Open Access Article
Open Access ArticleNon-leaching antibacterial cotton fabrics based on lipidated peptides†
A.
Opitakorn
a,
M.
Rauytanapanit
a,
R.
Waditee-Sirisattha
b and
T.
Praneenararat
 *a
*a
aDepartment of Chemistry, Faculty of Science, Chulalongkorn University, Phayathai Rd., Pathumwan, Bangkok, 10330, Thailand. E-mail: Thanit.P@chula.ac.th
bDepartment of Microbiology, Faculty of Science, Chulalongkorn University, Phayathai Rd., Pathumwan, Bangkok, 10330, Thailand
First published on 7th July 2017
Abstract
A set of lipidated peptides was in situ synthesised on cotton fabrics using Fmoc-based peptide synthesis. The resulting modified surfaces exhibit highly potent antibacterial activities in two Gram-negative bacteria including Escherichia coli (ATCC 8739) and Pseudomonas aeruginosa (ATCC 9027), along with moderate antibacterial activities in a Gram-positive bacterium Staphylococcus aureus (ATCC 6538P). As anticipated from covalently bound antimicrobial moieties, a representative membrane disintegration assay suggested that the modified surfaces inhibited bacteria by disrupting the integrity of bacterial cell membranes. Without non-natural D-peptides used in previous studies, this new class of non-leaching antibacterial surfaces offer potent activities while having more practical and economical productions. Furthermore, biocompatibility studies suggest that a selected set of surfaces show no toxicity, thus rendering them suitable for biomedical applications.
1. Introduction
Bacterial infections and their associated resistances to antibiotics have been a serious issue in public health and related disciplines over the past few decades despite some success in the development of new treatments and antibiotics. This is mainly due to the fact that bacterial resistances to drugs occurred at a far greater rate than that of the discovery of novel, effective drugs.1,2 Although selective and strategic uses of antibiotics could partially solve the problem, preventive measures are considered as a more effective approach to alleviate the resistance crisis. For example, the development of antimicrobial surfaces such as those used in medical devices gained a lot of attention recently3–5 as a practical way to prevent infections that would result in high cost and great effort to eradicate the bacteria.In order to fabricate active antibacterial surfaces, two main strategies were usually utilised.4–6 First, biocidal agents such as human antibiotics can be directly impregnated or physically adsorbed on desired materials. Although this strategy offers some advantages such as multiple modes of action and more controllable potency, a major drawback is leaching of active agents, which consequently leads to a limited activity and concern of toxicity in the case of sensitive environments. Therefore, another strategy aiming to permanently (usually covalently) immobilise an active moiety onto surfaces to provide contact-killing abilities was extensively investigated. In this method, the most likely mechanism was believed and occasionally confirmed to be perturbation and disintegration of bacterial cell membranes leading to cell lysis and eventually cell death. Several studies suggested that certain optimum ratios of hydrophobicity and hydrophilicity in the active molecule is crucial for high antibacterial activities7,8 – most active molecular scaffolds contained permanent positive charges that were thought to attract negatively charged molecules on bacterial cell surfaces such as teichoic acid in Gram-positive bacteria and lipopolysaccharides in Gram-negative bacteria.
Antimicrobial peptides (AMP) are a class of short peptides that are created by a number of organisms across different kingdoms as a host defence mechanism.9–11 Since most AMP were known to act mainly via disruption of cell membranes, they were utilised by some researchers as active moieties for antimicrobial surfaces.12,13 For instance, Hancock and coworkers reported a systematic study highlighting a screening of >100 surface-tethered cationic peptides on cellulose membrane.14 They found that good activities usually came from those surfaces that have cationic residues close to the surface, while hydrophobic residues near the N-terminus (the outside terminus) promoted antimicrobial activities. Another example is a work reported by Cleophas et al.,15 which was a preparation of antibacterial hydrogel based on HHC10, an AMP previously found to be active against multidrug resistant pathogens.16 Inverso HHC10, which consisted of only D-amino acids, was found to exert similar activity to the original HHC10 although it showed much slower degradation rate in human serum. Interestingly, several AMP were found to exhibit no cytotoxicity compared to some other classes of highly positively charged scaffolds, thus establishing a niche in the field of antimicrobial surfaces.
