Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enhancing menaquinone-7 production in recombinant Bacillus amyloliquefaciens by metabolic pathway engineering†
Jian-Zhong Xu *ab,
Wei-Liu Yanb and
Wei-Guo Zhangb
*ab,
Wei-Liu Yanb and
Wei-Guo Zhangb
aThe Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, 1800# Lihu Road, Wuxi 214122, People’s Republic of China. E-mail: xujianzhong@jiangnan.edu.cn; Fax: +86-510-85329312; Tel: +86-510-85329312
bThe Key Laboratory of Industrial Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, 1800# Lihu Road, Wuxi 214122, People’s Republic of China
First published on 30th May 2017
Abstract
Here, we compared the amino acid sequence of the head structure biosynthetic enzymes in the menaquinone-7 (MK-7) biosynthetic pathway between Bacillus amyloliquefaciens Y-2 and B. amyloliquefaciens W-21 that are distinct in MK-7 production, and investigated the effect of these enzymes on MK-7 production. Sequence analysis showed that six enzymes had undergone non-synonymous substitutions: MenA, MenC, MenD, MenE, MenH and HepS. Overexpression of these enzymes from strain Y-2 in B. subtilis 168 significantly increased the corresponding enzyme activity (increased by ≥500%), which was higher than that from overexpressing these enzymes from strain W-21 (increased by ≈200%). Moreover, the MK-7 content in B. subtilis 168 or B. amyloliquefaciens Y-2 was enhanced by the overexpression of these enzymes from strain Y-2. Note that the overexpression of MenA in B. subtilis 168 increased the MK-7 content up to 1.6-fold, whereas the overexpression of HepS in B. amyloliquefaciens Y-2 led to a greater increase in MK-7 production than that of other enzymes, both in the stilling culture (increased by 93.62%) and in the shaking culture (increased by 93.29%). It follows that the high enzyme activity and high-traffic biosynthetic pathway are beneficial to improve MK-7 production. These results provide a definite theoretical foundation for breeding MK-7 high-yielding strains via metabolic engineering.
1 Introduction
Menaquinones (MK-n, where n represents the number of isoprene units), a type of vitamin K2, have significant effects on the electron transfer of the respiratory chain.1 In prokaryotes, MK-n plays an important role in shuttling electrons between membranes bound to protein complexes in the electron transport chain.2 For human beings and other mammals, MK-n is an essential vitamin for blood coagulation, bone metabolism and cell-cycle regulation processes.3,4 Menaquinone-7 (MK-7) is part of a family of MK-ns with a side chain of seven isoprene units.1 As recent research shows, MK-7 can activate osteocalcin in bone marrow to promote the formation of bone, and is able to improve the bone mineral density (BMD) in the aged so as to reduce the risk of haunch bone fracture.5 In addition, it can also lower blood pressure in rats.6 Therefore, MK-7 is receiving increasing attention in the domain of nutritional supplements for humans.Many microorganisms can be used to biosynthesize MK-7, such as Escherichia coli,7 Sphingobacterium multivorum,8 Bacillus amyloliquefaciens9,10 and Bacillus subtilis.11–13 However, commercial production of MK-7 is mainly done through extraction from natto, a type of food, rather than via microbial fermentation.14 This is because the fermentative production of MK-7 is too low.9,11–13 Fortunately, the biosynthetic pathway of MK-7 has been clarified at present (Fig. 1). As can be seen in Fig. 1, there are nine genes, designated as menA, menB, menC, menD, menE, menH, menG, hepS and hepT, encoding the head structure biosynthesis enzymes that are involved in the MK-7 biosynthetic pathway starting from isochorismate. However, some enzymes in the biosynthetic pathway are feedback-regulated by intermediates. For example, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase is feedback-inhibited by 1,4-dihydroxy-2-naphthoate (DHNA), L-tyrosine, L-phenylalanine and L-tryptophan.15 To obtain mutants with removing feedback-regulation, the wild-type strains were treated with the conventional mutation-inducing methods, such as ultraviolet (UV) light and N-methyl-N-nitro-N-nitroso-guanidine (NTG).16–18 In addition, Kwon et al.19 pointed out that the coding levels of the enzymes involved in MK-7 biosynthesis are present in extremely low quantities. However, there are few genetic engineering models for developing a microbial producer of MK-7 via increasing the coding level of the enzymes.
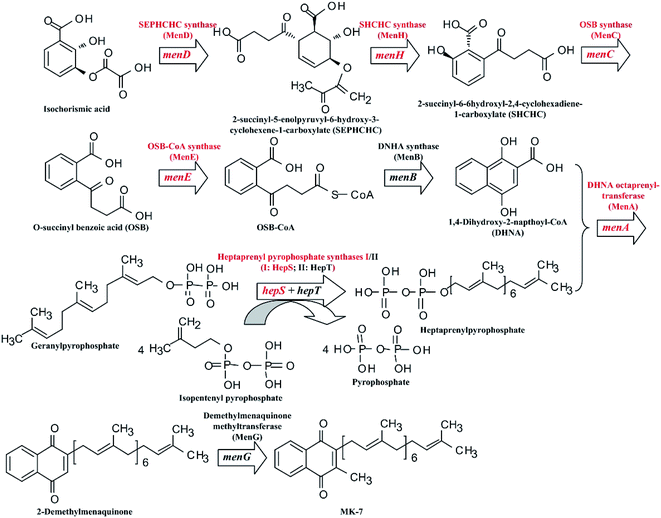 | ||
| Fig. 1 Schematic diagram of the MK-7 biosynthetic pathway in Bacillus sp. The red font represents the modified genes and the corresponding enzymes. | ||
In this study, the endogenous MK-7 biosynthetic pathway was metabolically engineered to construct an efficient B. amyloliquefaciens platform to produce MK-7. After comparing the genetic difference between strains with different MK-7 productivity, six genes in the MK-7 biosynthetic pathway were selected to evaluate the effect on MK-7 production. The study shows the first time that different enzymes in the MK-7 biosynthetic pathway have differing roles in MK-7 production, for example in cell growth as well as affecting the MK-7 yield.
