Open Access Article
Open Access ArticlePhotocontrollable fluorogenic probes for visualising near-membrane copper(II) in live cells†
Buxiang Chena,
Liulin Wanga,
Yanfei Zhaoa,
Yun Nia,
Chenqi Xina,
Chengwu Zhanga,
Jinhua Liua,
Jingyan Geb,
Lin Li *a and
Wei Huang*ac
*a and
Wei Huang*ac
aKey Laboratory of Flexible Electronics (KLOFE), Institute of Advanced Materials (IAM), Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM), Nanjing Tech University (Nanjing Tech), 30 South Puzhu Road, Nanjing, 211816, P. R. China. E-mail: iamlli@njtech.edu.cn; iamwhuang@njtech.edu.cn
bKey Laboratory of Bioorganic Synthesis of Zhejiang Province, College of Biotechnology and Bioengineering, Zhejiang University of Technology, Hangzhou 310014, P. R. China
cKey Laboratory for Organic Electronics and Information Displays, Institute of Advanced Materials (IAM), Nanjing University of Posts & Telecommunications, 9 Wenyuan Road, Nanjing 210023, P. R. China
First published on 15th June 2017
Abstract
In this work, we have described the design, synthesis, and characterisation of photocontrollable fluorogenic probe, Mem-5, which is equipped with a photolabile group (nitrobenzyl group), high brightness reporter (fluorescein), and membrane-anchoring unit (cholesterol or long aliphatic chain). This probe shows an intense fluorescence enhancement in response to copper(II) without interference from 45 other analytes, including metal cations, amino acids, and anions, under biological conditions (pH 6–9). The hydrolysis reaction is second-order, with k = 9.01 × 10−5 M−1 s−1. The detection limit is 3.3 × 10−7 M in HEPES buffer (25 mM). The confocal fluorescence imaging results demonstrated that Mem-5 can visualise near-membrane copper(II) in live mammalian cells. The clear advantage of our photocontrollable method is that it successfully avoids the influence of chemical species outside cells during near-membrane specific detection.
Introduction
Cell plasma membranes are regarded as asymmetric due to their inner and outer leaflets having different compositions, based on the classic Singer–Nicolson model.1 This prevents foreign substances from entering the cell barrier freely, ensuring a relatively stable environment within the cell,2 but absorbs and digests material inside and outside by endocytosis, phagocytosis, or exocytosis.3,4 Therefore, this lipid bilayer with embedded protein membranes is important for cell recognition, signal transduction, cellulose synthesis, microfibril assembly, and other mechanisms,5 and can regulate cell life activities as turntables for crucial processes in neurobiology, muscle contraction, and cell signalling.6,7 Given the pivotal role of cell plasma membranes, the development of membrane-specific fluorogenic probes to detect near-membrane elements has attracted increasing attention.8–15Copper(II) content in the human body is the second most common trace element, and plays an essential role in human metabolism.16 Copper(II) overload has been shown to cause a range of neurological diseases, such as Wilson's and Alzheimer's diseases.17,18 High-affinity copper transporter 1 (Ctr1) drives the entry of copper(II) into cells,19 while cells must eject excess copper(II) through the plasma membrane to maintain a low concentration of cytosolic free copper(II).20 Effective methods for evaluating free copper(II) near or across the plasma membrane will aid understanding of the copper(II) transportation-related biological functions of cells. Regrettably, tools to precisely detect near-membrane copper(II) in live cells without interference by external copper(II) have still not been developed.
With this in mind, herein, we have successfully developed two new photocontrollable fluorogenic probes (Fig. 1) with different membrane anchors to image near-membrane copper(II) in live mammalian cells with good spatial and temporal controls using efficiently photolysis by ultraviolet (UV) light irradiation. Mem-1/2 (structures shown in Fig. 2), which contain a cholesterol or 12-carbon aliphatic chain, served as membrane tracers, and their membrane staining ability was compared using this “piggyback” strategy.21–23 As shown in Fig. 1, fluorescein hydrazide hydrolysed by copper(II) would normally be blocked due to the existence of a caged unit after membrane staining,24 facilitating the complete accumulation of Mem-5/6 with no fluorescence.25 Subsequent UV irradiation will force removal of the caged group, releasing uncaged Mem-3/4 (structures shown in Fig. 2),26,27 which would then be hydrolysed by copper(II) to generate a specific localised and strong fluorescent signal.28 Notably, the modularity of our probe design should enable this strategy to be readily applicable to the detection of other targets near the membrane.