Nevertheless, a major disadvantage of making AMP was the complexity and the cost of peptide production. Although the technology of peptide synthesis has significantly advanced and allowed syntheses of peptides with substantially reduced cost, the mass production of peptides at the length of several AMP, ca. 20–50 mers, is still considered impractical and/or expensive.17 Moreover, some AMP consist of the unnatural, more expensive D-amino acids, leading to drastic increase of the synthesis cost of the peptides. Therefore, several studies instead focused on truncated amino acid sequences, and it was indeed found that this approach could retain significant amount of the antimicrobial activities in certain studies.14 In addition, to further improve the potency of truncated sequences, the addition of hydrophobic fatty acyl moieties to the peptides was recently exploited. This broad concept has been proved to be effective in selected models. For example, Radzishevsky et al. showed that acyl-lysine oligomers were potent antibacterial agents for Escherichia coli, which were also confirmed in mice.18 Likewise, ultrashort lipopeptides, consisting of a 4-mer peptide coupled with a long fatty acid (C12–C16) were studied and found to be active in some bacterial species, depending on the type of the amino acids and the fatty acid side chain.19 The same class of compounds was also immobilised on polydopamine-modified surfaces and found to be fairly active in both E. coli and Staphylococcus aureus.20
In this study, we sought to investigate the effect of hydrophobic alkyl chains on the antibacterial activity of peptide-immobilised cotton fabrics, a class of cellulosic materials. Since cellulose is a highly abundant, widely-used material, strategies for chemical modifications of cellulose are well documented.21–23 In particular, peptide synthesis (SPOT synthesis)24 and macroarray synthesis25 on cellulose membrane offer an attractive route to synthesising bioactive molecules. Therefore, our adaptation of in situ peptide synthesis with a coupling of a fatty acyl chain was deemed feasible. As a proof of concept, the first five amino acids of an AMP with a modified sequence derived from a beetle defensin,26–28 were individually appended on cellulose surface, followed by capping with a variety of fatty acids. Thereafter, we determined the antibacterial activities of these modified cottons, and showed that cotton fabrics with certain lipidated peptides could provide strong inhibitory activities in a selected set of bacterial species. With shorter sequences, lipidated peptides required much shorter time to produce, and the use of the naturally occurring L-isomers greatly reduced the cost of fabrication compared to previous work using only non-natural D-peptides. Furthermore, a selected set of lipidated peptide-modified cottons was also tested for cytotoxicity in a fibroblast cell and found to be non-toxic in all cases, thus showcasing the possibility of practical uses of these lipidated peptide-modified cotton fabrics.
2. Experimental
2.1 Materials
Chemicals were purchased from Sigma-Aldrich, Acros, or Chemimpex Ltd. Solvents (Analytical grade) were purchased from RCI Labscan (Thailand) and used without further purification. Cotton fabric was purchased from Thai Textile Development and Finishing Co., Ltd. Bacterial media (nutrient broth and agar powder) were purchased from Himedia (India).2.2 Synthesis of lipidated peptides on cellulose
Lipidated peptides were in situ generated by using the well-established Fmoc-based peptide synthesis on cellulose (so called “SPOT” synthesis). The overview steps are presented herein while all detailed procedures can be found in the ESI.† First, cotton fabrics (1 × 1 cm2) were washed with N,N-dimethylformamide (DMF), methanol, and CH2Cl2, followed by air drying. Subsequently, 4,7,10-trioxatridecane-1,13-diamine (the spacer) was then attached by two-step modifications: tosylation by 2 M tosyl chloride and incubation with 1 M spacer in DMF. After converting the surface to the amino functional group, each Fmoc-protected amino acid was subsequently immobilised using N,N′-diisopropylcarbodiimide (DIC) and 1-hydroxybenzotriazole (HOBt), followed by capping with 5% propionic anhydride in DMF. Thereafter, a new amino group was exposed by deprotection with 2% 1,8-diazabicycloundec-7-ene (DBU)/5% piperidine, rendering it ready for a new amino acid addition. It is worth noting that all modification steps were always followed by washing with various solvents and conditions – all details were provided in the ESI.† In the case of modified cottons with only amino acids, the addition of the final amino acid was directly followed by capping as above, an Fmoc deprotection with 2% DBU/20% piperidine, and a side-chain acid deprotection (30 min incubation in 90% TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1% phenol
1% phenol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2% H2O
2% H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3% triisopropylsilane (TIPS)
3% triisopropylsilane (TIPS)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4% CH2Cl2 and then 3 h incubation in 50% TFA
4% CH2Cl2 and then 3 h incubation in 50% TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1% phenol
1% phenol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2% H2O
2% H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3% TIPS
3% TIPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 44% CH2Cl2). After the final washing step (CH2Cl2, methanol and then H2O), the cotton surface was ready for biological assays. On the other hand, lipidated surfaces could be created by including an additional coupling step (after the final amino acid addition) where a linear fatty acid (with varying carbon atoms including 4, 6, 10, 12, 16, or 18 atoms) was coupled using the same coupling condition as the amino acid coupling protocol. Also, the same conditions of the final deprotections (Fmoc and side chains), and washing were also used, thereby completing all modification steps.
44% CH2Cl2). After the final washing step (CH2Cl2, methanol and then H2O), the cotton surface was ready for biological assays. On the other hand, lipidated surfaces could be created by including an additional coupling step (after the final amino acid addition) where a linear fatty acid (with varying carbon atoms including 4, 6, 10, 12, 16, or 18 atoms) was coupled using the same coupling condition as the amino acid coupling protocol. Also, the same conditions of the final deprotections (Fmoc and side chains), and washing were also used, thereby completing all modification steps.