2 Materials and methods
2.1 Strains, growth media and culturing conditions
The strains used in this study are listed in Table 1. B. amyloliquefaciens Y-2 and B. amyloliquefaciens W-21 were isolated from Chinese fermented beans. A phylogenetic tree based on the 16S rDNA sequence homology of strains Y-2 and W-21 is shown in Fig. S1.† Strains Y-2 and W-21 show high fibrinogenase activity (>1450 IU mL−1), but only strain Y-2 can be used to produce MK-7 (7.1 ± 0.5 mg L−1) with maize meal hydrolysate as the carbon source (Table S1†). Luria-Bertani (LB) was used as a standard medium for cultivating all strains. A tryptone–yeast extract–glucose (TYG) medium composed of 5 g L−1 tryptone, 2.5 g L−1 yeast extract and 5 g L−1 glucose was used as the seed medium. A glycerol–yeast extract–soy peptone (GYS) medium was used as the basal medium for MK-7 production.20 The Spizizen minimal (SM) medium or low SM (LSM) medium was used as the basal medium for preparing competent cells or isolating transformants.21,22 All media were adjusted to pH 7.2 ± 0.2 with 20% (w/v) NaOH. When appropriate, E. coli and Bacillus sp. were cultured with kanamycin (Km, 50 μg mL−1) at 100 rpm.| Strain | Relevant characteristic(s) | Source |
|---|---|---|
| E. coli JM109 | recA1 endA1 gyrA96 thi-1 hsdR17 e14−(mcrA−) supE44 relA1 Δ(lac-proAB)/F′ [traD36 proB+ laclq lacZΔM15] | Takara |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| B. subtilis strain | ||
| 168 | trpC2 sfp0 degQ0, IT− SF− PL− | ATCC |
| 168/pMA5 | B. subtilis 168 harboring plasmid pMA5 | This work |
| 168/pMA5-AY-2 | B. subtilis 168 harboring plasmid pMA5-AY-2 | This work |
| 168/pMA5-CY-2 | B. subtilis 168 harboring plasmid pMA5-CY-2 | This work |
| 168/pMA5-DY-2 | B. subtilis 168 harboring plasmid pMA5-DY-2 | This work |
| 168/pMA5-EY-2 | B. subtilis 168 harboring plasmid pMA5-EY-2 | This work |
| 168/pMA5-HY-2 | B. subtilis 168 harboring plasmid pMA5-HY-2 | This work |
| 168/pMA5-SY-2 | B. subtilis 168 harboring plasmid pMA5-SY-2 | This work |
| 168/pMA5-AW-21 | B. subtilis 168 harboring plasmid pMA5-AW-21 | This work |
| 168/pMA5-CW-21 | B. subtilis 168 harboring plasmid pMA5-CW-21 | This work |
| 168/pMA5-DW-21 | B. subtilis 168 harboring plasmid pMA5-DW-21 | This work |
| 168/pMA5-EW-21 | B. subtilis 168 harboring plasmid pMA5-EW-21 | This work |
| 168/pMA5-HW-21 | B. subtilis 168 harboring plasmid pMA5-HW-21 | This work |
| 168/pMA5-SW-21 | B. subtilis 168 harboring plasmid pMA5-SW-21 | This work |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| B. amyloliquefaciens strain | ||
| Y-2 | China Center for Type Culture Collection (CCTCC) M2013493 | 10 |
| W-21 | China Information Center of Industrial Microbial (CICIM) B4503 | Our lab |
| Y-2/pMA5 | B. amyloliquefaciens Y-2 harboring empty plasmid pMA5 | This work |
| Y-2/pMA5-AY-2 | B. amyloliquefaciens Y-2 harboring expression plasmid pMA5-AY-2 | This work |
| Y-2/pMA5-CY-2 | B. amyloliquefaciens Y-2 harboring expression plasmid pMA5-CY-2 | This work |
| Y-2/pMA5-DY-2 | B. amyloliquefaciens Y-2 harboring expression plasmid pMA5-DY-2 | This work |
| Y-2/pMA5-EY-2 | B. amyloliquefaciens Y-2 harboring expression plasmid pMA5-EY-2 | This work |
| Y-2/pMA5-HY-2 | B. amyloliquefaciens Y-2 harboring expression plasmid pMA5-HY-2 | This work |
| Y-2/pMA5-SY-2 | B. amyloliquefaciens Y-2 harboring expression plasmid pMA5-SY-2 | This work |
2.2 DNA manipulations
The plasmids and oligonucleotides used in this study are listed in Tables 2 and S2,† respectively. DNA was extracted from B. amyloliquefaciens Y-2 using the Ezup-pillar Bacterial Genomic DNA Extraction Kit (Sangon, Shanghai, China), and it was used as a template to amplify the tested genes. The tested genes from the strains Y-2 and W-21 were sequenced by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). Plasmid construction and transformation were performed according to the descriptions in previous reports.23 The build processes of the plasmids are illustrated in the ESI (Fig. S2†). The plasmids were extracted from E. coli or Bacillus sp. using the SanPrep Mini Plasmid Kit (Sangon, Shanghai, China) and confirmed by the restriction endonuclease reaction.| Plasmid | Relevant characteristic(s) | Source |
|---|---|---|
| pMA5 | HpaII promoter, E. coli-B. subtilis shuttle vector, Ampr and Kmr | Invitrogen |
| pMA5-AY-2 | pMA5 constitutively expressing the menA gene from strain Y-2 | This work |
| pMA5-CY-2 | pMA5 constitutively expressing the menC gene from strain Y-2 | This work |
| pMA5-DY-2 | pMA5 constitutively expressing the menD gene from strain Y-2 | This work |
| pMA5-EY-2 | pMA5 constitutively expressing the menE gene from strain Y-2 | This work |
| pMA5-HY-2 | pMA5 constitutively expressing the menH gene from strain Y-2 | This work |
| pMA5-SY-2 | pMA5 constitutively expressing the hepS gene from strain Y-2 | This work |
| pMA5-AW-21 | pMA5 constitutively expressing the menA gene from strain W-21 | This work |
| pMA5-CW-21 | pMA5 constitutively expressing the menC gene from strain W-21 | This work |
| pMA5-DW-21 | pMA5 constitutively expressing the menD gene from strain W-21 | This work |
| pMA5-EW-21 | pMA5 constitutively expressing the menE gene from strain W-21 | This work |