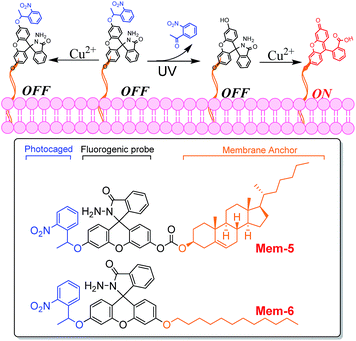 | ||
| Fig. 1 Overall strategy of our photocontrollable fluorogenic probes, Mem-5 and Mem-6 (structures shown in box), for imaging membrane copper(II) in live cells. | ||
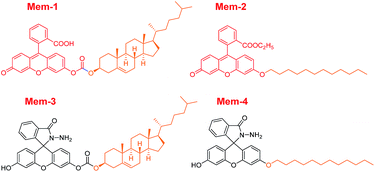 | ||
| Fig. 2 Structures of membrane tracers, Mem-1 and Mem-2, with different anchors, and unphotocontrollable fluorogenic probes, Mem-3 and Mem-4, for imaging membrane copper(II) in live cells. | ||
Experimental
Materials and instruments
All reagents and solvents were purchased from commercial suppliers and used without further purification, unless otherwise noted. N,N-Dimethylformamide (DMF) was distilled over CaH2. Petroleum ether (PE, 60–90 °C), DCM, and ethyl acetate (EA) were used as eluents for flash column chromatography with Merck silica gel (0.040–0.063). 1-(1-Bromoethyl)-2-nitrobenzene was synthesised according to a literature procedure, while fluorescein and hydrazine hydrate were purchased from Adamas.25 Cholesterol chloride and bromododecane were purchased from Tokyo Chemical Industry. Nuclear (Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342) and membrane (CM orange) trackers were purchased from Invitrogen. Human hepatocellular carcinoma (HepG2) cells and fetal bovine serum (FBS) were purchased from ATCC (HB-8065™) and Biological Industries (BI, 04-001-01ACS), respectively. Other reagents used in the experiments were purchased from Sigma-Aldrich and used without further purification unless otherwise noted. Distilled water was used throughout the experiments. Absorption spectra were recorded using SynergyHTX microplate reader or a Shimadzu UV-3600 UV-Vis-NIR spectrophotometer. Photoluminescence spectra were recorded using a HITACHI F4600 fluorescence spectrophotometer with excitation slit widths of 5 nm and emission slit widths of 10 nm. 1H and 13C NMR spectra were collected in CDCl3 or DMSO-d6 at 25 °C using an Avance AV-300 spectrometer. All photochemical reactions were conducted in an ultraviolet analyser ZF-20D. Ultrapure water was used to prepare all aqueous solutions. Fluorescein in 0.1 M NaOH (Φ = 0.85) was used as the standard for quantum yield measurements. All spectroscopic measurements were performed in 25 mM HEPES buffer (pH 7.4) at 37 °C. All images were acquired in the same way at 20 °C on Zeiss LSM880 NLO (2 + 1 with BIG) confocal microscope system equipped with an objective LD C-Apochromat 63×/1.15 W Corr M27, 405 nm diode laser, argon ion laser (458, 488, and 514 nm), HeNe laser (543 and 594 nm), and 633 nm laser, with 8 AOTF channels for simultaneous control of 8 laser lines. A PMT detector ranging from 420 nm to 700 nm was used for steady-state fluorescence. Internal photomultiplier tubes were used to collect the signals in 8 bit unsigned 1024 × 1024 pixels at a scan speed of 200 Hz. Images were processed with Zeiss User PC Advanced for LSM system (BLUE).