2.3 Characterisations
The loading of each step of amino acid addition could be monitored by Fmoc quantification. Briefly, fulvene cleaved from the base-promoted deprotection could be quantified using UV spectroscopy at 296 nm. With previously prepared calibration curve, the loading amount could then be calculated. For the identification of final molecules, Rink linker was inserted after the spacer addition but before the addition of the first amino acid (see ESI for full chemical steps and details†). An acid cleavage would then release the lipidated peptide into solution, which would then be characterised by Matrix-Assisted Laser Desorption/Ionisation-Time-of-Flight (MALDI-TOF) mass spectrometry.2.4 Antibacterial assays
An adaptation of the Japanese Industrial Standard (JIS) L 1902 testing for antimicrobial activity was combined with the drop-plating technique (sterility controls were performed by autoclaving all involving materials except the modified surfaces, which were washed multiple times by organic solvents). First, an overnight culture of Staphylococcus aureus subsp. aureus (ATCC 6538P), Escherichia coli (ATCC 8739), or Pseudomonas aeruginosa (ATCC 9027) was inoculated to 100 mL of sterile nutrient broth (NB, 1% inoculum). The liquid culture was then incubated at 37 °C at 200 rpm shaking. The growth of bacterial cells was monitored by measuring the absorbance at 620 nm with a BioMATE S3 spectrophotometer. When the growth reached to the mid-log phase, bacterial culture was diluted to approximately 1 × 107 colony forming unit (CFU) per mL with sterile normal saline solution (NSS, 0.85% w/v NaCl), and a 100 µL aliquot was dropped onto each cotton fabric (1 × 1 cm2) in 12-well plates. The soaked fabric was incubated at 37 °C. After 18 h, 10 mL of normal saline solution (NSS) was added to each well and the resulting solution was serially diluted to 10−4. All of the diluted solutions were then spotted on NB agar plates and incubated at 37 °C for 18 h (12 h for P. aeruginosa). Lastly, bacterial colonies were counted and the numbers were converted back to CFU. Inhibitory activities of each surface were calculated by comparing with a negative control, the unmodified cotton surface. All assays were performed in three independent replicates.2.5 Membrane disintegration assay
Escherichia coli (ATCC 8739) was used as a model for this assay. First, the bacterium was inoculated to 100 mL of sterile nutrient broth (NB, 1% inoculum). The liquid culture was then incubated at 37 °C at 200 rpm shaking. The growth of bacterial cells was monitored by measuring the absorbance at 620 nm with a BioMATE S3 spectrophotometer. When the growth reached to the mid-log phase, bacterial culture (2 mL) was centrifuged, and its supernatant was discarded. The remaining solid was diluted with 1 mL sterile normal saline solution (NSS, 0.85% w/v NaCl), and a 100 µL aliquot of this solution was dropped onto each cotton fabric (1 × 1 cm2) in 12-well plates. The soaked fabric was incubated at 37 °C. After 18 h, 0.5 mL of phosphate buffer saline (PBS) was added to each well, and the plates were shaken for 15 min. Thereafter, the solutions were transferred to centrifuge tubes, and another 0.5 mL PBS was added to the original plates with the washing protocol being repeated. The combined solutions from two washing were then centrifuged to remove supernatant, and the resulting solids were diluted with 100 µL PBS. Thereafter, each solution was added with fluorescein diacetate (20 µL of 6.0 mM, resulting in the final concentration of 0.22 mM), and propidium iodide (20 µL of 3.0 mM, resulting in the final concentration of 0.11 mM). Each solution was kept in the dark for 30 min. The solution was centrifuged, and PBS was added to wash the unbound dyes. This washing was performed twice, and 50 µL PBS was added to re-suspend the bacterial cells. This final solution was then spread onto a glass slide covered with a #1.5 coverslip. Lastly, the fluorescence images were taken with Olympus BX51 fluorescence microscope.2.6 Scanning electron microscope experiments
The experiments commenced with the same protocol as outlined in the membrane disintegration assay. After the exposure of bacterial cells to modified cotton surfaces for 18 h, each surface was soaked in 1 mL of 2.5% glutaraldehyde solution (in 0.1 M phosphate buffer pH 7.2) for 2 h. Thereafter, the surface was washed with the phosphate buffer twice, followed by sterile water, with 10 min interval for each solvent. The dehydration step was followed with the addition of ethanol at the concentrations of 30%, 50%, 70%, and 95%, respectively (10 min each before solvent removal). The addition (3 repetitions) of 100% ethanol was then followed with 5 min interval. The surfaces were dried with a critical point dryer (Leica, EM CPD300), and attached on a metal stub for gold sputtering (sputter coater, Balzers, SCD 040), making the substrates ready for the experiments. Thereafter, a scanning electron microscope (JEOL, JSM-6610LV) was used to probe the morphology of bacterial cells.2.7 Biocompatibility assay
