Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of a pigment-based whole-cell biosensor for the analysis of environmental copper†
Pei-Hsuan Chen,
Chieh Lin,
Kai-Hong Guo and
Yi-Chun Yeh *
*
Department of Chemistry, National Taiwan Normal University, 88, Section 4, Tingzhou Road, Taipei 11677, Taiwan. E-mail: yichuny@ntnu.edu.tw
First published on 5th June 2017
Abstract
Using engineered microorganisms to detect heavy metals in the environment has proven to be highly effective and robust. This paper reports on the development of a novel microbial sensor for the detection of copper ions. To develop this microbial sensor, we screened and characterized various biological parts, including promoters, output signals, and hosts. In addition, we used the plant pigment betaxanthin to output fluorescent signals in order to reduce the detection time. The resulting whole-cell biosensor presented a good sensitivity when detecting copper ions in environmental samples including freshwater pond and tap water.
Metal ions including iron, copper, zinc, and manganese play important roles as cofactors for enzymes involved in the catalysis of metabolic processes, and the maintenance of cell integrity, which is achieved through regulation by osmotic pressure.1,2 These ions are ubiquitous in all organisms; however, heavy metals can be toxic at high intracellular concentrations.3,4 For example, an excess of copper ions can cause protein dysfunction by interacting strongly with thiol groups and interfere with assembled iron–sulfur cofactors.5 This has led many microorganisms to develop the ability to sense small variations in metal concentrations as well as the means to control the influx and efflux of metal ions.1,6 Many molecular-based biosensors have been developed with high specificity, selectivity, and rapid reaction times for the detection of copper ions. However, these systems do not provide bioavailability of analytes.7–9 Compared with molecular-based biosensors, whole-cell based biosensors have disadvantages such as slower response and lower sensitivity; however, their ability to measure activities of analytes in a physiologically relevant manner to directly determine the bioavailability of the analytes. Cupriavidus metallidurans CH34 is a bacterium which has developed resistance to high concentrations of many heavy metals in the environment in which it is found.10,11 Therefore, C. metallidurans CH34 has recently been used in bioremediation processes and biosensors.12–15
In Gram-negative bacteria, there are three classes of copper-induced defence mechanisms including: (i) CueR-like activators,16,17 (ii) CusRS-like two-component systems,18–20 and (iii) TetR-like regulators.21 C. metallidurans utilizes the CusRS-like two-component regulatory system, which includes the sensor kinase CopS and the response regulator CopR. The CopS autophosphorylates at a histidine residue and subsequently relays the phosphoryl group to the response regulator, CopR. CopR contains two functional domains: an N-terminal CheY-like receiver domain and a C-terminal DNA-binding helix-turn-helix output domain.22 The phosphorylation of N-terminal domains induces conformational changes that affect the binding affinity between CopR and its recognition sites on chromosomal DNA.19,23 Therefore, CopR activates the transcription of cop gene clusters in the presence of Cu(I)/(II) ions.
In C. metallidurans, the megaplasmid and the pMOL30 plasmids encode a variety of genes which confer resistance to the toxic effects of heavy metals such as Cd(II), Zn(II), Pb(II), Ag(I), Au(III), and Cu(II).24–26 Two cop gene clusters (i.e. the cop clusters of pMOL30 and the megaplasmid) related to resistance mechanisms which protect against high copper concentrations. Under increasing concentrations of Cu(II), the second cop cluster on the pMOL30 regulated by CopSR, containing 19 ORFs (copVTMKNSRABCDIJGFLQHE) organized in nine operons (Fig. S1A, ESI†), showed much greater upregulation by quantitative-PCR and microarray data compared with the gene clusters on the megaplasmid.27 We therefore examined the regulations between CopSR and its regulators of pMOL30.
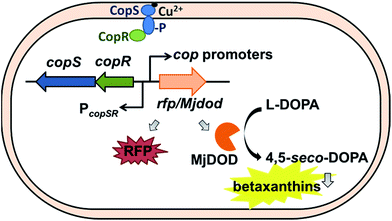
There were cop-based biosensors reported by Ng et al. and Leth et al.28,29 However, the CopSR regulatory system from C. metallidurans was not characterized. Previously, we designed a MerR-type CupR regulatory circuit in C. metallidurans to generate a fluorescence-based microbial sensor for the selective detection of gold ions.12 In current study, we employed a biosensor with a two-component regulatory system to detect bioavailable Cu(II). To optimize the performance of the Cu biosensor, we also examined the promoters of cop gene clusters and two reporter signals (red fluorescent proteins (RFP), and yellow fluorescent pigments, betaxanthin by expressing DOPA 4,5-dioxygenase from Mirabilis jalapa plant (Mjdod)) (Fig. 1). The biosensor plasmid was subsequently transformed into two hosts: the original host and a Ralstonia eutropha strain, which does not have copSR homolog and is widely used in industry for biotechnology applications.30,31
We began by comparing the CopR regulated promoters at the transcription level with the aim of achieving the better sensitivity for the analysis of Cu(II) (Fig. S1A and B†). Eight promoters of cop gene cluster of C. metallidurans (PcopA, PcopH, PcopT, PcopM, PcopF, PcopL, PcopQ, and PcopE) were constructed in order to drive the expression of RFP, which allowed the protein expression of individual promoters to be examined quantitatively. Previous reports indicate that the CopR homolog in E. coli, PcoR, was able to bind to a copper box containing the 25 bp motif AgxTtACaxaAxTGTaATxaxxxxG.32 We therefore searched for this motif in locations upstream from the transcription start codon of the cop gene cluster in C. metallidurans. However, the conservation of relatively weak consensus sequences in C. metallidurans suggests that the sequence recognition is species-dependent (Fig. S1C†).
Details as to the construction of the plasmids are provided as ESI.† CopSR sequences were introduced on the same plasmid (driven by its native promoter) to increase the number of copies of regulatory proteins that were produced (Fig. S1B†). Cells were then transformed and cultivated to exponential phase in lysogeny broth (LB) followed by a 24 h incubation with Cu(II) ions (ESI†). Following this, we quantified and compared the fluorescence of two hosts including C. metallidurans and R. eutropha cells in the presence of several copper concentrations (0, 10, 100, and 1000 μM) (Fig. S2†). We found that the fluorescence intensity of PcopA, PcopH, PcopT, PcopM, and PcopQ promoters increased in conjunction with an increase in the concentration of Cu(II). Interestingly, we observed a similar pattern of induction for both hosts. Nonetheless, no significant induction was observed for pcopF, PcopL, and PcopE promoters in the presence of Cu(II).
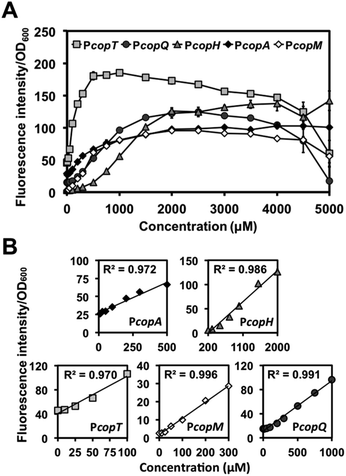
We then examined PcopA, PcopH, PcopT. PcopM, and PcopQ promoters to determine their fluorescence induction dynamic range of C. metallidurans in the presence of Cu(II). As shown in Fig. 2A, for all five promoters, fluorescence increased as the concentration of Cu(II) was increased in the media. Specifically, we observed a linear relationship between fluorescence response and the concentration of Cu(II) within various linear ranges (Fig. 2B). Among the five promoters, the calculated detection limit of PcopT was the lowest (i.e., presented the highest sensitivity) and PcopM, PcopA, PcopQ, and PcopH showed the second to the fifth lowest detection limits, respectively. Their limit of detection (LOD) was 24.3, 24.5, 99.4, 103.7 and 259.5 μM, respectively. Fig. 2B lists the details of linear regression, calculated LOD values, and the correlation coefficient (R2) for each promoter. Table S4† showed a comparison between linear ranges obtained with other whole-cell based biosensors.
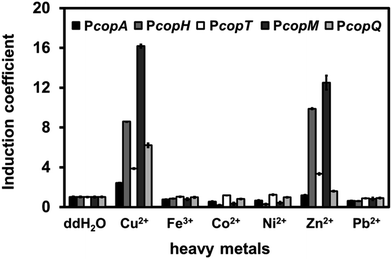
To distinguish among the various types of interference caused by different heavy metal salts, we monitored the fluorescence intensity of each promoter in the presence of Fe, Co, Ni, Zn, or Pb at 1 mM. Surprisingly, Zn(II) ions significantly induced RFP intensity for PcopH, PcopT, and PcopM at high concentration (Fig. 3). Unlike the other promoters, copQ promoter exhibited a Cu(II)-specific induction pattern. Therefore, we used PcopQ for all subsequent tests in this study.
Building a regulatory platform requires a mechanism that can output measurable signals in the presence of an input stimulus. Most of whole-cell biosensors involve the use of fluorescent proteins; therefore adopting a pigment-based reporter can be beneficial. Fluorescent proteins allow the easy detection of analytes; however, the production and maturation of these proteins can affect the time required for color to develop. Previously, the DOPA 4,5-dioxygenase, Mjdod, was shown to be involved in the synthesis of betaxanthin, a yellow betalain pigment that contains the chromophore betalamic acid, which is in-turn synthesized from l-dihydroxyphenylalanine (L-DOPA).33,34
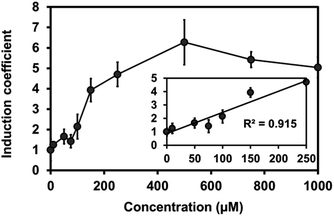
To test the expression of plant Mjdod in C. metallidurans, we constructed a plasmid containing pBAD-Mjdod. The production of yellow pigment was visible to the naked eye by inducing the exponential-phase of cells with a 0.2% arabinose and 1 mM L-DOPA for 2 h (Fig. S3†). However, the absence of either L-DOPA or arabinose failed to cause any change in color, indicating that the formation of betaxanthin depends on the presence of Mjdod as well as its own substrate, L-DOPA. We then investigated whether it would be possible to use betaxanthins as output signals for the proposed biosensor. For this, we replaced rfp with Mjdod and used dose–response curves to evaluate the response to Cu(II) at various concentrations. Specifically, cells were incubated with Cu(II) for 4 h and treated with L-DOPA for 2 h. In performing this investigation, we observed a similar pattern of induction and LOD (87.3 μM) with reduced detection time compared to RFP assays (Fig. 4).
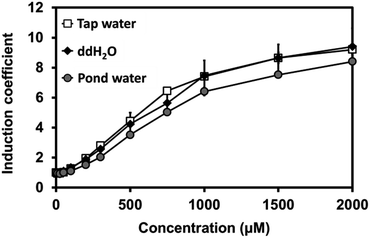
Our novel biosensor demonstrated great performance in the detection of Cu(II) in water under laboratory conditions. We also examined the efficacy of the biosensor using environmental samples, including pond and tap water. For this, sensor cells were grown at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of 2X LB to tap water, pond water, or pure water in the presence of Cu(II). Three samples presented a nearly identical increase in fluorescence signals, suggesting that the buffer matrix did not significantly affect the efficacy of the biosensors (Fig. 5).
1 ratio of 2X LB to tap water, pond water, or pure water in the presence of Cu(II). Three samples presented a nearly identical increase in fluorescence signals, suggesting that the buffer matrix did not significantly affect the efficacy of the biosensors (Fig. 5).
The main advantages that whole-cell microbial biosensors have over conventional analytical techniques are inexpensive, portability, and environmental compatibility. In this study, we developed a whole-cell biosensor for the detection of environmental Cu(II). We examined multiple promoters and identified their Cu(II) response concentrations. We found that betaxanthin was able to report both fluorescent and colorimetric signals within 6 h. These findings suggest that synthetic biology can be applied to improve the sensitivity and detection time of whole-cell biosensors for on-site environmental monitoring.
Acknowledgements
This work was funded by the Ministry of Science and Technology of Taiwan under the project number 103-2113-M-003 -002 -MY2 and 105-2113-M-003 -013 -MY2. We thank Dr Nobuhiro Sasaki for providing the plasmid (pDSET15: containing dioxygenase from Mirabilis jalapa).Notes and references
- W. Maret and A. Wedd, Binding, Transport and Storage of Metal Ions in Biological Cells, Royal Society of Chemistry, 2014 Search PubMed.
- D. H. Nies, Appl. Microbiol. Biotechnol., 1999, 51, 730–750 CrossRef CAS PubMed.
- B. Halliwell and J. M. Gutteridge, Biochem. J., 1984, 219, 1–14 CrossRef CAS PubMed.
- F. F. Xu and J. A. Imlay, Appl. Environ. Microbiol., 2012, 78, 3614–3621 CrossRef CAS PubMed.
- J. A. Lemire, J. J. Harrison and R. J. Turner, Nat. Rev. Microbiol., 2013, 11, 371–384 CrossRef CAS PubMed.
- C. Rademacher and B. Masepohl, Microbiology, 2012, 158, 2451–2464 CrossRef CAS PubMed.
- C. L. He, F. L. Ren, X. B. Zhang, Y. Y. Dong and Y. Zhao, Anal. Sci., 2006, 22, 1547–1551 CrossRef CAS PubMed.
- G.-Y. Lan, C.-C. Huang and H.-T. Chang, Chem. Commun., 2010, 46, 1257–1259 RSC.
- V. Chandrasekhar, S. Das, R. Yadav, S. Hossain, R. Parihar, G. Subramaniam and P. Sen, Inorg. Chem., 2012, 51, 8664–8666 CrossRef CAS PubMed.
- P. Vandamme and T. Coenye, Int. J. Syst. Evol. Microbiol., 2004, 54, 2285–2289 CrossRef PubMed.