342) and membrane (CM orange) trackers were purchased from Invitrogen. Human hepatocellular carcinoma (HepG2) cells and fetal bovine serum (FBS) were purchased from ATCC (HB-8065™) and Biological Industries (BI, 04-001-01ACS), respectively. Other reagents used in the experiments were purchased from Sigma-Aldrich and used without further purification unless otherwise noted. Distilled water was used throughout the experiments. Absorption spectra were recorded using SynergyHTX microplate reader or a Shimadzu UV-3600 UV-Vis-NIR spectrophotometer. Photoluminescence spectra were recorded using a HITACHI F4600 fluorescence spectrophotometer with excitation slit widths of 5 nm and emission slit widths of 10 nm. 1H and 13C NMR spectra were collected in CDCl3 or DMSO-d6 at 25 °C using an Avance AV-300 spectrometer. All photochemical reactions were conducted in an ultraviolet analyser ZF-20D. Ultrapure water was used to prepare all aqueous solutions. Fluorescein in 0.1 M NaOH (Φ = 0.85) was used as the standard for quantum yield measurements. All spectroscopic measurements were performed in 25 mM HEPES buffer (pH 7.4) at 37 °C. All images were acquired in the same way at 20 °C on Zeiss LSM880 NLO (2 + 1 with BIG) confocal microscope system equipped with an objective LD C-Apochromat 63×/1.15 W Corr M27, 405 nm diode laser, argon ion laser (458, 488, and 514 nm), HeNe laser (543 and 594 nm), and 633 nm laser, with 8 AOTF channels for simultaneous control of 8 laser lines. A PMT detector ranging from 420 nm to 700 nm was used for steady-state fluorescence. Internal photomultiplier tubes were used to collect the signals in 8 bit unsigned 1024 × 1024 pixels at a scan speed of 200 Hz. Images were processed with Zeiss User PC Advanced for LSM system (BLUE).
Synthetic procedures
All reactions were carried out under a dry nitrogen atmosphere. Reaction progress was monitored by TLC on pre-coated silica plates (thickness, 250 μm) and spots were visualised using ceric ammonium molybdate, basic KMnO4, UV light, or iodine.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) affording the product as a pink solid (yield 60%). 1H NMR (300 MHz, CDCl3): δ (ppm) 8.03 (d, J = 4.5 Hz, 1H), 7.93 (t, J = 4.5 Hz, 1H), 7.75 (d, J = 3 Hz, 1H), 7.59 (t, J = 7.5 Hz, 1H), 7.45 (m, 3H), 7.09 (s, 1H) 7.02 (d, J = 3 Hz, 1H), 6.52 (m, 4H), 6.82 (dd, J1 = 4.5 Hz, J2 = 1.5, 1H), 6.65 (d, J = 4.5 Hz, 2H), 6.52 (d, J = 3 Hz, 2H), 6.05 (dd, J1 = 3 Hz, J2 = 1.5 Hz, 1H), 5.42 (s, 1H), 4.60 (dd, J1 = 6 Hz, J2 = 3 Hz, 1H) 3.52 (bs, NH2), 2.48 (s, 2H), 2.00 (m, 5H), 1.56 (m, 10H), 1.33 (m, 4H), 0.91 (m, 22H), 0.68 (s, 3H). 13C NMR (75 MHz, CDCl3): δ (ppm) 166.41, 158.49, 152.49, 151.77, 150.66, 150.55, 139.16, 138.63, 134.26, 134.21, 133.05, 128.61, 128.43, 128.37, 128.30, 128.20, 127.50, 124.91, 123.94, 123.42, 123.37, 116.94, 116.51, 112.90, 112.66, 110.04, 103.36, 103.21, 79.31, 71.86, 56.80, 56.27, 50.11, 42.43, 39.83, 39.62, 37.98, 36.92, 36.65, 36.30, 35.87, 32.01, 31.96, 28.31, 28.09, 27.69, 24.38, 23.93, 23.67, 23.62, 22.90, 22.66, 21.16, 19.36, 18.83, 11.97.