L-929 mouse fibroblast cell line (ACC 2, DSMZ, Germany) was cultured in Roswell Park Memorial Institute (RPMI) medium with 10% Fetal Bovine Serum (FBS) at 37 °C in 5% CO2 humidified atmosphere. All cotton samples were sterilized by immersing in 70% EtOH overnight. After air-dried, cotton fabrics were placed in 24-well plates and 50 µL of L-929 fibroblast cells (4 × 105 cells per mL) was pipetted onto each sample, and the soaked fabrics were incubated for 2 h. 500 µL RPMI medium was subsequently added to each well, and the solution was transferred for further incubation for 24 h. Thereafter, 300 µL of 0.5 mg mL−1 MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) in 0.9% NSS was added. The solution was further incubated for 2 h, and then was added with 300 µL DMSO to dissolve formazan. The viability was determined by measuring 540 nm absorbance.3. Results and discussion
In order to improve the practicality of adopting AMP in antimicrobial surfaces, two key structural features were evaluated in this study. First, all amino acids were the natural L-isomer instead of the D-isomer used in the previous study.28 Second, only five amino acids (instead of nine) were appended and then a fatty acid of varying lengths was coupled to this chain. The hypothesis was that the fatty acyl chain, with its unique hydrophobicity profile, should be able to boost antibacterial activities of L-peptides that would otherwise be less active than the original D-peptides. Interestingly, as illustrated in a representative comparison in the ESI (page S-2†), most fatty acids are significantly cheaper than amino acids, especially D-amino acids. In an extreme case, this may lead to a cost reduction of 300–400 folds if a D-amino acid is omitted and replaced by a fatty acid. Therefore, it was also of interest to test whether these increased activities from the acyl chain would be sufficient that some amino acids may be additionally removed from the whole sequence, thereby reducing the cost of fabrication even further. Compared to the D-peptides with nine amino acids, these new surfaces not only represent an interesting class of potent antibacterial surfaces that can be developed further, but also result in a great reduction of fabrication cost, thereby making the application of AMPs on surfaces more practical.An overall synthetic plan is shown in Scheme 1. Cotton surface was initially converted to an amino-terminated surface by tosylation and a hydrophilic spacer attachment (see ESI†). Thereafter, a series of amino acid additions was performed using standard Fmoc peptide synthesis including a coupling by DIC/HOBt, a capping step to remove remaining amino groups, and a base deprotection to remove the Fmoc group and expose a new amino group. In the case of surfaces with only amino acids, the final step was the side-chain deprotection by acid solutions, thereby generating either D-9-mer, 9-mer, or 5-mer cotton fabrics. On the other hand, lipidated peptides could be synthesised by coupling the last amino acid with a long-chain carboxylic acid followed by the side-chain deprotection, thus generating 5-mer.R′ where R′ is a hydrophobic acyl chain with different lengths. In addition, certain cotton surfaces with directly coupled fatty acids to the amino spacer were also fabricated as controls (denoted R′). Washing protocols were incorporated between all synthetic steps to ensure that unreacted reagents were removed and could not complicate the subsequent reactions.
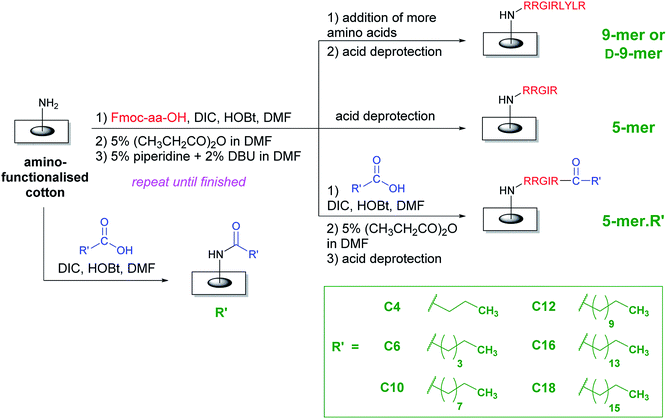 | ||
| Scheme 1 An overall synthetic plan of peptides and lipidated peptides on cotton surfaces using standard Fmoc peptide synthesis. | ||
After completing the fabrication of all modified surfaces, some characterisations were performed. First, the physical appearance of modified surfaces was found to be identical by naked eyes (examples shown in the ESI†), indicating that no major structural change occurred. The water contact angles of modified surfaces also confirmed successful chemical modifications (page S-5 in the ESI†). For example, an obvious change in contact angles was found when 5-mer was transformed into a hydrophobic 5-mer.C12. This resulted in a significant increase in contact angles (84 → 123°), indicating an increase in hydrophobicity. In addition, the loading of immobilised peptides could be quantified by determining the amount of cleaved fulvene from Fmoc deprotection. It was found that the loading was in the range of 0.2–0.4 µmol cm−2, with a general trend being that the loading amount decreased with longer chains of peptides due to less than 100% of coupling efficiency in each step (see ESI for experimental details†). Notably, the extent of the decrease of loading amounts was less towards the longer AMP chains. That is, the difference of loadings between 1-mer and 5-mer was higher than that of 5-mer and 9-mer. Since the loadings of 5-mer and 9-mer were not substantially different, it can be concluded that the antibacterial activities of these modified surfaces (see below) could be compared and analysed directly.
Moreover, lipidated peptides could also be analysed and characterised by HPLC and MALDI-TOF mass spectrometry – this was done by inserting Rink linker, an acid-labile linker, between the spacer and the first amino acid. The final acid deprotection then released free peptide molecules into solution for typical solution phase characterisations (details in the ESI†). HPLC analysis of 5-mer and 5-mer.C12 indicated that the synthesis was efficient, yielding more than 92% purity in 5-mer, and essentially 100% in 5-mer.C12 (page S-8 in the ESI†). On the other hand, MALDI-TOF MS showed good agreement between the observed m/z and the calculated m/z for all peptides (Fig. 1 for 5-mer.C16, and others in page S-9–S-15 of the ESI†). In essence, this clearly confirmed that all lipidated peptides could be successfully synthesised.