| pMA5-HW-21 | pMA5 constitutively expressing the menH gene from strain W-21 | This work |
| pMA5-SW-21 | pMA5 constitutively expressing the hepS gene from strain W-21 | This work |
2.3 Enzyme activity assay
Cell extracts were prepared as reported by Xu et al.24 After centrifugation at 4 °C for 30 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, the cell-free supernatants were immediately used to determine the enzyme activities. The DHNA octaprenyltransferase (MenA) assay was based on the protocol of Suvarna et al.25 and the o-succinylbenzoic acid (OSB) synthase (MenC) activity was determined as described by Palaniappan et al.26 Assays for 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic acid (SEPHCH) synthase (MenD),27 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid (SHCHC) synthase (MenH)26 and DHNA synthase (MenE)19 were performed as previously described. The separation and assay of heptaprenyl pyrophosphate synthetase I (HepS) activity were performed as described by Fujii et al.28 Protein concentrations were determined using a Bradford Protein Quantification Kit (Sangon, Shanghai, China) with bovine serum albumin as the standard.
000 × g, the cell-free supernatants were immediately used to determine the enzyme activities. The DHNA octaprenyltransferase (MenA) assay was based on the protocol of Suvarna et al.25 and the o-succinylbenzoic acid (OSB) synthase (MenC) activity was determined as described by Palaniappan et al.26 Assays for 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic acid (SEPHCH) synthase (MenD),27 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid (SHCHC) synthase (MenH)26 and DHNA synthase (MenE)19 were performed as previously described. The separation and assay of heptaprenyl pyrophosphate synthetase I (HepS) activity were performed as described by Fujii et al.28 Protein concentrations were determined using a Bradford Protein Quantification Kit (Sangon, Shanghai, China) with bovine serum albumin as the standard.
2.4 Construction of Bacillus sp. recombinant strains
Competent cells of B. subtilis 168 were prepared as described by Akamatsu and Taguchi.22 To prepare the competent cells of B. amyloliquefaciens Y-2, 4% (v/v) tween-80 must be added to medium I (SM medium supplemented with 200 mg L−1 casamino acids). After the cells were cultured in medium I for 4 h at 37 °C, 1 mL of culture was added to 9 mL of medium II (SM medium supplemented with 100 mg L−1 casamino acids), and cultivated for 1.5 h at 37 °C. Following this, 200 μL of 100× ethylene glycol tetraacetic acid (EGTA) solution (10 mmol L−1 EGTA, adjusted to pH 8.0 with NaOH) was added to the above-mentioned culture, and cultivated for 10 min at 37 °C and 80 rpm. An appropriate recombinant plasmid was added to 500 mL of competent-cell culture, and the mixture of plasmid and competent cells was incubated at 37 °C for 2 h. The cells were then plated on a LSM agar medium with appropriate Km.2.5 Fermentation for MK-7 production
Batch cultivations of MK-7 producing strains were carried out in shake flasks. Unless otherwise stated, all MK-7 fermentation was carried out in a 250 mL Erlenmeyer flask containing 50 mL of GYS media. All experiments were performed at a temperature of 37 °C with an agitation cycle of 100 rpm or standing incubation. The fermentation process was carried out at 37 °C for 6 days, and cell growth, pH and MK-7 accumulation were monitored over the course of the experiment.2.6 Analytical methods
MK-7 extraction was carried out according to the reports of Berenjian et al.11 using n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-propanol (2
2-propanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) as the extracting agent. In each run, a mixture of the culture supernatant and extracting agent (4
1, v/v) as the extracting agent. In each run, a mixture of the culture supernatant and extracting agent (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) was violently shaken with a vortex mixer for 10 min and then centrifuged at 5000 rpm for 10 min to collect the organic phase. The organic phase was then evaporated under vacuum to recover extracted MK-7. The concentration of MK-7 was detected using a high performance liquid chromatograph (HPLC) equipped with a diode array UV detector (248 nm). Separation was carried out on a Zorbax SB C18 column (250 × 4.6 mm, Agilent, USA) at 40 °C using a methanol–dichloromethane (9
1, v/v) was violently shaken with a vortex mixer for 10 min and then centrifuged at 5000 rpm for 10 min to collect the organic phase. The organic phase was then evaporated under vacuum to recover extracted MK-7. The concentration of MK-7 was detected using a high performance liquid chromatograph (HPLC) equipped with a diode array UV detector (248 nm). Separation was carried out on a Zorbax SB C18 column (250 × 4.6 mm, Agilent, USA) at 40 °C using a methanol–dichloromethane (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) solvent as the mobile phase with a flow rate of 1 mL min−1. The MK-7 calibration curve was set according to the reports of Berenjian et al.11 In addition, cell growth was reflected by the cell density, which was detected using a photometer at 600 nm after an appropriate dilution.