1) affording the product as a pink solid (yield 60%). 1H NMR (300 MHz, CDCl3): δ (ppm) 8.03 (d, J = 4.5 Hz, 1H), 7.93 (t, J = 4.5 Hz, 1H), 7.75 (d, J = 3 Hz, 1H), 7.59 (t, J = 7.5 Hz, 1H), 7.45 (m, 3H), 7.09 (s, 1H) 7.02 (d, J = 3 Hz, 1H), 6.52 (m, 4H), 6.82 (dd, J1 = 4.5 Hz, J2 = 1.5, 1H), 6.65 (d, J = 4.5 Hz, 2H), 6.52 (d, J = 3 Hz, 2H), 6.05 (dd, J1 = 3 Hz, J2 = 1.5 Hz, 1H), 5.42 (s, 1H), 4.60 (dd, J1 = 6 Hz, J2 = 3 Hz, 1H) 3.52 (bs, NH2), 2.48 (s, 2H), 2.00 (m, 5H), 1.56 (m, 10H), 1.33 (m, 4H), 0.91 (m, 22H), 0.68 (s, 3H). 13C NMR (75 MHz, CDCl3): δ (ppm) 166.41, 158.49, 152.49, 151.77, 150.66, 150.55, 139.16, 138.63, 134.26, 134.21, 133.05, 128.61, 128.43, 128.37, 128.30, 128.20, 127.50, 124.91, 123.94, 123.42, 123.37, 116.94, 116.51, 112.90, 112.66, 110.04, 103.36, 103.21, 79.31, 71.86, 56.80, 56.27, 50.11, 42.43, 39.83, 39.62, 37.98, 36.92, 36.65, 36.30, 35.87, 32.01, 31.96, 28.31, 28.09, 27.69, 24.38, 23.93, 23.67, 23.62, 22.90, 22.66, 21.16, 19.36, 18.83, 11.97.Photouncaging reactions
For in vitro analysis, different concentrations (2.0 μM for selectivity/kinetic experiments and 10.0 μM for pH titration experiments) of Mem-5 were placed in quartz glassware and submitted to UV irradiation (500 μJ cm−2 at 400 nm) for 5 min at room temperature to generate uncaged Mem-5 in HEPES buffer. In live cell imaging, cells were incubated with Mem-5 (2.0 μM) for 20 min at 37 °C, followed by further UV irradiation (500 μJ cm−2, 5 min) at room temperature in cover-free dishes to generate uncaged Mem-5.Cell culture and cytotoxicity activity assay
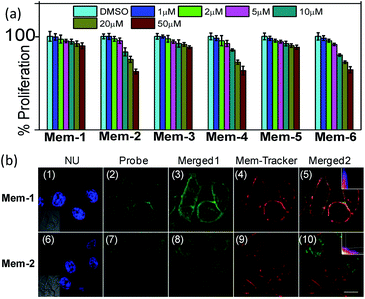
HepG2 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM), containing 10% FBS, 100.0 mg L−1 streptomycin, and 100 IU mL−1 penicillin. HepG2 cells were seeded in glass-bottom dishes (Mattek) and grown to 70–80% confluency. The cytotoxicity activities of the probes were determined using an XTT colorimetric cell proliferation kit (Roche), following the manufacturer guidelines. Briefly, different cells were grown to 20–30% confluency (they would reach 80–90% confluency within 48–72 h in the absence of compounds) in 96-well plates under the conditions described above. The medium was aspirated, washed with PBS, and then treated, in duplicate, with 0.1 mL of the medium containing different concentrations of Mem-1/2/3/4/5/6 (1–50 μM). Probes were applied from DMSO stocks, whereby DMSO never exceeded 1% in the final solution. The same volume of DMSO was used as a negative control, while the same volume of staurosporine (STS, 200 nM) was used as a positive control. After a total treatment time of 24 h, proliferation was assayed using the XTT colorimetric cell proliferation kit (Roche), following manufacturer guidelines (Fig. 4a).Live cells fluorescence imaging
For organelle-specific imaging experiments, the cells were incubated with Mem-1 and Mem-2 (1 μM prepared in fresh medium) for 20 min at 37 °C, followed by washing three times with PBS. To accurately visualise membrane copper, the cells were incubated with Mem-3 and Mem-5 (5 μM prepared in fresh medium) for 20 min at 37 °C. After washing three times with PBS, the selected cells were irradiated for 2 min with UV light through an ultraviolet analyser. The cells were then incubated with copper(II) (10 equiv.) for a further 60 min at 37 °C. Next, the cells were washed three times with PBS, then imaged with the Zeiss LSM880 NLO (2 + 1 with BIG) confocal microscope system (Fig. 2 and 3). Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342 (250 nM, Ex = 405 nm, PMT range = 420–460 nm) and CM orange (100 nM, Ex = 543 nm, PMT range = 550–650 nm) were used as nuclear and membrane trackers, respectively. Meanwhile, another identical set of samples was treated as described above, but without UV irradiation, as the negative control. Background signals of all images were verified to be nearly zero by imaging the same cells treated with a negative control.
342 (250 nM, Ex = 405 nm, PMT range = 420–460 nm) and CM orange (100 nM, Ex = 543 nm, PMT range = 550–650 nm) were used as nuclear and membrane trackers, respectively. Meanwhile, another identical set of samples was treated as described above, but without UV irradiation, as the negative control. Background signals of all images were verified to be nearly zero by imaging the same cells treated with a negative control.