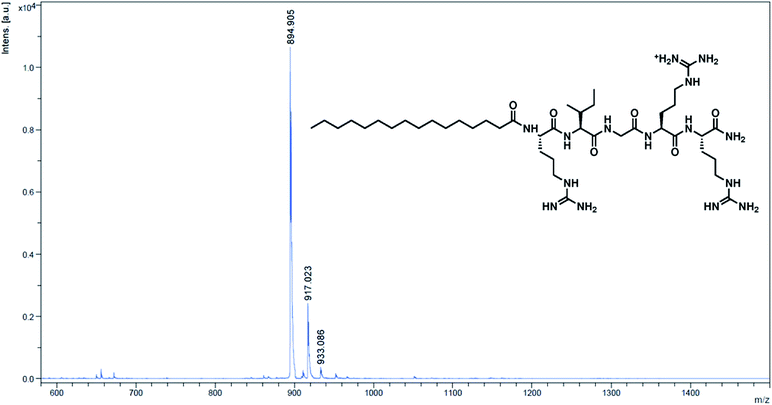 | ||
| Fig. 1 MALDI-TOF MS spectrum of the cleaved peptide from 5-mer.C16. Calculated m/z [M + H]+ = 895.21; observed m/z = 894.91. | ||
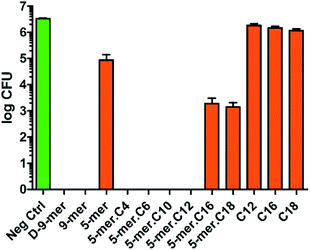
After successfully synthesising a set of modified cotton surfaces, antibacterial assays based on the Japanese Industrial Standard (JIS) L 1902 test were performed to evaluate modified cotton fabrics for their antibacterial properties. Three bacterial species were selected in this study, namely Escherichia coli (ATCC 8739), Pseudomonas aeruginosa (ATCC 9027), and Staphylococcus aureus (ATCC 6538P). In the case of E. coli (Fig. 2), almost all peptides tested gave complete killing, meaning that no CFU was observed after the culture exposed to the cotton fabrics was diluted and transferred onto nutrient agar plates and incubated for 18 h. It is noteworthy that 5-mer alone did not completely kill the bacterium, demonstrating a benefit of additional hydrophobicity. Interestingly, excessive hydrophobicity (5-mer.C16 and 5-mer.C18) did not always result in better activities and thus suggested again that optimum hydrophobicity with short peptides is needed for strong inhibitory activities. Nevertheless, even the incomplete inhibitors were still considered to exhibit significantly high inhibitory effects – growth reduction was found to be over 3–4 orders of magnitude (>99.9% inhibition) compared to the negative controls. Interestingly, both D-9-mer and 9-mer showed very strong inhibition with no colony found at the end. This alone suggested that D-peptides may not be necessary in developing antibacterial cottons for Gram-negative bacteria.
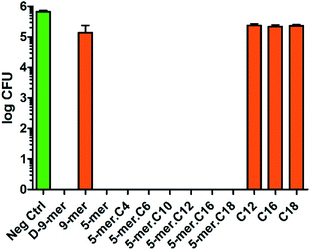
Likewise, P. aeruginosa showed strong inhibition profiles in most of the tested surfaces, but some differences from E. coli could be discerned (Fig. 3). First, among all peptides and lipidated peptides tested, 9-mer was the only one that could not provide complete killing, while 5-mer was confirmed to possess such capability. While the mechanism of this finding is currently unclear and will be investigated in due course, the experiments were repeated several times and confirmed to be valid. Therefore, this finding may suggest that peptides with D-amino acids or higher numbers of amino acids may not always be critical for obtaining high antibacterial activities. On the other hand, in contrast to the results in S. aureus, all cotton fabrics with a directly coupled fatty acid (C12, C16, and C18) did not show significant activity in both Gram-negative bacteria tested, confirming that unique structural features of peptides and lipidated peptides are crucial for antibacterial activities. Also, it is worth mentioning that the inhibitory effects found in this study were likely a bactericidal effect, since further incubation (up to 4 days) of the cultures on nutrient agar after being exposed to modified cotton fabrics did not give any colony at all (data not shown). This indicated that all bacterial cells in “complete killing” samples had been permanently destroyed and could not reproduce more cells even with favorable growth condition and prolonged time. In brief, it was found that great antibacterial activities in Gram-negative bacteria can come from short peptides (with or without a lipid chain) without the need of D-amino acids – a finding of which would be of tremendous significance for further developments.