- M. Vaneechoutte, P. Kampfer, T. De Baere, E. Falsen and G. Verschraegen, Int. J. Syst. Evol. Microbiol., 2004, 54, 317–327 CrossRef PubMed.
- H.-W. Tseng, Y.-J. Tsai, J.-H. Yen, P.-H. Chen and Y.-C. Yeh, Chem. Commun., 2014, 50, 1735–1737 RSC.
- L. Fairbrother, B. Etschmann, J. Brugger, J. Shapter, G. Southam and F. Reith, Environ. Sci. Technol., 2013, 47, 2628–2635 CrossRef CAS PubMed.
- X. Jian, E. C. Wasinger, J. V. Lockard, L. X. Chen and C. He, J. Am. Chem. Soc., 2009, 131, 10869–10871 CrossRef CAS PubMed.
- R. Biondo, F. A. da Silva, E. J. Vicente, J. E. Souza Sarkis and A. C. Schenberg, Environ. Sci. Technol., 2012, 46, 8325–8332 CrossRef CAS PubMed.
- F. W. Outten, D. L. Huffman, J. A. Hale and T. V. O'Halloran, J. Biol. Chem., 2001, 276, 30670–30677 CrossRef CAS PubMed.
- A. Changela, K. Chen, Y. Xue, J. Holschen, C. E. Outten, T. V. O'Halloran and A. Mondragón, Science, 2003, 301, 1383–1387 CrossRef CAS PubMed.
- G. P. Munson, D. L. Lam, F. W. Outten and T. V. O'Halloran, J. Bacteriol., 2000, 182, 5864–5871 CrossRef CAS PubMed.
- S. A. Gudipaty and M. M. McEvoy, Biochim. Biophys. Acta, Bioenerg., 2014, 1844, 1656–1661 CrossRef CAS PubMed.
- S. A. Gudipaty, A. S. Larsen, C. Rensing and M. M. McEvoy, FEMS Microbiol. Lett., 2012, 330, 30–37 CrossRef CAS PubMed.
- M. Mermod, D. Magnani, M. Solioz and J. V. Stoyanov, BioMetals, 2012, 25, 33–43 CrossRef CAS PubMed.
- M. Y. Galperin, J. Bacteriol., 2006, 188, 4169–4182 CrossRef CAS PubMed.
- R. Gao, Y. Tao and A. M. Stock, Mol. Microbiol., 2008, 69, 1358–1372 CrossRef CAS PubMed.
- G. Grass, C. Große and D. H. Nies, J. Bacteriol., 2000, 182, 1390–1398 CrossRef CAS PubMed.
- S. Juhnke, N. Peitzsch, N. Hübener, C. Große and D. H. Nies, Arch. Microbiol., 2002, 179, 15–25 CrossRef CAS PubMed.
- S. Taghavi, M. Mergeay and D. van der Lelie, Plasmid, 1997, 37, 22–34 CrossRef CAS PubMed.
- S. Monchy, M. A. Benotmane, R. Wattiez, S. van Aelst, V. Auquier, B. Borremans, M. Mergeay, S. Taghavi, D. van der Lelie and T. Vallaeys, Microbiology, 2006, 152, 1765–1776 CrossRef CAS PubMed.
- S. P. Ng, E. A. Palombo and M. Bhave, World J. Microbiol. Biotechnol., 2012, 28, 2221–2228 CrossRef CAS PubMed.
- S. Leth, S. Maltoni, R. Simkus, B. Mattiasson, P. Corbisier, I. Klimant, O. S. Wolfbeis and E. Csöregi, Electroanalysis, 2002, 14, 35–42 CrossRef CAS.
- D. Hu, A. L. Chung, L. P. Wu, X. Zhang, Q. Wu, J. C. Chen and G. Q. Chen, Biomacromolecules, 2011, 12, 3166–3173 CrossRef CAS PubMed.
- C. J. Brigham, E. N. Reimer, C. Rha and A. J. Sinskey, AMB Express, 2012, 2, 26 CrossRef PubMed.
- D. A. Rouch and N. L. Brown, Microbiology, 1997, 143, 1191–1202 CrossRef CAS PubMed.
- N. Sasaki, Y. Abe, Y. Goda, T. Adachi, K. Kasahara and Y. Ozeki, Plant Cell Physiol., 2009, 50, 1012–1016 CrossRef CAS PubMed.
- W. C. DeLoache, Z. N. Russ, L. Narcross, A. M. Gonzales, V. J. Martin and J. E. Dueber, Nat. Chem. Biol., 2015, 11, 465–471 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supporting Tables S1–S3 and Supporting Fig. S1–S3. See DOI: 10.1039/c7ra03778c |
| This journal is © The Royal Society of Chemistry 2017 |