Results and discussion
Synthesis and characterisation
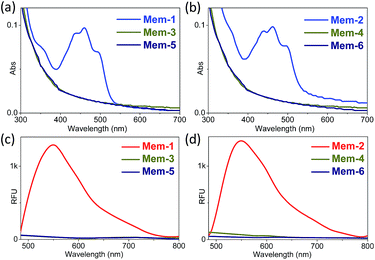
To validate our design, two photocontrollable fluorogenic probes (Mem-5/6) and four additional control probes (Mem-1/2/3/4) were synthesised following modified literature methods,29–31 as summarised in Scheme S1.† Two membrane tracers (Mem-1/2) with different membrane anchors were obtained from the condensation of fluorescein/fluorescein esters (reporter, a dye with high brightness) and cholesteryl chloroformate/bromododecane in ∼50% yield. Fluorescein hydrazide, a copper(II)-responsive unit, was synthesised in one step with an overall yield of 92% from commercially available hydrazine hydrate and fluorescein, followed by coupling with membrane-targeting moieties to generate Mem-3/4, both in 50% yield. Nitrobenzyl-protected fluorescein hydrazide was prepared by the condensation of fluorescein hydrazide and 1-(1-bromoethyl)-2-nitrobenzene in 54% yield in the presence of Cs2CO3. Mem-5/6 (∼60% yields) were prepared using a procedure similar to that described above for Mem-3/4. The compound structures were fully characterised by 1H and 13C NMR, with detailed synthetic procedures and original spectra described in the experimental section and ESI.†To systemically demonstrate the potential utility of our newly developed probes as good fluorogenic systems for live cell imaging, we first evaluated their chemical, photophysical, and biological properties. As shown in Fig. 3a and b, under physiological conditions (HEPES buffer, pH 7.4), Mem-1/2, the membrane tracer references of Mem-5/6, had maximum absorptions at 460 nm, with two shoulder peaks at 440 and 460 nm, and an emission at 550 nm (Fig. 3c and d, Φ = ∼0.15 with fluorescein in 0.1 M NaOH as the reference). Mem-1/2/3/4 showed nearly no absorption or emission (Φ = 0.001–0.0015) at around the same wavelength, based on the closed-loop structure. A similar outcome was observed in organic solvent (ethanol, Fig. S1–S4†). Furthermore, two different membrane anchors had no effect on the photophysical properties of the probes, and the caged probes had very low fluorescence backgrounds or high S/N ratios for live cell imaging.
Subsequently, the cytotoxicity assays showed no obvious cell toxicity at concentrations up to 50 μM for Mem-1/3/5 and 20 μM for Mem-2/4/6, in human hepatocellular carcinoma (HepG2) cells (Fig. 4a). The results showed that Mem-1/3/5 with a cholesterol unit were more suitable for the living cell assay than the group a long aliphatic chain at high concentrations. Theoretically, a probe/plasma membrane lipid ratio must be lower than 1/100 for there to be no significant perturbation of the biomembrane properties in cell membrane staining.32 Therefore, both Mem-1/3/5 and Mem-2/4/6 were suitable for living cell imaging at low working concentrations, because we incorporated a high brightness reporter in the probe design. Therefore, to establish that Mem-1/2 was indeed a good membrane tracer for live cell imaging unequivocally, HepG2 cells were treated with Mem-1/2, followed by one-photon fluorescence imaging (background control shown in Fig. S5†). We observed fluorescent staining exclusively within the plasma membrane of the cells for Mem-1 (Fig. 4b(2/3), 1 μM treatment for 20 min). Otherwise, Mem-2, the probe with a 12-carbon aliphatic chain, stained whole cells without specific localisation on the plasma membrane (Fig. 4b(7/8)). As shown in Fig. 4b(5), the green fluorescence signal of Mem-1 overlapped well with the red fluorescence signal of the commercially available membrane tracker (CM-orange) with a high contrast ratio. Prolonged incubation of the cells with Mem-1 did not lead to the staining of other lipid-like environments in intracellular organelles, indicating minimal probe internalisation, and confirming Mem-1 as cell-membrane specific.