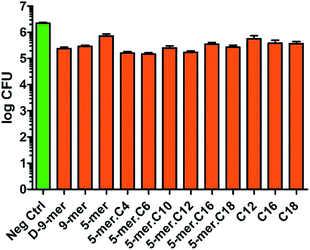
Since Gram-negative bacteria and Gram-positive bacteria have substantial differences in chemical compositions and structural formats of the cell walls and cell membranes, we also sought to test our modified cotton surfaces in a Gram-positive bacterium S. aureus. It is worth noting that cotton modified with a 9-mer D-peptide (denoted D-9-mer) was previously found to exhibit strong antibacterial activities in methicillin-resistant S. aureus.28 Overall, the results showed that all tested modified cotton surfaces possessed some inhibitory activities (Fig. 4), with the most active surface (5-mer.C6) being able to reduce the bacterial growth by about 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log CFU, or about 93% inhibition. Interestingly, appending a fatty acyl chain onto 5-mer (which was significantly less active than 9-mer) obviously increased the potency of the cotton surface. In fact, all lipidated peptides exerted stronger inhibitory effects than did 5-mer, with certain modified surfaces (5-mer.C4, 5-mer.C6 and 5-mer.C12) surpassing even D-9-mer under this condition. Similar to the case of E. coli, excessively long chains like 5-mer.C16 and 5-mer.C18, however, seemed to decrease the activities. Also, despite the fact the strain of S. aureus used in our work is different from that of the previous study by Nakamura et al.,28 it is still deemed surprising that D-9-mer was a significantly weaker inhibitor in our condition – this was confirmed by several repeated experiments. In fact, its activity was not much higher than those cotton fabrics with only long-chain fatty acids (C12, C16, and C18), which showed significant inhibitions in the current study (about 70–80% growth reduction). The activities of these long-chain fatty acids were considered to be species-specific since the same surfaces showed much less inhibition in P. aeruginosa and E. coli (see above). In addition, it was previously reported that fatty acids impregnated in cellulose exhibited some antibacterial activity although a direct comparison is not possible due to the differences in methods used in chemical modification and antibacterial assays.29
log CFU, or about 93% inhibition. Interestingly, appending a fatty acyl chain onto 5-mer (which was significantly less active than 9-mer) obviously increased the potency of the cotton surface. In fact, all lipidated peptides exerted stronger inhibitory effects than did 5-mer, with certain modified surfaces (5-mer.C4, 5-mer.C6 and 5-mer.C12) surpassing even D-9-mer under this condition. Similar to the case of E. coli, excessively long chains like 5-mer.C16 and 5-mer.C18, however, seemed to decrease the activities. Also, despite the fact the strain of S. aureus used in our work is different from that of the previous study by Nakamura et al.,28 it is still deemed surprising that D-9-mer was a significantly weaker inhibitor in our condition – this was confirmed by several repeated experiments. In fact, its activity was not much higher than those cotton fabrics with only long-chain fatty acids (C12, C16, and C18), which showed significant inhibitions in the current study (about 70–80% growth reduction). The activities of these long-chain fatty acids were considered to be species-specific since the same surfaces showed much less inhibition in P. aeruginosa and E. coli (see above). In addition, it was previously reported that fatty acids impregnated in cellulose exhibited some antibacterial activity although a direct comparison is not possible due to the differences in methods used in chemical modification and antibacterial assays.29
Notably, the relationship of hydrophobicity and antibacterial activities is worth mentioning, especially in comparison to previous studies. In general, it can be concluded that increasing hydrophobicity of an AMP, i.e., by acylation with fatty acids, tends to increase antibacterial activities. Nevertheless, the increasing activities were usually found on only selected bacterial species depending on the sequence of the AMP, and the chain length of the hydrophobic chain. For example, both studies from Makovitzki et al. and Rotem et al. proved that the sequence and the acyl chain of lipopeptides both influenced the selectivity towards a variety of bacteria, mainly discriminating between Gram-positive and Gram-negative bacteria.19,30 In this regard, it was found that our study yielded similar finding, in that our modified surfaces were more active against Gram-negative bacteria than a Gram-positive bacterium. Nevertheless, it is important to note that a majority of studies involving lipidated peptides dealt with the soluble version of AMP. Therefore, the mode of actions of these AMP may differ drastically from our case. In a study by Shalev et al.,20 immobilised versions of a lipidated AMP were investigated, but no clear discrimination could be discerned between E. coli and S. aureus. However, it should be noted that the assay used was different from ours, and this may be the cause of the discrepancy of the results. In brief, further systematic investigations would be needed to fully understand structure–activity relationships, especially for immobilised peptides.