1, v/v) solvent as the mobile phase with a flow rate of 1 mL min−1. The MK-7 calibration curve was set according to the reports of Berenjian et al.11 In addition, cell growth was reflected by the cell density, which was detected using a photometer at 600 nm after an appropriate dilution.
3 Results and discussion
3.1 Nucleotide sequence analysis
According to our previous results,10 the fibrinogenase-producing strains Y-2 and W-21 isolated from Chinese douchi have very different capacities to produce MK-7. Strain Y-2 produces 7.1 ± 0.5 mg L−1 of MK-7 with maize meal hydrolysate as the carbon source, whereas strain W-21 cannot accumulate any MK-7 (≤0.1 mg L−1) (Table S1†). In order to analyze the difference based on the genetic level, the sequence of genes from strains Y-2 and W-21 was obtained by creating primers based on the conserved sequence of the genes (Table S2†). The nucleotide sequences and the deduced amino acid sequences of these tested genes (i.e. menA, menB, menC, menD, menE, menG, menH, hepT and hepS) are presented in Fig. S3.† As can be seen in Fig. S3,† there are different nucleotide sequences of the tested genes between strains Y-2 and W-21. When the results were compared with the corresponding amino acid sequence, only menB, menG and hepT were not different between strains Y-2 and W-21. Among these tested genes, menE and menH had thirteen and eight amino acid variations, respectively, whereas hepS only had one amino acid variation and the others had four amino acid variations (Tables 3 and S3†). Given that the MK-7 productivity is essentially different between strains Y-2 and W-21 (Table S1†), the late-stage study focused on analyzing the effect of different genes between strains Y-2 and W-21 on MK-7 production.| Content | menA | menB | menC | menD | menE | menG | menH | hepT | hepS |
|---|---|---|---|---|---|---|---|---|---|
| Fragment size (bp) | 936 | 819 | 1116 | 1737 | 1464 | 702 | 825 | 963 | 765 |
| Nucleotide sequence similarity (%) | 98.40 | 99.02 | 99.19 | 98.39 | 96.99 | 99.29 | 97.58 | 98.65 | 98.95 |
| Nucleotide variations quantum | 15 | 8 | 9 | 27 | 44 | 5 | 20 | 13 | 7 |
| Amino acid sequence similarity (%) | 98.71 | 100.00 | 98.92 | 99.31 | 97.54 | 100.00 | 97.08 | 100.00 | 99.61 |
| Amino acid variations quantum | 4 | 0 | 4 | 4 | 12 | 0 | 8 | 0 | 1 |
3.2 Analysis of enzymes activity
According to the above-mentioned results, there are six key enzymes (i.e. encoded by menA, menC, menD, menE, menH and hepS) with amino acid variations between strains Y-2 and W-21 (Table 3). However, previous research reported that gene allelic variation will affect the activity of the corresponding enzyme.29,30 In order to investigate whether these amino acid variations will lead to a difference in enzyme activity, the recombinant plasmids with the above genes from strain Y-2 or W-21 were constructed, and electro-transformed into B. subtilis 168 competent cells. Consistent with the expected molecular mass of the corresponding enzyme, the thick protein band that appeared when SDS-PAGE was performed was observed in the cell-free extracts from strains carrying the recombinant plasmid (data not shown). In addition, the specific activities of these six key enzymes are markedly increased by overexpression of the related genes in B. subtilis 168 (Table 4). These results are in agreement with the previous results.19,25,31 It should be noted that overexpression of the tested genes from strain Y-2 induced more than a five-fold increase in the related enzyme activity, which was higher than that of overexpressing genes from strain W-21. However, there was no obvious difference in the activity of MenH during the overexpression of menH from strains Y-2 and W-21 (Table 4). Among the six genes, the overexpression of menA and menE induced much difference between strains Y-2 and W-21: 3.54 ± 0.15 nmol per h per mg protein vs. 1.01 ± 0.23 nmol per h per mg protein, and 294.3 ± 21.5 nmol per h per mg protein vs. 103.8 ± 15.7 nmol per h per mg protein, respectively. These results may be used to determine the reason why only strain Y-2 can be used to produce MK-7,10 and indicated that the overexpression of genes from strain Y-2 is beneficial to increase the related enzyme activity involved in MK-7 production.| B. subtilis strains | Specific activitya | |||||