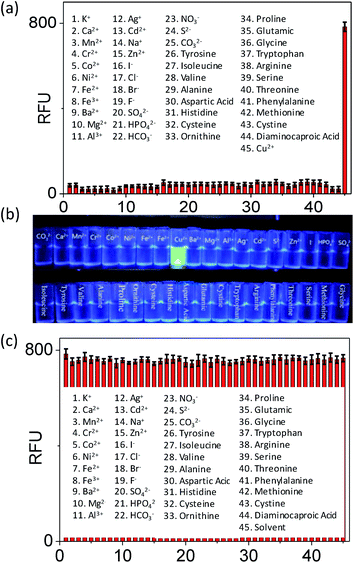
We next investigated whether Mem-5 serves as an effective photocontrollable fluorogenic probe for visualising near-membrane copper(II) in live cells. Upon photolysis with UV irradiation (500 μJ cm−2 for 5 min), non-fluorescent Mem-5 generated non-fluorescent uncaged Mem-5, according to a literature method.24 To evaluate the selectivity of Mem-5 toward copper(II) among a series of metal cations, amino acids and anions, 45 analytes (10 eq.), including K+, Ca2+, Mn2+, Cr2+, Co2+, Ni2+, Fe2+, Fe3+, Ba2+, Mg2+, Al3+, Ag+, Cd2+, Na+, Cu2+, tyrosine, 2,6-diaminocaproic acid, isoleucine, valine, alanine, aspartic acid, histidine, cysteine, ornithine, proline, glutamic acid, glycine, tryptophan, arginine, serine, threonine, phenylalanine, methionine, cystine, CO32−, I−, Cl−, Br−, F−, SO42−, HPO42−, HCO3−, S2−, and NO3−, were incubated with uncaged Mem-5 in HEPES buffer at 37 °C for 1 h, as shown in Fig. 5. A significant fluorescence enhancement in Mem-5 was observed when copper(II) was added only, while other species had a negligible fluorescent intensity (Fig. 5a) with little interference (Fig. 5c). These results illustrate that Mem-5 had a high selectivity for copper(II) recognition.
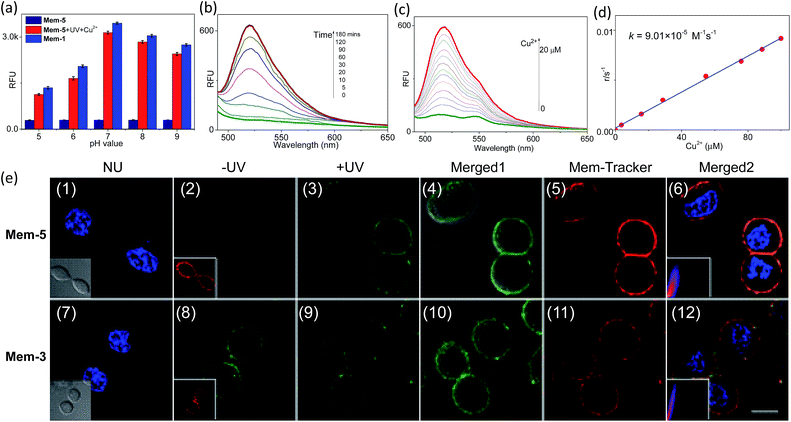
As the pH of the cell cytosol is between 6.8 and 7.4,33 we attempted to determine whether the probe could emit fluorescence and be stable under these conditions. Therefore, the effect of pH on the fluorescence response of Mem-5 to copper(II) was tested in a pH titration experiment. Mem-5 was found to be stable in a wide pH range of 5–9, and showed a better response to copper(II) in the region pH 6–9 (Fig. 6a) after photolysis. When copper(II) (10 equiv.) was added to uncaged Mem-5 (2 μM) in HEPES buffer, the fluorescence intensity increased gradually with time (Fig. 6b). The 1H-NMR titration experiments of Mem-3 with different equivalents of copper(II) indicated that fluorescein hydrazide was hydrolysed by copper(II) under these conditions (Fig. S6†). Next, the introduction of an increasing copper(II) concentration into the solution of uncaged Mem-5 (2 μM) induced a gradual increase in the emission at around 525 nm (Fig. 6c). The slopes of the plots had a pseudo-first-order rate constant. A plot of the rate constants vs. copper(II) concentrations was a straight line passing through the origin, indicating that the reaction is second-order overall, with k = 9.01 × 10−5 M−1 s−1 (Fig. 6d).34 Fluorescence intensities also exhibited a linear relationship with the copper(II) concentration, as shown in Fig. S7† (R2 = 0.97). A linear regression curve was fitted to these normalised fluorescent intensity data, and the point at which the regression line crossed the axis was considered the detection limit (3.3 × 10−7 M).