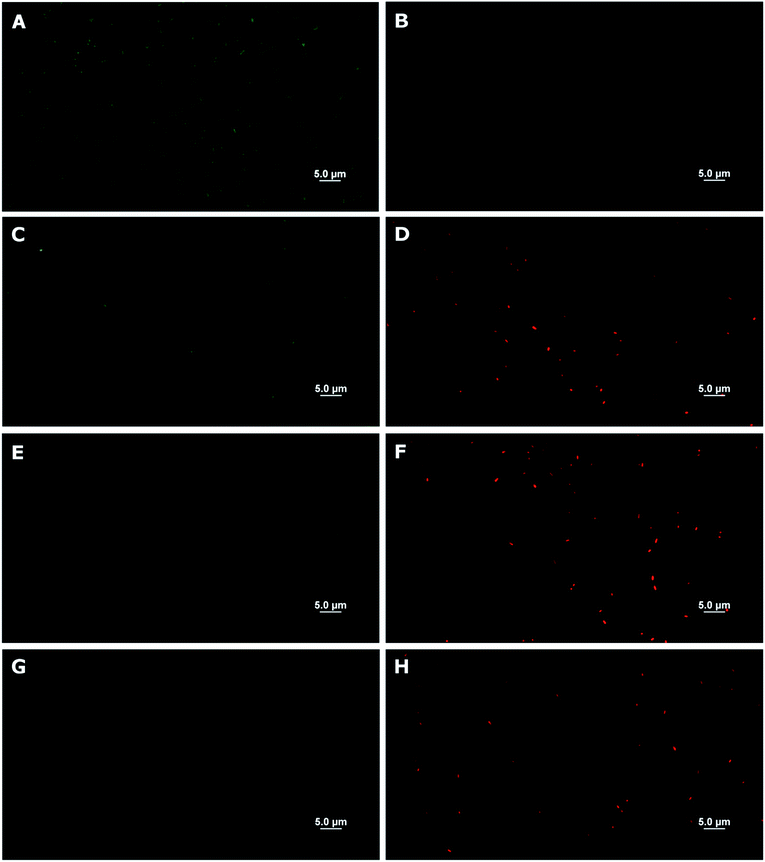
To obtain further insight in the inhibitory activities, a membrane disintegration assay against E. coli was also conducted to confirm whether the killing activities were ascribed to the destruction of bacterial membranes, which is the most well-documented cause for the activities of AMPs.12 In this assay, fluorescein diacetate was used to detect live cells, which could hydrolyse the ester groups in the compound and release the green fluorescent product. On the other hand, propidium iodide, a known membrane-impermeable DNA-binding fluorescent dye, was employed for the detection of dead cells, which released DNA materials after cell leakage. Thus, a relative increase in the red fluorescent cells in comparison to green cells suggests that the bacterial membrane has been compromised. In our case, this was found to be true for all samples tested (5-mer, 5-mer.C4, and 5-mer.C12), where there was an obvious increase in dead cells (Fig. 5C–H) compared to the unmodified surface (Fig. 5A and B). In addition, to provide alternative information about the loss of membrane integrity, scanning electron microscope (SEM) was also used to probe the morphology of bacterial cells after exposure to 5-mer.C12. The results indicated that both S. aureus and E. coli exhibited no morphological change after exposure to unmodified cotton (Fig. 6). On the other hand, 5-mer.C12 caused significant change in morphology of both bacteria (rough and shrunk bacterial surface in S. aureus, and irregular cell shape and incomplete cell division in E. coli). Given the fact that all bioactive moieties could not be released from support due to covalent immobilisation, other modes of action that involve intracellular targets were thus deemed unlikely. Hence, the results in these experiments corroborated well with this theoretical consideration.
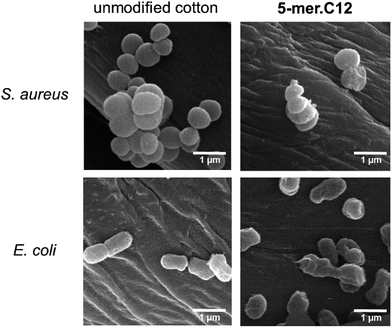 | ||
| Fig. 6 SEM experiments to probe the morphology of bacterial cells (S. aureus and E. coli) after exposure to unmodified cotton, and 5-mer.C12. | ||
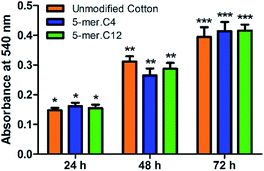
Lastly, in order to verify the versatility of our modified cotton fabrics for more sensitive applications such as materials with direct contact to human cells, two selected modified fabrics (5-mer.C4 and 5-mer.C12) were tested for biocompatibility with L-929 mouse connective tissue fibroblast cell line (ACC 2, DSMZ, Germany). The data suggested that these lipidated peptides with highly potent antibacterial activities showed no toxicity in mammalian cells for over three days (Fig. 7), indicating that the killing mechanism is unique to bacterial cell membrane systems. Hence, this showcased that cotton surface modified with lipidated peptides could be a promising material for biomedical applications.
4. Conclusions
We reported herein a fabrication of cotton fabrics modified with antimicrobial peptides with various acyl moieties. As anticipated, acyl chains with different lengths could improve the antibacterial activity, albeit with differing degrees – this suggested that there may be advantageous effect of hydrophobicity to the activities of modified surfaces. The study suggested that cotton fabrics modified with lipidated peptides were much more active in Gram-negative bacteria than in Gram-positive bacteria although it should be noted that statistically significant antibacterial activities could also be observed in S. aureus, thus qualifying as broad-spectrum antibacterial surfaces. With lower cost of synthesis and shorter fabrication time, this strategy offers a great improvement in the production of antibacterial cotton surfaces with low cytotoxicity. Evaluation of this strategy in other lipid structures, peptide sequences and other types of surfaces will be investigated and reported in due course.Acknowledgements
The authors would like to thank the Ratchadaphiseksomphot Endowment Fund of Chulalongkorn University (RES560530078-AM) for funding of this work. Ms Songchan Puthong is acknowledged for her guidance and assistance in biocompatibility assays. Dr Tana Taechalertpaisarn is recognised for his guidance and assistance in experiments involving fluorescence microscope. Ms Rujiporn Prateepasen is acknowledged for her assistance in SEM experiments.Notes and references
- K. Lewis, Nature, 2012, 485, 439–440 CrossRef CAS PubMed.
- J. Davies and D. Davies, Microbiol. Mol. Biol. Rev., 2010, 74, 417–433 CrossRef CAS PubMed.