|---|---|---|---|---|---|---|
| MenA | MenC | MenD | MenE | MenH | HepS | |
| a The unit of specific activity is nanomoles per hour per milligram of protein. | ||||||
| 168 | 0.62 ± 0.21 | 5.3 ± 1.5 | 8.6 ± 2.2 | 47.7 ± 5.6 | 34.6 ± 5.4 | 0.27 ± 0.12 |
| 168/pMA5-AY-2 | 3.54 ± 0.15 | 5.9 ± 0.7 | 8.9 ± 1.4 | 55.3 ± 6.4 | 36.3 ± 1.7 | 0.46 ± 0.21 |
| 168/pMA5-AW-21 | 1.01 ± 0.23 | 5.4 ± 2.0 | 8.5 ± 2.3 | 49.1 ± 7.3 | 35.5 ± 3.1 | 0.35 ± 0.15 |
| 168/pMA5-CY-2 | 0.83 ± 0.16 | 27.7 ± 3.4 | 9.9 ± 1.1 | 60.2 ± 6.9 | 40.1 ± 4.3 | 0.38 ± 0.13 |
| 168/pMA5-CW-21 | 0.65 ± 0.24 | 11.4 ± 2.1 | 8.7 ± 1.5 | 51.3 ± 7.1 | 37.3 ± 7.4 | 0.31 ± 0.05 |
| 168/pMA5-DY-2 | 0.67 ± 0.13 | 6.1 ± 1.2 | 41.1 ± 2.9 | 51.5 ± 4.3 | 46.2 ± 5.9 | 0.35 ± 0.11 |
| 168/pMA5-DW-21 | 0.61 ± 0.27 | 5.6 ± 1.6 | 20.3 ± 3.2 | 48.3 ± 5.2 | 40.4 ± 4.7 | 0.28 ± 0.03 |
| 168/pMA5-EY-2 | 0.90 ± 0.25 | 6.3 ± 1.1 | 8.9 ± 0.8 | 294.3 ± 21.5 | 38.2 ± 2.6 | 0.39 ± 0.08 |
| 168/pMA5-EW-21 | 0.71 ± 0.21 | 5.5 ± 0.5 | 8.7 ± 1.3 | 103.8 ± 15.7 | 36.1 ± 3.5 | 0.31 ± 0.04 |
| 168/pMA5-HY-2 | 0.78 ± 0.15 | 6.7 ± 1.7 | 10.3 ± 2.0 | 57.7 ± 10.3 | 181.3 ± 9.3 | 0.38 ± 0.13 |
| 168/pMA5-HW-21 | 0.65 ± 0.21 | 5.6 ± 1.5 | 9.1 ± 1.7 | 49.3 ± 6.9 | 109.6 ± 8.5 | 0.29 ± 0.07 |
| 168/pMA5-SY-2 | 1.06 ± 0.29 | 5.5 ± 1.4 | 8.7 ± 1.4 | 49.0 ± 5.5 | 35.7 ± 1.4 | 1.86 ± 0.39 |
| 168/pMA5-SW-21 | 0.69 ± 0.24 | 5.3 ± 1.9 | 8.4 ± 2.1 | 48.3 ± 6.7 | 35.1 ± 5.2 | 1.07 ± 0.14 |
All data are mean values of three determinations of three independent experiments with errors (±SD).
3.3 Growth performance and MK-7 production of genetically defined B. subtilis strains
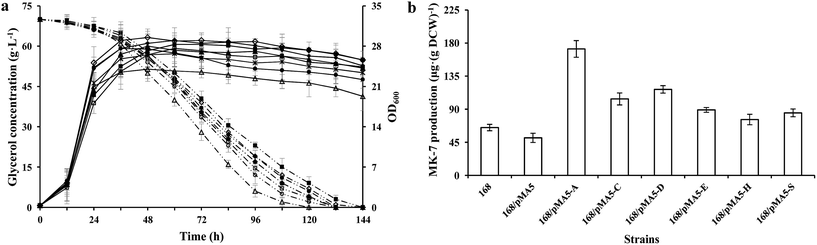
To test the influences of overexpression of these six genes from strain Y-2 in B. subtilis for growth performance and MK-7 production, we analyzed the cell growth (OD600), glycerol metabolism and MK-7 production of these recombinant strains, and compared them with those of B. subtilis 168. There were no obvious differences in cell growth among these tested strains except for strain 168/pMA5-AY-2 (Fig. 2a), which is consistent with the previous result.7 The strain 168/pMA5-AY-2 showed poor cell growth (maximum OD600 = 24 ± 4.1 g L−1 at 32 h) during overexpression of the menA gene from strain Y-2. Although Shaw et al.32 have reported that a relatively low level of MK-n biosynthesis enzyme activity is sufficient to provide adequate levels of MK-n for cell growth, in fact, the specific glycerol uptake rate of strain 168/pMA5-AY-2 was increased (Fig. 2a). The reduced OD600 value was a consequence of the increased MK-7 production during menA-overexpression (increased by 164.62%, Fig. 2b). Debnath et al.33 pointed out that MenA is one of the key enzymes in MK-n biosynthesis, which is catalyzed by the conversion of the soluble bicyclic naphthalenoid compound, DHNA, to membrane-bound 2-demethylmenaquinone (Fig. 1). In addition, Suvarna et al.25 also pointed out that the inactivation of MenA in E. coli led to the cut off of MK-8 production, whereas increasing the enzyme activity of MenA will improve the MK-8 synthetic capacity of the strains. Thus, the overexpression of MenA did not influence the uptake of glycerol into the cells, instead influencing the intracellular distribution of glycerol, with a shift from biomass toward MK-7 production. In addition, the increasing production of MK-7 was also significant for the overexpression of menC (i.e. strain 168/pMA5-CY-2, increased by 60%) and menD (i.e. strain 168/pMA5-DY-2, increased by 80%) (Fig. 2b). Compared with strain 168, however, the MK-7 production decreased during the introduction of empty plasmid pMA-5 into B. subtilis 168 cells (Fig. 2b). Many studies reported that although some microorganisms (e.g. E. coli and B. subtilis) possess the MK-7 biosynthesis enzyme-coding genes, the native enzymes are present at extremely low levels.31,32 Moreover, MenD catalyzes the first committed step of the classical MK-n biosynthetic pathway,34 and it is the essential enzyme for producing MK-n in B. subtilis.35 Therefore, the significant effect of MenD is reasonable. Interestingly, overexpression of MenE did not significantly increase MK-7 production (i.e. strain 168/pMA5-EY-2, increased by 36.92%), even though the activity of MenE was greatly increased during overexpression of menE compared to that of strain 168 (Fig. 2b and Table 4). This may be because the activity of enzymes downstream of the OSB-CoA pool is too low to provide enough carbon flux for MK-7 production, for example, the activity of MenA.363.4 Growth performance of genetically defined B. amyloliquefaciens strains
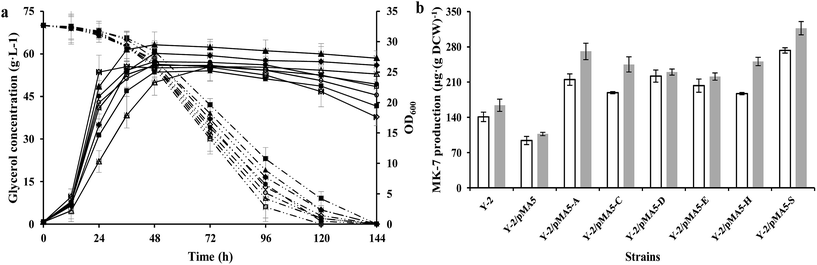
According to our previous studies, B. amyloliquefaciens Y-2 can be used to produce MK-7 (7.1 ± 0.5 mg L−1) with maize meal hydrolysate as the carbon source (Table S1†10). Therefore strain Y-2 has the potential to produce MK-7 with maize meal hydrolysate as the carbon source in an industrial process, and is worthy of further work in genetic breeding. In order to investigate whether the overexpression of six structure biosynthesis enzymes will cause any adverse effect on growth performance, the DCW and glycerol consumption of these recombinant strains were investigated and compared with the parental strain Y-2 during cultivation at a constant temperature and humidity in a shaker. As can be seen in Fig. 3a, the cell growth rate was slightly retarded during the introduction of the pMA-5 or recombinant plasmid to the tested gene, whereas overexpression of these six genes led to an increase in biomass yield, and could prevent cell lysis to some extent (Fig. 3a). This is notably different from the reports of Kong and Lee,7 who reported that overexpression of the head structure biosynthesis enzymes and the tail structure biosynthesis enzymes in E. coli slightly affected the cell growth compared with the control, E. coli JM109. There is little research to investigate the effect of enzymes involved in MK-n production on cell growth and MK-n production, thus it is hard to clarify what reason led to this phenomenon in the field of MK-n production. In previous studies done with Corynebacterium glutamicum, increasing the carbon flux into the amino acid biosynthetic pathway (e.g. L-valine and L-leucine) would retard the cell growth due to the redirection of carbon from cell growth toward amino acid production.37,38 With this in mind, we conclude that the decreased cell growth is the result of increasing the MK-7 production. This assumption is fully backed by research into the MK-7 production of genetically defined B. amyloliquefaciens strains in different cultivating methods (Fig. 3b).In addition, we found that the glycerol consumption rate did not correspond to the cell growth rate, which proved our assumption once again (Table 5). Strain Y-2/pMA5-CY-2 showed the bigger cell growth rate and biomass yield, but its glycerol consumption rate (qs = 28.6 ± 1.3 mg OD600−1 h−1) was lower than that of the other strains except for strain Y-2/pMA5 (qs = 25.7 ± 1.9 mg OD600−1 h−1). Among these strains, strain Y-2/pMA5-SY-2 had the highest glycerol consumption rate (qs = 42.1 ± 2.3 mg OD600−1 h−1), followed by strains Y-2/pMA5-AY-2 (qs = 41.4 ± 3.5 mg OD600−1 h−1) and Y-2/pMA5-DY-2 (qs = 39.0 ± 2.4 mg OD600−1 h−1). It should be noted that strain Y-2 showed a high glycerol consumption rate in the initial stage of fermentation, and then rapidly decreased because of fast cell lysis.
| B. amyloliquefaciens strains | Final OD600 | μmaxa (h−1) | qs, max (mg OD600−1 h−1) | MK-7 yield (μg (g DCW)−1) |
|---|---|---|---|---|
| a ΔOD600 per units of bacteria per hour. | ||||
| Y-2 | 17.6 ± 2.6 | 1.71 ± 0.19 | 31.4 ± 2.2 | 141 ± 9.3 |
| Y-2/pMA5 | 19.4 ± 1.7 | 0.95 ± 0.16 | 25.7 ± 1.9 | 94 ± 8.2 |
| Y-2/pMA5-AY-2 | 24.7 ± 1.2 | 0.68 ± 0.23 | 41.4 ± 3.5 | 215 ± 11.3 |
| Y-2/pMA5-CY-2 | 27.3 ± 2.1 | 1.58 ± 0.25 | 28.6 ± 1.3 | 189 ± 2.1 |
| Y-2/pMA5-DY-2 | 21.2 ± 1.4 | 1.37 ± 0.17 | 39.0 ± 2.4 | 222 ± 12.5 |
| Y-2/pMA5-EY-2 | 23 ± 1.8 | 1.45 ± 0.13 | 34.9 ± 1.4 | 203 ± 13.3 |
| Y-2/pMA5-HY-2 | 26.1 ± 1.5 | 1.11 ± 0.21 | 32.3 ± 3.0 | 187 ± 2.6 |
| Y-2/pMA5-SY-2 | 22.6 ± 1.3 | 1.31 ± 0.24 | 42.1 ± 2.3 | 273 ± 5.4 |
All data are mean values of three determinations of three independent experiments with errors (±SD).
3.5 MK-7 production of genetically defined B. amyloliquefaciens strains with different cultivating methods
To investigate the effect of overexpressing six structure biosynthesis enzymes on MK-7 production, the parental strain Y-2 and derived strains (i.e. Y-2/pMA5, Y-2/pMA5-AY-2, Y-2/pMA5-CY-2, Y-2/pMA5-DY-2, Y-2/pMA5-EY-2, Y-2/pMA5-HY-2 and Y-2/pMA5-SY-2) were cultivated in GYS media as described in “Materials and methods”. Moreover, the different cultivating methods (i.e. stilling culture and shaking culture) were also investigated. Consistent with the overexpression of six structure biosynthesis enzymes in B. subtilis 168 and the previous results,7 the overexpression of six key enzymes in strain Y-2 increased the MK-7 production, either in shaking culture or in stilling culture (Fig. 3b). However, MK-7 production decreased during the introduction of pMA-5 into B. amyloliquefaciens Y-2 cells, which is in line with the introduction of pMA-5 into B. subtilis 168 cells (Fig. 2b and 3b). As can be seen from Fig. 3b, the MK-7 production in the stilling culture is much higher than that in the shaking culture. Compared with the MK-7 production of parental strain Y-2 (141 ± 9.3 μg (g DCW)−1 in the shaking culture and 164 ± 12.1 μg (g DCW)−1 in the stilling culture), the highest increase was observed for strain Y-2/pMA5-SY-2, both in the stilling culture (increased by 93.62%) and in the shaking culture (increased by 93.29%) (Fig. 3b). This result is unlike that from the overexpression of the structure biosynthesis enzymes in B. subtilis 168, in which the overexpression of menA plays the most important role in increasing MK-7 production (Fig. 2b and 3b). Zhang et al.31 pointed out that heptaprenyl diphosphate synthase, including component I (i.e. HepS) and component II (i.e. HepT), is an essential enzyme for catalyzing the biosynthesis of the side chain of MK-7. It may be that the activity of HepS in B. amyloliquefaciens Y-2 is too low to support much MK-7 production, whereas the overexpression of HepS solves the problem of the low activity of HepS and then increases the activity of heptaprenyl diphosphate synthase. It is worth pointing out that the same enzyme has a different effect on MK-7 production during different cultivating methods (Fig. 3b). Culture conditions affect the gene expression and the viability of the protein,39,40 and this may be why the same enzyme has different effects on MK-7 production.4 Conclusions
MK-7 is part of a family of MK-ns with a side chain of seven isoprene units, and has unique physiological functions, such as promoting the formation of bone and reducing blood pressure.1,5,6 In the present study, we revealed the difference in the head structure biosynthesis enzymes involved in the MK-7 biosynthetic pathway starting from isochorismate based on the genetic level between strains Y-2 and W-21 that are distinct in MK-7 production, and demonstrate the importance of six enzymes with amino acid variations for increasing MK-7 production. Moreover, the present work points out that the same enzyme has different effects on MK-7 production for different cultivating methods. On the basis of these results and the previous ones,7,10,15–17 we reveal that strategies which lead to an increased flux of the chorismate precursor pathway and the isoprenoid pathway could increase MK-7 production or the production of other MK-ns. The results provide a good example of how genetic engineering can be combined with an effective way to rationally modify the production of strains.Abbreviations
| MK-n | Menaquinones |
| DHNA | 1,4-Dihydroxy-2-naphthoate |
| DAHP | 3-Deoxy-D-arabino-heptulosonate-7-phosphate |
| DHNA | 1,4-Dihydroxy-2-naphthoate |
| OSB | o-Succinylbenzoic acid |
| SEPHCH | 2-Succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic acid |
| SHCHC | 2-Succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid |
| EGTA | Ethylene glycol tetraacetic acid |
| LB | Luria-Bertani medium |
| TYG | Tryptone–yeast extract–glucose medium |
| GYS | Glycerol–yeast extract–soy peptone medium |
| SM | Spizizen minimal medium |
| DCW | Dry cell weight |
| CCTCC | China Center for Type Culture Collection |
| CICIM | China Information Center of Industrial Microbiol |
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 31601459), Natural Science Foundation of Jiangsu Province (No. BK20150149), China Postdoctoral Science Foundation Grant (No. 2016M590410), the 111 project (No. 111-2-06) and Fundamental Research Funds for the Central Universities (No. JUSRP115A19).References
- B. Walther, J. P. Karl, S. L. Booth and P. Boyaval, Adv. Nutr., 2013, 4, 463–473 CrossRef CAS PubMed.
- T. Darri, J. Antibiot., 2009, 62, 347–352 CrossRef PubMed.
- D. W. Lamson and S. M. Plaza, Altern. Med. Rev., 2003, 8, 303–318 Search PubMed.
- E. C. Cranenburg, L. J. Schurger and C. Vermeer, Thromb. Haemostasis, 2007, 98, 120–125 CAS.
- M. H. J. Knapen, N. E. Drummen, E. Smit, C. Vermeer and E. Theuwissen, Osteoporosis Int., 2013, 24, 2499–2507 CrossRef CAS PubMed.
- Y. K. Kim, S. M. Kim, J. Y. Kim and O. Kwon, J. Korean Soc. Appl. Biol. Chem., 2011, 54, 959–965 CrossRef CAS.
- M. K. Kong and P. C. Lee, Biotechnol. Bioeng., 2011, 108, 1997–2002 CrossRef CAS PubMed.
- S. Rosa-Putra, A. Hemmerlin, J. Epperson, T. J. Bach, L. H. Guerra and M. Rohmera, FEMS Microbiol. Lett., 2001, 204, 347–353 CrossRef CAS PubMed.
- W. J. Wu and B. Y. Ahn, J. Korean Soc. Appl. Biol. Chem., 2011, 54, 783–789 CrossRef CAS.
- J. Z. Xu and W. G. Zhang, J. Zhejiang Univ., Sci., B, 2017 DOI:10.1631/jzus.B1600127.
- A. Berenjian, R. Mahanama, A. Talbot, R. Biffin, H. Regtop and P. Valtchev, et al., New Biotechnol., 2011, 28, 665–672 CrossRef CAS PubMed.
- A. Berenjian, N. L. C. Chan, R. Mahanama, A. Talbot, H. Regtop, J. Kavanagh and F. Dehghani, Mol. Biotechnol., 2013, 54, 371–378 CrossRef CAS PubMed.
- A. Ebrahiminezhad, V. Varma, S. Y. Yang and A. Berenjian, Appl. Microbiol. Biotechnol., 2016, 100, 173–180 CrossRef CAS PubMed.
- L. J. Schurgers, K. J. Teunissen, K. Hamulyák, M. H. Knapen, H. Vik and C. Vermeer, Blood, 2007, 109, 3279–3283 CrossRef CAS PubMed.
- Y. Tsukamoto, M. Kasai and H. Kakuda, Biosci., Biotechnol., Biochem., 2001, 65, 2007–2015 CrossRef CAS PubMed.
- T. Sato, Y. Yamada, Y. Ohtani, N. Mitsui, H. Murasawa and S. Araki, J. Biosci. Bioeng., 2001a, 91, 16–20 Search PubMed.
- T. Sato, Y. Yamada, Y. Ohtani, N. Mitsui, H. Murasawa and S. Araki, J. Ind. Microbiol. Biotechnol., 2001b, 26, 115–120 Search PubMed.
- Y. Tani, S. Asani and H. Yamada, Agric. Biol. Chem., 1985, 49, 111–115 CAS.
- O. Kwon, D. K. Bhattacharyya and R. Meganathan, J. Bacteriol., 1996, 178, 6778–6781 CrossRef CAS PubMed.
- F. Fernandez and D. M. Collin, FEMS Microbiol. Lett., 1987, 41, 175–180 CrossRef CAS.
- J. Spizizen, Proc. Natl. Acad. Sci. U. S. A., 1958, 44, 304–310 CrossRef.
- T. Akamatsu and H. Taguchi, J. Biosci. Bioeng., 2012, 114, 138–143 CrossRef CAS PubMed.
- J. Z. Xu, X. H. Xia, J. L. Zhang, Y. F. Guo and W. G. Zhang, Plasmid, 2014a, 72, 9–17 Search PubMed.
- J. Z. Xu, M. Han, J. L. Zhang, Y. F. Guo and W. G. Zhang, Amino Acids, 2014b, 46, 2165–2175 Search PubMed.
- K. Suvarna, D. Stevnson, R. Meganathan and M. E. S. Hudspeth, J. Bacteriol., 1998, 180, 2782–2787 CAS.
- C. Palaniappan, H. Taber and R. Meganathan, J. Bacteriol., 1994, 176, 2648–2653 CrossRef CAS PubMed.
- M. Jiang, X. L. Chen, Z. F. Guo, Y. Cao, M. J. Chen and Z. H. Guo, Biochemistry, 2008, 47, 3426–3434 CrossRef CAS PubMed.
- H. Fujii, H. Sagami, T. Koyama, K. Ogura and S. Seto, Biochem. Biophys. Res. Commun., 1980, 96, 1648–1653 CrossRef CAS PubMed.
- M. Jiang, X. L. Chen, X. H. Wu, M. J. Chen, Y. D. Wu and Z. H. Guo, Biochemistry, 2009, 48, 6921–6931 CrossRef CAS PubMed.
- B. Teng, C. Zhang, Y. Zhang, J. D. Wu, Z. F. Li, Z. X. Luo and J. B. Yang, Plant Growth Regul., 2015, 77, 117–124 CrossRef CAS.
- Y. W. Zhang, T. Koyama, D. M. Marecak, G. D. Prestwich, Y. Maki and K. Ogura, Biochemistry, 1998, 37, 13411–13420 CrossRef CAS PubMed.
- D. J. Shaw, J. R. Guest, R. Meganathan and R. Bentley, J. Bacteriol., 1982, 152, 1132–1137 CAS.
- J. Debnath, S. Siricilla, B. Wan, D. C. Crick, A. J. Lenaerts, S. G. Franzblau and M. Kurosu, J. Med. Chem., 2012, 55, 3739–3755 CrossRef CAS PubMed.
- A. Dawson, M. Chen, P. K. Fyfe, Z. Guo and W. N. Hunter, J. Mol. Biol., 2010, 401, 253–264 CrossRef CAS PubMed.
- K. Kobayashi, S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen and M. Arnaud, et al., Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4678–4683 CrossRef CAS PubMed.
- R. Meganathan, R. Bentley and H. Taber, J. Bacteriol., 1981, 145, 328–332 CAS.
- M. Vogt, S. Haas, S. Klaffl, T. Polen, L. Eggeling, J. van Ooyen and M. Bott, Metab. Eng., 2014, 22, 40–52 CrossRef CAS PubMed.
- C. Chen, Y. Y. Li, J. Y. Hu, X. Y. Dong and X. Y. Wang, Metab. Eng., 2015, 29, 66–75 CrossRef CAS PubMed.
- H. L. Zhao, C. Xue, Y. Wang, X. Q. Yao and Z. M. Liu, Appl. Microbiol. Biotechnol., 2008, 81, 235–241 CrossRef CAS PubMed.
- C. Stöckmann, T. G. Palmen, K. Schroer, G. Kunze, G. Gellissen and J. Büchs, J. Ind. Microbiol. Biotechnol., 2014, 41, 965–976 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra03388e |
| This journal is © The Royal Society of Chemistry 2017 |