Finally, encouraged by these results, we determined whether Mem-5 worked as a specific probe for imaging near-membrane copper(II). As shown in Fig. 6e, HepG2 cells were incubated with Mem-3/5 in the presence of copper(II). Cells treated with Mem-5 without UV irradiation showed no fluorescence, indicating temporal control of the “caging” strategy (Fig. 6e(2)). However, significant fluorescent enhancement of Mem-3-treated cells was observed using the same procedure (Fig. 6e(8)). Upon UV irradiation, a gradual increase in green fluorescence was observed both in Mem-5 (Fig. 6e(3)) and Mem-3-treated cells (Fig. 6e(9)). The same cells were simultaneously treated with commercially available Mem-tracker to independently verify the localisation results. Both Mem-5 and Mem-3-treated cells showed similar membrane-specific images by comparing the colocalisation analysis in Fig. 6e(6) and (12), indicating the strategy was amenable to the original design principle. Therefore, our photocontrollable method was successfully used to qualitatively detect near-membrane copper(II).
Conclusions
In this work, we have described the designed photocontrollable fluorogenic probe, Mem-5, which can visualise near-membrane copper(II) via fluorescence imaging, and is equipped with a photolabile group, high brightness reporter, and membrane-anchoring unit (cholesterol or long aliphatic chain). This probe shows an intense fluorescence enhancement in response to copper(II) without interference from 45 other analytes, including metal cations, amino acids and anions, under biological conditions. Live-cell imaging results indicate that the probe can detect near-membrane copper(II) after membrane anchoring using a photo-labile spatial and temporal control releasing method. The probe could be very useful for monitoring the homeostasis of near-membrane copper(II). A clear advantage of our photocontrollable method is that it successfully avoids the influence of chemical species outside cells during near-membrane specific detection. An important unresolved issue from this study is the lack of sensitivity for recording copper(II) across the membrane.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (81672508, 61505076), Natural Science Foundation of Jiangsu Province (BK20140951), Natural Science Foundation of Zhejiang Province (LQ16B020003), and Key University Science Research Project of Jiangsu Province (Grant 16KJA180004).Notes and references
- S. J. Singer and G. I. Nicolson, Science, 1972, 175, 720–731 CAS.
- H. Lodish, A. Berk, S. L. Zipursky, P. Matsudaira, D. Baltimore and J. Darnell, Molecular Cell Biology, Scientific American Books, New York, 4th edn, 2004 Search PubMed.
- L. Wu, E. Hamid, W. Shin and H. Chiang, Annu. Rev. Physiol., 2014, 76, 301–331 CrossRef CAS PubMed.
- G. Meer, D. R. Voelker and G. W. Feigenson, Nat. Rev. Mol. Cell Biol., 2008, 9, 112–124 CrossRef PubMed.
- Z. Darwich, A. S. Klymchenko, D. Dujardin and Y. Mély, RSC Adv., 2014, 4, 8481–8488 RSC.
- D. Lingwood and K. Simons, Science, 2010, 327, 46–50 CrossRef CAS PubMed.
- E. Sezgin, I. Levental, S. Mayor and C. Eggeling, Nat. Rev. Mol. Cell Biol., 2017, 18, 361–374 CrossRef CAS PubMed.
- J. L. Pincus, C. Y. Jin, W. J. Huang, H. K. Jacobs, A. S. Gopalan, Y. J. Song, J. A. Shelnutta and D. Y. Sasaki, J. Mater. Chem., 2005, 15, 2938–2945 RSC.
- L. Li, X. Q. Shen, Q.-H. Xu and S. Q. Yao, Angew. Chem., Int. Ed., 2013, 52, 424–428 CrossRef CAS PubMed.
- Y. Xia and L. Peng, Chem. Rev., 2013, 113, 7880–7929 CrossRef CAS PubMed.
- Z. Liu, T. P. Yang, X. Li, T. Peng, H. C. Hang and X. D. Li, Angew. Chem., Int. Ed., 2015, 54, 1149–1152 CrossRef CAS PubMed.
- T. Peng and H. C. Hang, J. Am. Chem. Soc., 2015, 137, 556–559 CrossRef CAS PubMed.
- M. D. Molin, Q. Verolet, A. Colom, R. Letrun, E. Derivery, M. Gonzalez-Gaitan, E. Vauthey, A. Roux, N. Sakai and S. Matile, J. Am. Chem. Soc., 2015, 137, 568–571 CrossRef PubMed.
- I. A. Karpenko, M. Collot, L. Richert, C. Valencia, P. Villa, Y. Mély, M. Hibert, D. Bonnet and A. S. Klymchenko, J. Am. Chem. Soc., 2015, 137, 405–412 CrossRef CAS PubMed.
- R. J. Radford, W. Chyan and S. J. Lippard, Chem. Sci., 2013, 4, 3080–3084 RSC.
- D. Strausak, J. F. B. Mercer, H. H. Dieter, W. Stremmel and G. Multhaup, Brain Res. Bull., 2001, 55, 175–185 CrossRef CAS PubMed.
- A. Robert, Y. Liu, M. Nguyen and B. Meunier, Acc. Chem. Res., 2015, 48, 1332–1339 CrossRef CAS PubMed.
- O. Bandmann, K. H. Weiss and S. G. Kaler, Lancet Neurol., 2015, 14, 103–113 CrossRef CAS PubMed.
- B. E. Kim, T. Nevitt and D. J. Thiele, Nat. Chem. Biol., 2008, 4, 176–185 CrossRef CAS PubMed.
- B. J. McCranor, H. Szmacinski, H. H. Zeng, A. K. Stoddard, T. Hurst, C. A. Fierke, J. R. Lakowicz and R. B. Thompson, Metallomics, 2014, 6, 1034–1042 RSC.
- M. R. Krause and S. L. Regen, Acc. Chem. Res., 2014, 47, 3512–3521 CrossRef CAS PubMed.
- E. Ikonen, Nat. Rev. Mol. Cell Biol., 2008, 9, 125–138 CrossRef CAS PubMed.
- C. S. Lim, M. Y. Kang, J. H. Han, I. A. Danish and B. R. Cho, Chem.–Asian J., 2011, 6, 2028–2033 CrossRef CAS PubMed.
- L. Li, J. Y. Ge, H. Wu, Q.-H. Xu and S. Q. Yao, J. Am. Chem. Soc., 2012, 134, 12153–12160 Search PubMed.
- L. Yuan, W. Y. Lin, Z. M. Cao, L. L. Long and J. Z. Song, Chem.–Eur. J., 2011, 17, 689–696 CrossRef CAS PubMed.
- Y. R. Zhao, Q. Zheng, K. Dakin, K. Xu, M. L. Martinez and W. H. Li, J. Am. Chem. Soc., 2004, 126, 4653–4663 CrossRef CAS PubMed.
- I. Aparici-Espert, M. C. Cuquerella, C. Paris, V. Lhiaubet-Vallet and M. A. Miranda, Chem. Commun., 2016, 52, 14215–14218 RSC.
- C. Wu, Q. N. Bian, B.-G. Zhang, X. Cai, S.-D. Zhang, H. Zheng, S.-Y. Yang and Y.-B. Jiang, Org. Lett., 2012, 14, 4198–4201 CrossRef CAS PubMed.
- V. Dujols, F. Ford and A. W. Czarnik, J. Am. Chem. Soc., 1997, 119, 7386–7387 CrossRef CAS.
- R. L. Zhang, J. Zhao, G. M. Han, Z. J. Liu, C. Liu, C. Zhang, B. H. Liu, C. L. Jiang, R. Y. Liu, T. T. Zhao, M. Y. Han and Z. P. Zhang, J. Am. Chem. Soc., 2016, 138, 3769–3778 CrossRef CAS PubMed.
- J. F. Hulvat, M. Sofos, K. Tajima and S. I. Stupp, J. Am. Chem. Soc., 2005, 127, 366–372 CrossRef CAS PubMed.
- M. Collot, R. Kreder, A. L. Tatarets, L. D. Patsenker, Y. Melya and A. S. Klymchenko, Chem. Commun., 2015, 51, 17136–17139 RSC.
- J. Llopis, J. Michael McCaffery, A. Miyawaki, M. G. Farquhar and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6803–6808 CrossRef CAS.
- J. H. Lee, C. S. Lim, Y. S. Tian, J. H. Han and B. R. Cho, J. Am. Chem. Soc., 2010, 132, 1216–1217 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis and characterisation, photophysical spectra and background control images. See DOI: 10.1039/c7ra03559d |
| This journal is © The Royal Society of Chemistry 2017 |