- Q. Yu, Z. Q. Wu and H. Chen, Acta Biomater., 2015, 16, 1–13 CrossRef CAS PubMed.
- F. Siedenbiedel and J. C. Tiller, Polymers, 2012, 4, 46–71 CrossRef CAS.
- K. Bazaka, M. V. Jacob, W. Chrzanowski and K. Ostrikov, RSC Adv., 2015, 5, 48739–48759 RSC.
- J. C. Tiller, Adv. Polym. Sci., 2011, 240, 193–217 CrossRef CAS.
- K. Lewis and A. M. Klibanov, Trends Biotechnol., 2005, 23, 343–348 CrossRef CAS PubMed.
- L. Timofeeva and N. Kleshcheva, Appl. Microbiol. Biotechnol., 2011, 89, 475–492 CrossRef CAS PubMed.
- K. A. Brogden, Nat. Rev. Microbiol., 2005, 3, 238–250 CrossRef CAS PubMed.
- A. Bahar and D. Ren, Pharmaceuticals, 2013, 6, 1543–1575 CrossRef PubMed.
- G. Wang, B. Mishra, K. Lau, T. Lushnikova, R. Golla and X. Wang, Pharmaceuticals, 2015, 8, 123 CrossRef CAS PubMed.
- F. Costa, I. F. Carvalho, R. C. Montelaro, P. Gomes and M. C. L. Martins, Acta Biomater., 2011, 7, 1431–1440 CrossRef CAS PubMed.
- K. Glinel, P. Thebault, V. Humblot, C. M. Pradier and T. Jouenne, Acta Biomater., 2012, 8, 1670–1684 CrossRef CAS PubMed.
- K. Hilpert, M. Elliott, H. Jenssen, J. Kindrachuk, C. D. Fjell, J. Körner, D. F. H. Winkler, L. L. Weaver, P. Henklein, A. S. Ulrich, S. H. Y. Chiang, S. W. Farmer, N. Pante, R. Volkmer and R. E. W. Hancock, Chem. Biol., 2009, 16, 58–69 CrossRef CAS PubMed.
- R. T. C. Cleophas, M. Riool, H. C. Quarles van Ufford, S. A. J. Zaat, J. A. W. Kruijtzer and R. M. J. Liskamp, ACS Macro Lett., 2014, 3, 477–480 CrossRef CAS.
- A. Cherkasov, K. Hilpert, H. Jenssen, C. D. Fjell, M. Waldbrook, S. C. Mullaly, R. Volkmer and R. E. W. Hancock, ACS Chem. Biol., 2009, 4, 65–74 CrossRef CAS PubMed.
- G. Laverty, S. P. Gorman and B. F. Gilmore, Int. J. Mol. Sci., 2011, 12, 6566–6596 CrossRef CAS PubMed.
- I. S. Radzishevsky, S. Rotem, D. Bourdetsky, S. Navon-Venezia, Y. Carmeli and A. Mor, Nat. Biotechnol., 2007, 25, 657–659 CrossRef CAS PubMed.
- A. Makovitzki, D. Avrahami and Y. Shai, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15997–16002 CrossRef CAS PubMed.
- T. Shalev, A. Gopin, M. Bauer, R. W. Stark and S. Rahimipour, J. Mater. Chem., 2012, 22, 2026–2032 RSC.
- D. Roy, M. Semsarilar, J. T. Guthrie and S. Perrier, Chem. Soc. Rev., 2009, 38, 2046–2064 RSC.
- M. N. Belgacem, M. C. Salon-Brochier, M. Krouit and J. Bras, J. Adhes. Sci. Technol., 2011, 25, 661–684 CrossRef CAS.
- Y. Habibi, Chem. Soc. Rev., 2014, 43, 1519–1542 RSC.
- R. Volkmer, ChemBioChem, 2009, 10, 1431–1442 CrossRef CAS PubMed.
- H. E. Blackwell, Curr. Opin. Chem. Biol., 2006, 10, 203–212 CrossRef CAS PubMed.
- H. Saido-Sakanaka, J. Ishibashi, E. Momotani, F. Amano and M. Yamakawa, Peptides, 2004, 25, 19–27 CrossRef CAS PubMed.
- T. Iwasaki, H. Saido-Sakanaka, A. Asaoka, D. Taylor, J. Ishibashi and M. Yamakawa, J. Insect Biotechnol. Sericol., 2007, 76, 25–29 CAS.
- M. Nakamura, T. Iwasaki, S. Tokino, A. Asaoka, M. Yamakawa and J. Ishibashi, Biomacromolecules, 2011, 12, 1540–1545 CrossRef CAS PubMed.
- L. Adamopoulos, J. Montegna, G. Hampikian, D. S. Argyropoulos, J. Heitmann and L. A. Lucia, Carbohydr. Polym., 2007, 69, 805–810 CrossRef CAS.
- S. Rotem, I. Radzishevsky and A. Mor, Antimicrob. Agents Chemother., 2006, 50, 2666–2672 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed procedures of all steps of cotton modification, loading quantification of immobilised peptides, characterisation of immobilised peptides with MALDI-TOF. See DOI: 10.1039/c7ra03565a |
| This journal is © The Royal Society of Chemistry 2017 |