: 10.1039/c7ra04317a |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |