Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
iTRAQ-based proteomic technology revealed protein perturbations in intestinal mucosa from manganese exposure in rat models
Hui Wang *a,
Shengyi Wanga,
Dongan Cuia,
Shuwei Donga,
Xin Tuoa,
Zhiqi Liub and
Yongming Liu*a
*a,
Shengyi Wanga,
Dongan Cuia,
Shuwei Donga,
Xin Tuoa,
Zhiqi Liub and
Yongming Liu*a
aEngineering and Technology Research Center of Traditional Chinese Veterinary Medicine of Gansu Province, Key Lab of New Animal Drug Project of Gansu Province, Key Lab of Veterinary Pharmaceutical Development of Ministry of Agriculture, Lanzhou Institute of Husbandry and Pharmaceutical Sciences of Chinese Academy of Agricultural Sciences, Lanzhou 730050, Gansu, China. E-mail: wanghui01@caas.cn; myslym@sina.com; Tel: +86-931-2115262
bInstitute of Agro-Products Processing Science and Technology, Chinese Academy of Agricultural Sciences, Beijing 100193, China
First published on 22nd June 2017
Abstract
Manganese (Mn) is an essential metal ion as a biological cofactor, but in excess, it is toxic; however, the homeostatic mechanisms of Mn at the cellular level have not been identified. We hypothesized that intracellular Mn can be modulated by proteins that could provide an insight into Mn regulatory mechanisms at the cellular level. Our experiment demonstrated that rats exposed to Mn (200 mg L−1 in water for 5 weeks) showed a significant Mn-induced decrease in total distance traveled, but daily body weight gain and feed intake were not significantly changed. The major organs showed a normal orientation and well defined histological structures without any signs of histopathological changes. We performed a qualitative and quantitative analysis of 3012 proteins for modifiers of cellular Mn content in intestinal mucosa cells by isobaric tags for relative and absolute quantitation (iTRAQ). Following stringent validation assays and bioinformatics, a total of 175 intestinal mucosa proteins were found that may regulate Mn levels under biologically relevant Mn exposures. These proteins are related to pancreatic secretion, protein digestion and absorption, fat digestion and absorption, biosynthesis of amino acids, glycerolipid metabolism, Alzheimer's and Parkinson's disease, mineral absorption associated protein, etc. The identified proteins and biological targets may have potential applications in vivo for modulating cellular Mn content, and will help us understand the homeostatic mechanisms of Mn at the cellular level.
1. Introduction
Metal ions have been demonstrated to play important roles in biological systems. These roles include the structural components of biomolecules, as signaling molecules, as catalytic cofactors in hydrolytic reactions and oxidation–reduction, and in the structural rearrangements of electron transfer chemistry and organic molecules.1 However, at high intracellular concentrations, metal ions are toxic because they can perturb cellular redox potential and produce highly reactive hydroxyl radicals.2 The study of metal ion homeostasis and their importance for cell viability through protein-coding and regulatory machinery has been extensive.3,4 A battery of regulatory proteins in all cells mediate metal homeostasis by regulating the expression of genes that encode intracellular chelators, metal transporters or other detoxification enzymes.5 The homeostasis mechanisms of metal ions are maintained by a concerted effect including metallochaperone proteins, metal transporters, transcription factors and metal responsive signaling pathways.6Manganese (Mn) is an essential metal ion in some important physiological processes such as immune function, development, energy metabolism, reproduction, and antioxidant defenses.7 Incorporation of Mn into metalloproteins has beneficial effects, as these metalloproteins are involved with transferases, oxidoreductases, hydrolases, etc.8 But in excess, Mn brings neurotoxicity, and as a result, the body will experience a variety of psychiatric and motor disturbances due to basal ganglia dysfunction.9,10 In our previous publication, we found that Mn-induced alterations are associated with metabolic responses to manganese poisoning, or manganism. Major alterations were observed in purine metabolism, amino acid metabolism and fatty acid metabolism.11 However, little is known about the complex regulatory biology involving the intracellular handling of Mn.
The critical function of the gastrointestinal tract is absorbing nutrients and water.12 Gastrointestinal changes could be detrimental to the performance, health and welfare of mammals. Mn uptake is regulated by the gastrointestinal tract by responding to dietary Mn levels; however, the molecular mechanisms are not fully understood. The action of the transporters involved in regulating uptake and efflux of Mn into cells is not well established and signaling pathways responsive to physiological Mn concentrations have not been described.13
Proteomics is a mechanism-based high throughput screening approach used to identify proteins expressed that may lead to pathology or disease outcomes in tissue types or certain cells at a given moment.14 Isobaric tags for relative and absolute quantitation (iTRAQ) is an isotope labeling technique, characterized by high throughput, high sensitivity and high accuracy-based proteomic technology.15 iTRAQ coupled with liquid chromatography-tandem mass spectrometry (LC-MS) can produce a full set of information through simultaneously isolating and identifying hundreds of proteins.16,17 Proteomics technologies have the potential ability to solve ecotoxicological issues and elucidate the underlying molecular mechanisms through screening critical proteins based on the protein expression signature (PES).18,19 The PES is defined as the expression signature or expression pattern of a set of proteins as biomarkers.20 Mn-specific signaling pathways or binding proteins responsive to physiological Mn concentrations in vivo have not been systematically described. To identify the differentially expressed proteins that modify cellular Mn content, the proteomics of iTRAQ was performed to identify the modifiers of intracellular and cellular regulation of Mn homeostasis and trafficking. Understanding the targets of PES could provide an insight into the regulatory mechanisms of Mn at the cellular level.
2. Materials and methods
2.1 Ethics statement
All experiments were carried out with respect and humanely for alleviation of suffering following the protocols approved by the Institutional Animal Care and Use Committee of Lanzhou Institute of Husbandry and Pharmaceutics Sciences of the Chinese Academy of Agricultural Sciences (animal use permit: SCXK20008-0013). We received approval for the specific procedures used in this study. Animals were maintained and experiments were conducted following the recommendations in the Guide for the Use and Care of Laboratory Animals. The weight, health status and body temperature of the rats were monitored on a daily basis. They were anesthetized using sodium pentobarbital (50 mg kg−1) by intraperitoneal injection at the end point of the study, and then mounted in a stereotaxic device with heads held in a horizontal plane. All efforts were made to minimize suffering using analgesia, anesthesia, and post-injury care and monitoring. We confirm that the rats did not sustain any injury in our study.2.2 Animals and experimental design
The experiment was performed on male Sprague–Dawley rats. Sixteen rats (148 ± 2 g) were randomly allocated to either the control or Mn-treated group (eight rats per treatment group) and were housed in plastic cages (5 days adaptation and 5 weeks of feeding with experimental diets). All of the rats were housed under pathogen-free conditions on a 12 h light–dark cycle at 20 ± 3 °C and 40–60% relative humidity. During the first 5 days, all groups were fed the basal diet (AIN-93G). This was prepared according to guidelines described elsewhere (35, 10, and 6 mg kg−1 of Fe, Mn, and Cu, respectively).21 The rats in the control group received the AIN-93G diet and non-ionic water, and those in the Mn-treated group were fed the AIN-93G diet and Mn-rich water (200 mg L−1) for 5 weeks. According to WHO guidelines, the risk concentration for Mn in drinking water is 0.5 mg L−1.22 All animals had free access to their diets and water. The body weight (BW) of all rats in each group was measured weekly.2.3 Behavior analysis
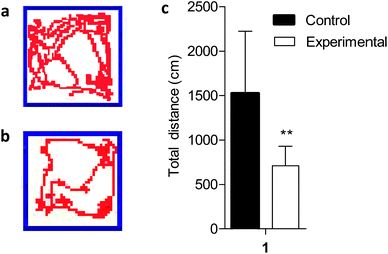
An open-field measurement system was used to measure rats' activity at the end of the experiment; this system can automatically record all activities of the animal. Rats were transported (within their home cage) to acclimatize to the testing room for 1 hour before testing started. Control and Mn-treated rats were put in the separated testing open field chamber (60 cm × 60 cm × 60 cm), and 6 min locomotor activity was automatically recorded by video surveillance. Cameras were mounted on tripods and placed parallel to the shoebox cages. The experiments were repeated three times in a 1 hour period. End points for analysis were the total distance travelled.2.4 Tissue collection
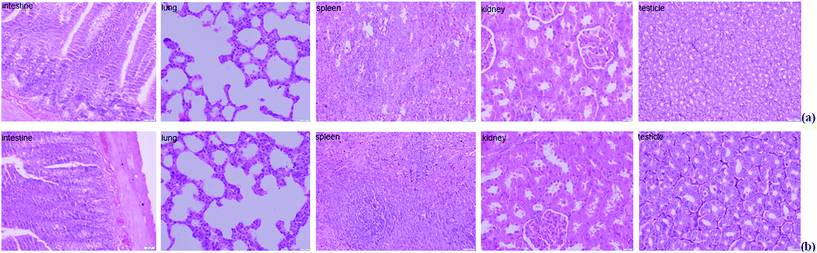
At the end point of the study, rats were anesthetized using sodium pentobarbital. The small intestines were obtained immediately and thoroughly rinsed using normal saline then cut into 1 cm length segments and fixed in 10% neutral formalin used for morphological analysis. Mucosa was obtained from the remaining segment as described previously and snap frozen using liquid nitrogen then stored at −80 °C until biochemical and molecular analyses.23The intestines, lung, spleen, kidney and testicles were harvested and rinsed using normal saline, then immersed immediately in 10% neutral buffered formalin for 24 h. A gradient of alcohols was used for dehydration, and then the tissues were embedded in paraffin using conventional methods.11,24 The tissues were sectioned into 4 μm thick slices; slices were collected and mounted on microscope slides. For observation of morphological changes, hematoxylin–eosin staining was performed and a light microscope (Olympus x51 model, Tokyo, Japan) equipped with a camera (Olympus 20, Tokyo, Japan) was used.
2.5 Intestinal mucosa protein extraction
Total intestinal mucosa proteins were extracted according to Wang et al.23 with slight modifications. In brief, small samples of mucosal scrapings from the intestine were crushed using a mortar with liquid nitrogen. Approximately 100 mg of each powdered sample was transferred to sterile tubes with 600 μL of lysis buffer (SDT; containing 4% SDS (w/v), 100 mM Tris–HCl pH 7.6, 0.1 M DTT).Each mixture was then sonicated in a boiling water bath using an ultrasonicator (Sonics & Materials, Newtown, CT, USA) with 100 W power output. They were sonicated 10 times with 10 s on and 15 s off cycles. After that, each lysed cell suspension was sonicated in a boiling water bath for 15 min to solubilize the proteins, then the suspension was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 rpm for 25 min. The supernatant protein was collected and the protein concentration measured using the BCA (bicinchoninic acid) protein assay. The protein concentration was 23.64 ± 4.07 μg μL−1.
400 rpm for 25 min. The supernatant protein was collected and the protein concentration measured using the BCA (bicinchoninic acid) protein assay. The protein concentration was 23.64 ± 4.07 μg μL−1.
2.6 Protein digestion and peptide quantitation
First, DTT was added to 40 μL of protein from each sample (until the DTT concentration was 100 mM) in a boiling water bath for 5 min, then it was cooled to room temperature. Next, 200 μL of UA buffer (150 mM Tris–HCl pH 8.0, 8 M urea) was used to remove the DTT and detergent components by ultrafiltration (Microcon 10 kDa centrifugal filter units), followed by centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min. Subsequently, 100 μL of 0.05 M iodoacetamide (IAA) in UA buffer was added to the concentrate followed by centrifugation at 14
000g for 15 min. Subsequently, 100 μL of 0.05 M iodoacetamide (IAA) in UA buffer was added to the concentrate followed by centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. Then 100 μL UA buffer was added to the resulting concentrate with centrifugation at 14
000g for 10 min. Then 100 μL UA buffer was added to the resulting concentrate with centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min and repeating this step a further two times. Afterwards, 100 μL of Dissolution buffer was added, followed centrifugation at 14
000g for 10 min and repeating this step a further two times. Afterwards, 100 μL of Dissolution buffer was added, followed centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min, and this step was repeated two times. Finally, the protein suspensions were digested overnight at 37 °C with 40 μL trypsin buffer (3 μg trypsin in 40 μL dissolution buffer). The peptides were quantified using UV light at 280 nm.25
000g for 10 min, and this step was repeated two times. Finally, the protein suspensions were digested overnight at 37 °C with 40 μL trypsin buffer (3 μg trypsin in 40 μL dissolution buffer). The peptides were quantified using UV light at 280 nm.25
2.7 iTRAQ labeling
The iTRAQ Reagent 8-plex Multiplex Kit (Applied Biosystems, Foster City, CA, USA) was used to label each sample according to the manufacturer's instructions. The controls of normal small intestine tissue samples were labeled as (Sample 1)-113, (Sample 2)-114, (Sample 3)-115 and (Sample 4)-116, while Mn-treated samples were labeled as (Sample 5)-117, (Sample 6)-118, (Sample 7)-119 and (Sample 8)-121.2.8 Peptide fractionation and LC-MS/MS analysis
Strong cation exchange (SCX) chromatography was used to fractionate iTRAQ labeled mixed peptides on a 20AD HPLC system (Shimadzu; Kyoto, Japan) using a polysulfoethyl column (5 μm, 200 Å, 2.1 × 100 mm, The Nest Group, MA, USA).Then the samples were analyzed by Q Exactive mass spectrometer (Thermo Finnigan). The samples were added to a C18-reversed phase column in buffer A, then separated with buffer B. Peptides were separated using a flow rate of 300 nL min−1. The gradient profile was: 0–50% B in 0 min to 220 min, 50–100% B in 220 min to 228 min and 100% B in 228 to 240 min.
Tandem mass spectrometry was used to identify the different components on the top 15 ions. Full MS scans were applied at 70k resolution with a 3e6 AGC target and a scan range of 300–1800 m/z. The 10 fragment patterns were collected after each full scan. MS2 scans were monitored with a 2.0 m/z isolation window, 17.5k resolution with a 1e5 AGC target, 60 ms maximum injection time, and a normalized collisional energy (NCE) of 30 eV.
2.9 Data analysis
MS/MS spectra were embedded in the software of Proteome Discoverer 1.4 (thermo) against the uniprot_rat_33675_ 20141101.fasta (33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 675 sequences, downloaded on 11 November 2014) and decoy databases and analyzed using Mascot 2.2 (Matrix Science). The following options were used for protein identification: peptide mass tolerance = 20 ppm, enzyme = trypsin, missed cleavage = 2, and fragment MS/MS tolerance = 0.1 Da; carbamidomethyl (C), iTRAQ8plex (K) and iTRAQ8plex (N-term) were set as fixed modifications; oxidation (M), iTRAQ8plex (Y), and false discovery rate (FDR) ≤ 0.01 were set as variable modifications.26 The peak intensity was extracted using Proteome Discoverer 1.4 from each fragmentation spectrum.
675 sequences, downloaded on 11 November 2014) and decoy databases and analyzed using Mascot 2.2 (Matrix Science). The following options were used for protein identification: peptide mass tolerance = 20 ppm, enzyme = trypsin, missed cleavage = 2, and fragment MS/MS tolerance = 0.1 Da; carbamidomethyl (C), iTRAQ8plex (K) and iTRAQ8plex (N-term) were set as fixed modifications; oxidation (M), iTRAQ8plex (Y), and false discovery rate (FDR) ≤ 0.01 were set as variable modifications.26 The peak intensity was extracted using Proteome Discoverer 1.4 from each fragmentation spectrum.
The peptides identified that expressed changes in comparison with the control were calculated based on the iTRAQ reporter ion intensities. Twofold changed cut-offs were selected as significantly changed to categorize proteins based on statistical analysis and relative quantification, that is, the proteins were considered to be up-regulated if iTRAQ ratios > 1.2, whereas they were considered to be down-regulated if iTRAQ ratios < 0.83.
2.10 Bioinformatics
Proteins were further analyzed for biological and functional relevance based on statistically significant differences. These proteins were classified using their gene function and the freely available Gene Ontology (GO) by biological pathways. NCBI BLAST+ client software was used to identify homologous proteins based on the functional annotation to transfer to the targeted proteins. The top 10 blast hits for each query protein with an E-value less than 1 × 10−3 were retrieved and loaded into Blast2GO (Version 2.7.2) for GO pathway annotation and mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Automatic Annotation Server (http://www.genome.jp/tools/kaas/). In addition, a functional and differentially expressed protein association network was built using Search Tool of the Retrieval of Interacting Genes/Proteins (STRING; http://string.embl.de/).2.11 Statistical analysis
Statistical analyses were conducted using SPSS version 19.0 software (SPSS, Inc., Chicago, IL, USA) and a difference at P < 0.05 was considered to be statistically significant. Data were expressed as mean ± S.D.3. Results and discussion
3.1 Mn exposure significantly decreased locomotion
The open field test is a widely used procedure for examining the behavioral effects of drugs.27 Because altered locomotion is associated with Mn exposure, we conducted behavioral analysis during the fifth week of Mn exposure using video surveillance to monitor Mn-induced changes in activity. Depicting behaviors revealed a significant Mn-induced decrease in total activity in the Mn-exposed group (P < 0.01), measured as total distance traveled at the end of the experiment (Fig. 1). Excess Mn in the brain is neurotoxic and known as manganism.28 Previous studies have reported that excess Mn will produce a variety of psychiatric and motor disturbances including decreased motor skills, memory deficits and psychotic behavior resembling Parkinson's disease due to basal ganglia dysfunction.9,10,28 The results indicated that a model of Mn poisoning was established and the behavioral patterns displayed significant changes in the body produced by motor disturbances due to excess Mn which could lead to the accumulation of neurofibrillary tangles (striatum, globus pallidus and substantia nigra),29 a hallmark of Parkinson's disease.3.2 Effect of manganese exposures on body weight of rats
Body weights (BW) of rats during each week of the experiment are shown in Fig. 2. The BW increased gradually, but the doses and period of Mn exposures had no significant effect on the BW of Mn-treated rats compared with the control group in this study (P > 0.05). Our experiment demonstrated that longer Mn exposure time influenced the behavioral pattern of animals, but feed intake and daily BW gain were not significantly different.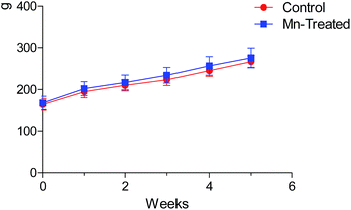 | ||
| Fig. 2 Body weight of rats during the experimental period. No statistically significant difference was observed in the body weight of rats between Mn treated and control groups (P > 0.05). | ||
3.3 Effect of manganese exposure on histopathology
The histopathology of the major organs was studied. No histopathological changes were observed in the intestine, lung, spleen, kidney and testicle of the rats after exposure to Mn at 200 mg L−1 for 5 weeks (Fig. 3). The small intestine of mammalians provides an extensive contact surface for ingested compounds. Absorption and metabolism of xenobiotics, nutrients and drugs are enabled by the unique morphological and functional properties of the small intestine.30 Horning et al. reported that the nervous system is the primary target for excessive Mn, and that Mn exposure has not been linked with damage to the kidney, liver, skin, blood, or stomach.7 But in our previous study, we found extensive necrosis and dissolved nuclei in hepatic tissue in the Mn-treated group of rats.11 The reason for this phenomenon is because the liver plays a major role in Mn excretion, and impairment of liver function results in excessive retention of Mn.31 Our results indicated that Mn exposure did not damage every organ in the body.3.4 Proteomic alterations of small intestine in response to Mn-treated
The homeostasis of transition metal ions in all cells are mediated by a list of regulatory proteins through modulating the expression of genes that encode intracellular chelators, metal transporters and/or other detoxification enzymes.5 Small intestinal enterocytes contain a large variety of regulatory proteins, plasma membrane transporters, intracellular binding proteins, metabolic enzymes, and others within the basolateral membrane.30,32 The enterocytes have a rapid turnover, which allows quick cellular adaptation and quick replacement of cells in response to diets or exposure to xenobiotics.30 The advantages of proteomics technology are high throughput, high sensitivity, rapid testing of small molecules, and discovery of new proteins that are related to disease.33 iTRAQ along with LC-MS is a robust protein discovery technique, allowing the determination of which proteins are affected in intestinal mucosa cells under biologically relevant Mn exposures. Our application of iTRAQ-based proteomic technology targeted specific enzymatic or protein processes with rapid testing of small molecules. This approach would be expected to lead to many potential protein or biological targets.In the current study, rats were treated with Mn for 5 weeks, and at the end of the experiment, the small intestine mucosa were collected and used for protein extraction, protein digestion and iTRAQ labeling to provide more information about proteomes on exposure to Mn. A total of 3012 proteins in intestinal mucosa cells were quantitatively and qualitatively analysed by iTRAQ in this study. The statistically significant proteins were detected using the ANOVA test (P < 0.05), and if the iTRAQ ratio was less than 1.2, the proteins were discarded. A total of 175 intestinal mucosa proteins passed the criteria, and may modulate intracellular Mn levels based on their statistically significant abundance (P < 0.05) (Table 1). Table 1 shows that the expression of 108 different proteins in rat small intestinal mucosa were down-regulated, whereas 67 were up-regulated. The 175 proteins were assigned to seven functional groups, i.e. metabolism (amino acid, lipid and sugar metabolism), binding proteins, disease (Alzheimer's disease, Parkinson's disease, amyotrophic lateral sclerosis, etc.), cellular component and process, oxidative stress, signaling pathway, and enzymatic activity based on Gene Ontology (GO) classification (Table 1). This small group of proteins will help us understand the homeostatic processes mediating normal cellular Mn content.
| Accession | Description | Coverage | Unique peptides | MW [kDa] | Calc. pI | Fold change | P-Value |
|---|---|---|---|---|---|---|---|
| Metabolism | |||||||
| P00774 | Chymotrypsin-like elastase family member 2A | 21.03 | 5 | 28.9 | 8.60 | 0.421 | 0.00 |
| D3ZAM3 | Carboxypeptidase B | 43.37 | 13 | 47.5 | 5.58 | 0.475 | 0.00 |
| G3V976 | Carboxypeptidase A2 | 35.49 | 12 | 46.9 | 5.33 | 0.480 | 0.00 |
| G3V7G2 | Bile salt-activated lipase | 24.02 | 11 | 67.1 | 5.57 | 0.481 | 0.00 |
| G3V7Q8 | Cationic trypsinogen | 35.22 | 7 | 26.3 | 7.49 | 0.558 | 0.00 |
| P00762 | Anionic trypsin-1 | 21.14 | 3 | 25.9 | 4.89 | 0.599 | 0.00 |
| P00731 | Carboxypeptidase A1 | 31.74 | 10 | 47.2 | 5.76 | 0.621 | 0.00 |
| P00763 | Anionic trypsin-2 | 10.16 | 2 | 26.2 | 4.97 | 0.629 | 0.00 |
| G3V844 | Alpha-amylase | 57.48 | 17 | 57.1 | 7.80 | 0.372 | 0.00 |
| Q5I0L0 | Alpha-amylase | 13.11 | 2 | 57.5 | 6.42 | 0.362 | 0.00 |
| G3V8A7 | Pancreatic lipase | 36.56 | 1 | 51.4 | 6.73 | 0.486 | 0.00 |
| P27657 | Pancreatic triacylglycerol lipase | 36.56 | 1 | 51.4 | 6.79 | 0.492 | 0.00 |
| G3V8J3 | Chymotrypsin-like | 23.48 | 5 | 28.1 | 8.19 | 0.678 | 0.00 |
| D3ZFG3 | Elastase 3B | 29.74 | 5 | 28.9 | 5.53 | 0.719 | 0.00 |
| D3ZQV0 | Protein LOC100365995 | 8.13 | 1 | 26.2 | 5.33 | 0.806 | 0.01 |
| B1WC02 | CTP synthase | 2.71 | 1 | 66.6 | 6.58 | 1.296 | 0.00 |
| B1WBV8 | Pld4 protein | 3.18 | 1 | 55.9 | 6.96 | 1.326 | 0.00 |
| G3V8A9 | Inactive pancreatic lipase-related protein 1 | 30.87 | 10 | 52.3 | 6.11 | 0.620 | 0.00 |
| P50442 | Glycine amidinotransferase | 18.68 | 6 | 48.2 | 7.49 | 0.573 | 0.00 |
| P54318 | Pancreatic lipase-related protein 2 | 6.41 | 2 | 52.5 | 6.34 | 0.543 | 0.00 |
| P18757 | Cystathionine gamma-lyase | 20.10 | 7 | 43.6 | 7.96 | 0.678 | 0.00 |
| P49088 | Asparagine synthetase | 6.42 | 3 | 64.2 | 6.46 | 0.750 | 0.00 |
| P02692 | Fatty acid-binding protein | 73.23 | 8 | 14.3 | 8.07 | 0.798 | 0.01 |
| D4A746 | GDP-mannose pyrophosphorylase B | 5.56 | 2 | 39.9 | 6.74 | 0.819 | 0.02 |
| P43427 | Solute carrier family 2 | 2.19 | 1 | 55.5 | 6.60 | 0.824 | 0.02 |
| Q6P784 | Branched-chain-amino-acid aminotransferase | 17.27 | 5 | 43.6 | 8.31 | 0.820 | 0.02 |
| F1LNA9 | Colipase | 34.82 | 3 | 12.2 | 7.80 | 0.572 | 0.00 |
| E9PSV5 | Phosphoserine aminotransferase | 5.68 | 2 | 40.5 | 7.97 | 0.761 | 0.00 |
| D3ZXI0 | Pyrroline-5-carboxylate reductase | 4.21 | 1 | 32.2 | 6.84 | 0.769 | 0.00 |
| Q562C3 | Glycine C-acetyltransferase | 28.85 | 7 | 45.2 | 7.65 | 0.692 | 0.00 |
| G3V960 | Guanidinoacetate N-methyltransferase | 2.97 | 1 | 26.4 | 6.27 | 0.726 | 0.00 |
| Q7M0C4 | Cytochrome P450 2B4 | 9.54 | 3 | 41.4 | 7.09 | 0.736 | 0.00 |
| F1LSR9 | Alcohol dehydrogenase 1 | 24.00 | 8 | 39.7 | 8.56 | 0.772 | 0.00 |
| Q3B7D0 | Oxygen-dependent coproporphyrinogen-III oxidase | 4.97 | 2 | 49.2 | 8.53 | 0.782 | 0.00 |
| P04797 | Glyceraldehyde-3-phosphate dehydrogenase | 64.56 | 1 | 35.8 | 8.03 | 0.787 | 0.00 |
| P24329 | Thiosulfate sulfurtransferase | 10.44 | 2 | 33.4 | 7.85 | 0.790 | 0.00 |
| Q5BJY6 | Putative N-acetylglucosamine-6-phosphate deacetylase | 3.42 | 1 | 43.5 | 6.39 | 0.799 | 0.01 |
| P12336 | Solute carrier family 2 | 6.32 | 4 | 57.0 | 6.54 | 0.806 | 0.01 |
| P52873 | Pyruvate carboxylase | 3.23 | 3 | 129.7 | 6.81 | 0.812 | 0.01 |
| Q91XT9 | Neutral ceramidase | 1.97 | 1 | 83.4 | 7.01 | 0.817 | 0.01 |
| P51538 | Cytochrome P450 3A9 | 15.51 | 6 | 57.8 | 8.82 | 0.825 | 0.02 |
| Q5HZE4 | Methylthioribose-1-phosphate isomerase | 5.42 | 2 | 39.6 | 6.05 | 0.828 | 0.02 |
| D4A7D7 | Hexose-6-phosphate dehydrogenase | 3.76 | 2 | 89.8 | 7.23 | 1.221 | 0.01 |
| P15337 | Cyclic AMP-responsive element-binding protein 1 | 4.11 | 1 | 36.6 | 5.57 | 1.275 | 0.00 |
| P29975 | Aquaporin-1 | 7.43 | 1 | 28.8 | 7.83 | 1.230 | 0.01 |
| B2GV54 | Neutral cholesterol ester hydrolase 1 | 6.13 | 1 | 45.8 | 7.05 | 1.241 | 0.00 |
| M0R451 | Glyceraldehyde-3-phosphate dehydrogenase | 34.34 | 1 | 35.7 | 8.19 | 1.208 | 0.01 |
| Q921A4 | Cytoglobin | 5.79 | 1 | 21.5 | 6.80 | 1.212 | 0.01 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Binding proteins | |||||||
| Q4KLG2 | GTPase IMAP family member 8 | 2.18 | 1 | 77.0 | 8.19 | 0.565 | 0.00 |
| E9PU29 | Protein Rnf31 | 2.35 | 1 | 119.2 | 6.62 | 0.600 | 0.00 |
| P02634 | Protein S100-G | 48.10 | 3 | 9.0 | 5.01 | 0.612 | 0.00 |
| A0A068F1Y2 | Beta-actin | 53.23 | 1 | 20.4 | 5.91 | 0.652 | 0.00 |
| G3V8I8 | Alkaline phosphatase | 22.67 | 7 | 59.4 | 6.28 | 0.692 | 0.00 |
| D3ZXR2 | Protein Wdr52 | 0.33 | 1 | 209.4 | 5.44 | 0.694 | 0.00 |
| Q2XTB0 | Metallothionein 1 | 44.44 | 1 | 1.7 | 7.62 | 0.749 | 0.00 |
| D4AA47 | ATPase | 1.03 | 1 | 144.7 | 7.15 | 0.766 | 0.00 |
| P60905 | DnaJ homolog subfamily C member 5 | 12.63 | 1 | 22.1 | 5.07 | 1.204 | 0.01 |
| Q63862 | Myosin-11 | 57.72 | 1 | 152.4 | 6.18 | 1.205 | 0.01 |
| G3V9F3 | Myosin phosphatase Rho-interacting protein | 1.65 | 1 | 116.9 | 6.21 | 1.205 | 0.01 |
| Q9EPF2 | Cell surface glycoprotein MUC18 | 10.19 | 6 | 71.3 | 6.09 | 1.210 | 0.01 |
| G3V9E3 | Caldesmon 1 | 39.17 | 20 | 60.6 | 6.58 | 1.210 | 0.01 |
| Q99N97 | H-Caldesmon | 27.75 | 6 | 26.6 | 4.98 | 1.212 | 0.01 |
| P47875 | Cysteine and glycine-rich protein 1 | 67.36 | 11 | 20.6 | 8.57 | 1.213 | 0.01 |
| F1M265 | Palladin | 22.63 | 12 | 79.0 | 7.17 | 1.220 | 0.01 |
| Q62896 | BET1 homolog | 15.25 | 1 | 13.2 | 9.07 | 0.798 | 0.01 |
| F1LP76 | Elongator complex protein 1 | 0.83 | 1 | 149.1 | 6.34 | 0.801 | 0.01 |
| Q8VIL3 | ZW10 interactor | 9.02 | 1 | 30.1 | 6.10 | 0.802 | 0.01 |
| D4A929 | WD repeat-containing protein 81 | 1.03 | 1 | 212.1 | 5.81 | 0.799 | 0.01 |
| Q9QYF3 | Unconventional myosin-Va | 0.66 | 1 | 211.6 | 8.69 | 0.805 | 0.01 |
| G3V8M5 | Serine/threonine-protein phosphatase | 8.79 | 1 | 35.1 | 5.06 | 0.808 | 0.01 |
| F1MAD9 | Structural maintenance of chromosomes protein | 1.01 | 1 | 146.7 | 6.96 | 1.232 | 0.01 |
| A2NB82 | Productively rearranged V-lambda-2 | 9.91 | 1 | 12.2 | 6.27 | 1.233 | 0.01 |
| Q62951 | Dihydropyrimidinase-related protein 4 | 2.84 | 1 | 61.0 | 6.77 | 1.237 | 0.00 |
| D3ZHB7 | Protein Ube3c | 1.57 | 1 | 124.3 | 6.64 | 1.249 | 0.00 |
| Q5U2Z5 | Cap-specific mRNA-methyltransferase 1 | 1.79 | 1 | 95.6 | 7.02 | 1.258 | 0.00 |
| B2GUV0 | Cyth1 protein | 5.29 | 1 | 46.2 | 5.63 | 1.260 | 0.00 |
| P0C0R5 | Phosphoinositide 3-kinase regulatory subunit 4 | 1.18 | 1 | 152.4 | 7.18 | 1.269 | 0.00 |
| F1LRJ0 | Protein Kcmf1 | 4.52 | 1 | 41.1 | 5.66 | 1.282 | 0.00 |
| Q5U214 | Protein Snrpa | 2.85 | 1 | 31.2 | 9.83 | 1.337 | 0.00 |
| P68136 | Actin | 71.88 | 2 | 42.0 | 5.39 | 1.357 | 0.00 |
| B2RYP0 | Protein Rhoc | 38.34 | 1 | 22.0 | 6.58 | 1.598 | 0.00 |
| P62747 | Rho-related GTP-binding protein RhoB | 20.92 | 1 | 22.1 | 5.24 | 1.666 | 0.00 |
| Q6IFV3 | Keratin | 11.19 | 1 | 48.8 | 4.86 | 2.109 | 0.00 |
| P00684 | Ribonuclease pancreatic beta-type | 15.13 | 2 | 16.8 | 8.29 | 0.589 | 0.00 |
| F1M9X2 | Glycoprotein GP2 | 3.58 | 2 | 58.8 | 5.07 | 0.667 | 0.00 |
| P15865 | Histone H1.4 | 49.32 | 5 | 22.0 | 11.11 | 1.212 | 0.01 |
| A8QIC3 | DnaJ (Hsp40) homolog | 11.97 | 2 | 30.0 | 7.33 | 1.212 | 0.01 |
| F1LSL1 | Transcription factor Pur-beta | 8.52 | 1 | 33.5 | 5.33 | 1.356 | 0.00 |
| M0R6T4 | Protein Mms19l | 2.23 | 1 | 112.9 | 6.24 | 1.784 | 0.00 |
| M0R5F9 | Protein LOC100910864 | 6.38 | 1 | 33.7 | 9.82 | 1.432 | 0.00 |
| F1M1U0 | Protein Mxra7 | 9.59 | 1 | 15.9 | 4.20 | 1.321 | 0.00 |
| D3ZA93 | Protein Acot13 | 8.57 | 1 | 15.3 | 8.84 | 0.806 | 0.01 |
| D4A9L9 | Protein Erp27 | 8.46 | 2 | 30.7 | 5.03 | 0.555 | 0.00 |
| D4A970 | Protein Ovca2 | 6.61 | 1 | 24.3 | 5.99 | 0.724 | 0.00 |
| D3ZHM7 | Protein Dnttip2 | 1.71 | 1 | 84.7 | 5.81 | 0.755 | 0.00 |
| D4A8G0 | Protein Lsm12 | 4.10 | 1 | 21.7 | 7.74 | 0.826 | 0.02 |
| B2RYE6 | Uncharacterized protein | 13.51 | 1 | 20.7 | 9.52 | 1.256 | 0.00 |
| Q38PG1 | AHNAK 1 | 17.13 | 1 | 58.7 | 5.45 | 1.320 | 0.00 |
| P63055 | Purkinje cell protein 4 | 41.94 | 2 | 6.8 | 6.71 | 1.281 | 0.00 |
| Q499T8 | Intelectin 1 | 4.47 | 1 | 34.5 | 5.78 | 1.582 | 0.00 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Diseases | |||||||
| D4A0X4 | Elongation factor 1-alpha | 27.37 | 1 | 48.7 | 8.72 | 0.697 | 0.00 |
| M0RE01 | Uncharacterized protein | 17.65 | 1 | 36.9 | 6.34 | 0.750 | 0.00 |
| Q569A6 | ER lumen protein-retaining receptor 1 | 3.77 | 1 | 24.5 | 8.62 | 0.766 | 0.00 |
| Q4FZS2 | Budding uninhibited by benzimidazoles 3 homolog | 10.74 | 3 | 36.9 | 6.84 | 0.796 | 0.01 |
| D4A5L9 | Protein LOC679794 | 54.29 | 6 | 11.6 | 9.58 | 0.811 | 0.01 |
| Q06645 | ATP synthase F(0) complex subunit C1 | 5.15 | 1 | 14.2 | 9.91 | 0.827 | 0.02 |
| D4A817 | Histone H2B | 67.46 | 1 | 13.9 | 10.32 | 1.205 | 0.01 |
| Q4QQV6 | Lymphocyte specific 1 | 17.22 | 5 | 36.5 | 4.67 | 1.210 | 0.01 |
| Q9R066 | Coxsackievirus and adenovirus receptor homolog | 6.58 | 1 | 39.9 | 7.27 | 1.214 | 0.01 |
| Q6P725 | Desmin | 73.77 | 27 | 53.4 | 5.27 | 1.229 | 0.01 |
| G3V913 | Heat shock 27 kDa protein 1 | 50.49 | 7 | 22.8 | 6.21 | 1.241 | 0.00 |
| D3ZJ08 | Histone H3 | 60.29 | 2 | 15.4 | 11.27 | 1.356 | 0.00 |
| Q5FVG5 | Similar to tropomyosin 1 | 69.37 | 10 | 32.9 | 4.67 | 1.279 | 0.00 |
| Q5U1W8 | High-mobility group nucleosome binding domain 1 | 8.33 | 1 | 10.1 | 9.76 | 1.264 | 0.00 |
| Q91XN7 | Tropomyosin alpha isoform | 50.00 | 2 | 28.5 | 4.74 | 1.313 | 0.00 |
| D3ZBP2 | Protein Sin3a | 2.13 | 1 | 127.7 | 6.61 | 1.458 | 0.00 |
| P49793 | Nuclear pore complex protein Nup98–Nup96 | 0.83 | 1 | 197.2 | 6.23 | 1.761 | 0.00 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Cellular component and process | |||||||
| Q5XI41 | Translocating chain-associated membrane protein 1 | 2.94 | 1 | 43.0 | 9.67 | 0.747 | 0.00 |
| B2RZ37 | Receptor expression-enhancing protein 5 | 13.76 | 2 | 21.4 | 8.12 | 0.743 | 0.00 |
| D4AC92 | Sulfotransferase | 9.66 | 2 | 33.5 | 6.95 | 0.751 | 0.00 |
| Q5U3Y7 | Transmembrane protein 97 | 13.07 | 2 | 20.9 | 9.29 | 0.752 | 0.00 |
| D3ZE21 | Protein Tmed11 | 7.94 | 1 | 24.6 | 5.57 | 0.771 | 0.00 |
| Q5U3Z3 | Isochorismatase domain-containing protein 2 | 7.62 | 1 | 23.1 | 7.83 | 0.772 | 0.00 |
| P19944 | 60S acidic ribosomal protein P1 | 14.04 | 1 | 11.5 | 4.32 | 0.786 | 0.00 |
| B5DEQ0 | Translocon-associated protein subunit beta | 3.83 | 1 | 20.0 | 8.35 | 0.792 | 0.01 |
| F1M8X9 | Protein Gbf1 | 0.97 | 1 | 206.4 | 5.80 | 0.797 | 0.01 |
| D3ZPJ2 | Protein RGD1564095 | 10.53 | 1 | 11.6 | 4.64 | 0.801 | 0.01 |
| F1LNE5 | Protein MEMO1 | 6.12 | 1 | 31.6 | 6.95 | 0.803 | 0.01 |
| Q5XIG4 | OCIA domain-containing protein 1 | 7.29 | 1 | 27.6 | 7.42 | 0.813 | 0.01 |
| Q6IG04 | Keratin | 3.65 | 1 | 56.8 | 7.36 | 0.814 | 0.01 |
| P0C5H9 | Mesencephalic astrocyte | 31.84 | 5 | 20.4 | 8.25 | 0.823 | 0.02 |
| D3Z952 | Microfibrillar-associated protein 2 | 5.41 | 1 | 20.8 | 4.89 | 0.824 | 0.02 |
| P62250 | 40S ribosomal protein S16 | 45.21 | 8 | 16.4 | 10.21 | 0.827 | 0.02 |
| P15693 | Intestinal-type alkaline phosphatase 1 | 33.15 | 11 | 58.4 | 6.00 | 0.832 | 0.02 |
| Q9JHW0 | Proteasome subunit beta type-7 | 3.25 | 1 | 29.9 | 7.97 | 1.204 | 0.01 |
| Q62713 | Neutrophil antibiotic peptide NP-3A | 19.54 | 1 | 9.3 | 7.65 | 1.211 | 0.01 |
| B1WC16 | BCL2-associated transcription factor 1 | 3.47 | 3 | 106.0 | 9.99 | 1.211 | 0.01 |
| F1LLV6 | Protein Ces1f | 5.35 | 1 | 62.4 | 7.12 | 1.215 | 0.01 |
| Q5BJS4 | FUN14 domain-containing protein 1 | 10.97 | 1 | 17.1 | 8.63 | 1.229 | 0.01 |
| Q6AYC8 | SH2 domain-containing protein 4A | 2.84 | 1 | 48.5 | 9.04 | 1.238 | 0.00 |
| M0R4Z0 | Protein Cbx1 | 41.33 | 2 | 17.2 | 5.03 | 1.254 | 0.00 |
| P24051 | 40S ribosomal protein S27-like | 28.57 | 1 | 9.5 | 9.45 | 1.254 | 0.00 |
| Q6IM78 | DNA topoisomerase I | 4.38 | 1 | 69.0 | 9.22 | 1.268 | 0.00 |
| Q6EIX2 | Mitochondrial import inner membrane translocase | 10.48 | 1 | 13.7 | 6.80 | 1.469 | 0.00 |
| D3Z9K7 | Protein Pdia2 | 41.18 | 17 | 58.4 | 4.82 | 0.524 | 0.00 |
| P00773 | Chymotrypsin-like elastase family member 1 | 12.78 | 2 | 29.0 | 8.47 | 0.565 | 0.00 |
| G3V7A3 | Protein Serpini2 | 5.68 | 1 | 45.9 | 6.15 | 0.652 | 0.00 |
| Q5XFW5 | Peptidyl-prolyl cis–trans isomerase | 10.42 | 1 | 10.4 | 4.70 | 0.764 | 0.00 |
| E9PTI2 | Putative lysozyme C-2 | 5.98 | 1 | 13.1 | 6.84 | 1.209 | 0.01 |
| D4A305 | Coiled-coil domain containing 58 | 5.56 | 1 | 16.7 | 8.16 | 0.819 | 0.02 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Oxidative stress | |||||||
| Q5M875 | 17-Beta-hydroxysteroid dehydrogenase 13 | 23.00 | 6 | 33.5 | 9.38 | 0.582 | 0.00 |
| D3ZTP0 | Protein Aldh1l2 | 2.26 | 1 | 102.9 | 6.39 | 0.618 | 0.00 |
| Q9Z0V5 | Peroxiredoxin-4 | 28.94 | 4 | 31.0 | 6.65 | 0.632 | 0.00 |
| Q6AYT0 | Quinone oxidoreductase | 3.65 | 1 | 35.0 | 8.22 | 0.735 | 0.00 |
| D3ZEN2 | Glyceraldehyde-3-phosphate dehydrogenase | 13.03 | 1 | 35.9 | 8.12 | 0.770 | 0.00 |
| Q5PPL3 | Sterol-4-alpha-carboxylate 3-dehydrogenase | 1.93 | 1 | 40.4 | 8.85 | 0.771 | 0.00 |
| D3Z7Y1 | Uncharacterized protein | 5.47 | 2 | 37.5 | 6.96 | 0.770 | 0.00 |
| Q6AYS8 | Estradiol 17-beta-dehydrogenase 11 | 20.81 | 4 | 32.9 | 8.63 | 0.799 | 0.01 |
| F1M201 | Dehydrogenase/reductase SDR family member 7B | 4.63 | 1 | 35.2 | 9.55 | 0.806 | 0.01 |
| Q5XIB4 | Ufm1-specific protease 2 | 3.25 | 1 | 52.3 | 6.54 | 0.753 | 0.00 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Signaling pathway | |||||||
| Q7TP21 | Cb1-727 | 1.47 | 1 | 75.0 | 8.10 | 1.217 | 0.01 |
| Q6AYK1 | RNA-binding protein with serine-rich domain 1 | 4.92 | 1 | 34.2 | 11.84 | 1.217 | 0.01 |
| G3V9A3 | Protein LOC679816 | 19.76 | 1 | 27.7 | 4.78 | 0.775 | 0.00 |
| Q63450 | Calcium/calmodulin-dependent protein kinase type 1 | 4.81 | 1 | 41.6 | 5.35 | 0.822 | 0.02 |
| Q5XI64 | Monoacylglycerol lipase ABHD6 | 13.95 | 4 | 38.3 | 8.62 | 0.830 | 0.02 |
| Q06486 | Casein kinase I isoform delta | 3.37 | 1 | 47.3 | 9.74 | 1.304 | 0.00 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Enzymatic activity | |||||||
| Q6AYS4 | Plasma alpha-L-fucosidase | 2.40 | 1 | 53.2 | 6.48 | 0.736 | 0.00 |
| D3ZD11 | Protein Spcs2 | 16.37 | 2 | 25.0 | 8.57 | 0.771 | 0.00 |
| D3ZP98 | Histocompatibility 13 | 12.09 | 4 | 40.2 | 6.71 | 0.797 | 0.01 |
| D4A6W4 | Solute carrier family 52 | 2.96 | 1 | 60.3 | 8.59 | 0.798 | 0.01 |
| D3ZL24 | Calpastatin | 6.95 | 3 | 71.7 | 5.08 | 1.244 | 0.00 |
| Q6MG88 | G-Protein-signaling modulator 3 | 11.39 | 1 | 17.5 | 5.85 | 1.257 | 0.00 |
| Q5GAM3 | Ribonuclease 16 | 15.38 | 2 | 17.7 | 9.22 | 1.493 | 0.00 |
| D3Z8M2 | Serine/threonine-protein kinase WNK4 | 2.13 | 2 | 173.0 | 6.33 | 0.762 | 0.00 |
Among these proteins, fatty acid-binding protein (P02692) was down-regulated after Mn exposure. Fatty acid-binding protein is an important cytosolic protein in small intestine epithelial cells, and participates in the uptake, intracellular metabolism and transport of long chain fatty acids.34 The down-regulation of fatty acid-binding protein in our study suggested that Mn exposure might have interrupted the lipid metabolism and fatty acid transport of the intestine. Apart from this protein, chymotrypsin (P00774, F1MA56, P00773 and G3V8J3) and trypsin (G3V7Q8, P00762 and P00763), two important proteolytic enzymes involved in the digestive systems of mammals,35 were significantly down-regulated after Mn exposure. Hence, we speculated that a high concentration of Mn ion might destroy the absorptive functions of the intestinal epithelium through inhibiting the activities of digestive enzymes.
The oxidation states of Mn2+ and Mn3+ are the two common species in the human body and can cycle through redox.36 The Mn redox cycle provides a ‘double-sword’ effect on nutritional metabolism and toxic effects on biological function. Mn serves as a cofactor for Mn superoxide dismutase (MnSOD) that catalyzes superoxide (O2˙−) to hydrogen peroxide (H2O2) and thereby detoxifies free radicals in the mitochondria to prevent oxidative stress through the Mn2+/Mn3+ cycle.28 But the Mn2+/Mn3+ cycle can also accelerate dopamine auto-oxidation to form cytotoxic quinones and produce reactive oxygen species (ROS).37 Oxidative stress and dysregulation of neurological signaling seem to be the mechanisms involved in the neurodegeneration caused by Mn, and they have been found as a consequence of Mn toxicity in the brain.38–40 Our proteomic analysis found that 10 proteins related to oxidative stress were down-regulated significantly in Mn-treated rats. Peroxiredoxin (Q9Z0V5) is a highly expressed antioxidant enzyme that catalyzes and scavenges hydrogen peroxide and other ROS.35 The present study also demonstrated that Mn exposure decreases the activities of 17β-hydroxysteroid dehydrogenase (Q5M875). Interactions between the antioxidant and steroidogenic enzyme systems are physiologically relevant.41 The data on antioxidant status in Mn-treated rats obtained in the present study are in agreement with the observed changes in the activities of steroidogenic enzymes indicating an imbalance in pro-oxidant and antioxidant systems, leading to oxidative stress in manganese-exposed rats. Heat shock proteins are highly regulated proteins that are involved in normal cellular activity, but when the cell is exposed to heat or excess ROS production, they will be up-regulated.35,42 Our results found that the level of heat shock protein (G3V913) increased after Mn exposure. This result indicated that Mn exposure might induce excess ROS production in the intestine, which enhances the expression of heat shock protein and depresses the expression of antioxidative enzymes, and subsequently destroys the antioxidant defense barrier of the intestines.
The altered expression of components of protein complexes that are involved in RNA processing and protein translation was also observed in our study. Although changed protein concentrations cannot be directly correlated with changed functions, the resulting changes strongly indicate altered digestion, absorption, and metabolism as well as oxidative stress within the small intestine.
3.5 Gene Ontology annotation
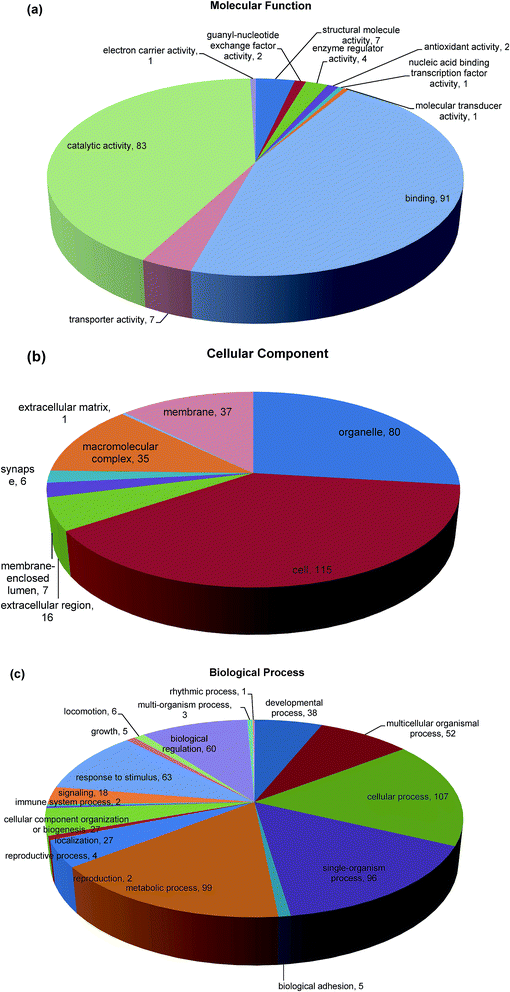
Gene Ontology (GO) functional annotation and enrichment analysis are useful for the analysis of large genomic and proteomic datasets.43 The biological events behind the data can be speculated through examining significantly overrepresented GO terms and provide a primary overview of the intestinal mucosa proteome. Blast2GO (Version 2.8.0) uses NCBI BLAST to find homologous sequences for each obtained hit. Functional classification of the identified proteins to the query sequence according to GO resulted in an assessment of the cellular compartments, molecular function, and biological process represented. Enrichment analysis of GO category was conducted using 175 differentially expressed proteins that represent the overall trends of the specific functional categories enriched in intestinal mucosa cells in order to gain more insight into the biological significance of the differentially expressed proteins. Here, 138 proteins (79.77%) from intestinal mucosa cells with 5079 annotation terms were involved in the GO categories molecular function, biological process, and cellular component (Fig. 4). In the category of molecular function, the majority of differential proteins were found in binding (45.7%), catalytic activity (41.7%), transporter activity (3.5%) and structural molecule activity (3.5%) (Fig. 4a). The binding functions included nucleic acid and nucleotide binding, ion binding and protein binding that were mainly at the intracellular level, while catalytic activities included hydrolase and transferase activities.44 The category cellular component involved locations, subcellular structures, and macromolecular complexes, such as telomere, nucleus, and origin recognition complex. The differentially abundant proteins in our study were related to cellular components in the intracellular space (38.7%), organelle (26.9%), membrane (12.5%), macromolecular complex (11.8%) and extracellular region (5.4%) (Fig. 4b). The final category, biological process included broad biological goals, such as purine metabolism or mitosis. The majority of proteins in our study were found in cellular process (17.4%), metabolic process (16.1%), single organism process (15.6%), response to stimulus (10.2%), biological regulation (9.8%) and multicellular organismal process (8.5%) (Fig. 4c). The functional annotation will help us to effectively analyze the differentially expressed unigenes caused by Mn exposure.3.6 KEGG automatic annotation server (KAAS)
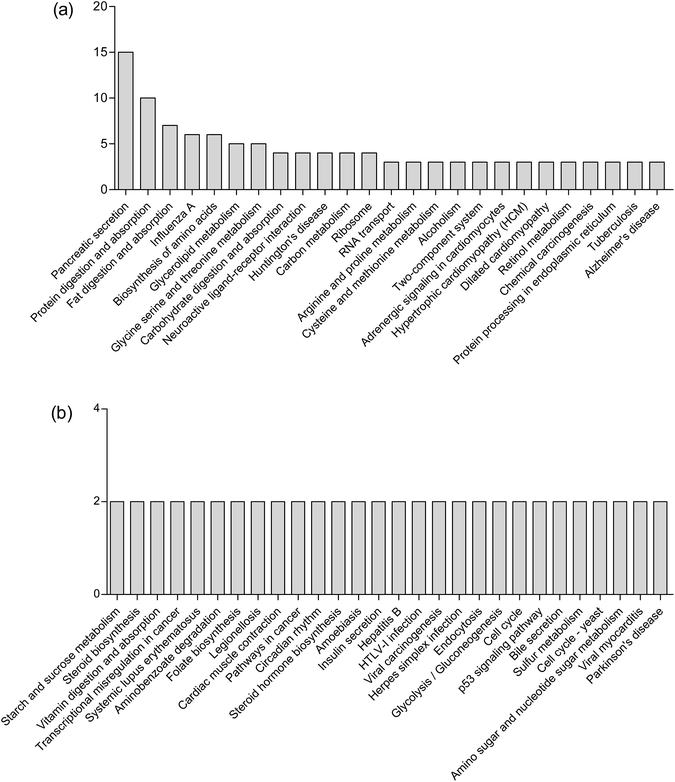
KEGG pathway enrichment can reveal protein functional information in the metabolic pathway through analyzing the differentially expressed proteins.45 The KEGG pathway is a collection of manually drawn pathway maps graphically displaying the distribution of differentially expressed genes among the biochemical pathways. In our study, intestinal mucosa proteins in Mn-treated rats were annotated to biochemical pathways from the KO number of the homologous/similar proteins through aligning with rat protein sequences in the KEGG GENES database using KAAS. The signal/metabolic pathway associations for 93 differential proteins with 140 unique KEGG orthologues were established. The most represented KEGG maps were protein processing in pancreatic secretion (15 members, path: ko04792), bile secretion (2 members, path: ko04976); protein digestion and absorption (10 members, path: ko04974), fat digestion and absorption (7 members, path: ko04975), biosynthesis of amino acids (6 members, path: ko01230), glycerolipid metabolism (5 members, path: ko00561), glycine, serine and threonine metabolism (5 members, path: ko00260), arginine and proline metabolism (3 members, path: ko00330), cysteine and methionine metabolism (3 members, path: ko00270), carbohydrate digestion and absorption (3 members, path: ko04973); Huntington's disease (4 members, path: ko05016), Alzheimer's disease (3 members, path: ko05010), Parkinson's disease (2 members, path: ko05012), etc. (Fig. 5). Upon Mn exposure, profound metabolic changes and activation/deactivation occur in a vast number of intracellular pathways.Mn2+ is taken up by glucose-activated β-cells, resulting in a robust signal increase in glucose-stimulated rodent β-cell lines and in islets.46 Mn2+ can be used to characterize isolated islet potency in vitro and assess the functionality of both grafted and endogenous pancreatic islets in vivo.47 Mn2+ is excreted from blood into bile to form complexes with bile acids.28 Our results showed that high concentrations of Mn2+ disturbed the secretion of pancreas and bile, which is consistent with the above theories. Mineral metabolism can interact with the metabolism of macronutrients such as proteins and fats.48 Our study also demonstrated that Mn exposure is associated with significantly perturbed amino acid metabolism, fatty acid metabolism and carbohydrate metabolism. These results are consistent with our previous finding that Mn exposure significantly altered the concentrations of 36 metabolites in the plasma of rats, which were associated with amino acid metabolism and fatty acid metabolism.11 Exposure to high Mn levels causes neurodegenerative diseases including Parkinson's disease, Huntington's disease and Alzheimer's disease,49 caused by Mn preferentially accumulating in the globus pallidus, striatum, substantia nigra and subthalamic nucleus.50 The results observed in this study were consistent with these important findings.
3.7 Protein–protein interaction analysis
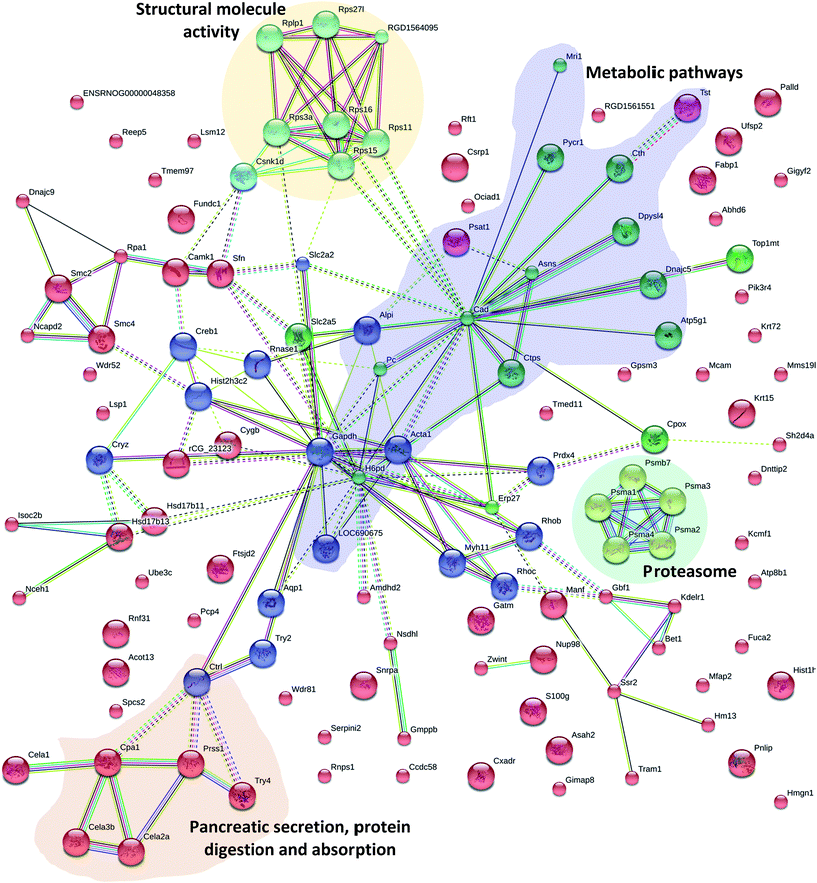
The elementary part of protein complexes is protein function in living cells. However, protein function does not act independently. In addition to KEGG, the protein–protein interactions (PPI) network was also constructed using STRING Database version 10.5. Fig. 6 shows the results that were in accordance with the Pathway and GO analysis highlighting different groups of PPI; 16 proteins have various relationships in PPI and were recognized as key nodes: anionic trypsin-1 (Prss1), chymotrypsin-like elastase family member 2A (Cela2a), chymotrypsin-like protease CTRL-1 precursor (Ctrl), chymotrypsin-like elastase family member 3B precursor (Cela3b), glyceraldehyde-3-phosphate dehydrogenase (Gapdh), peroxiredoxin-4 (Prdx4), chymotrypsin-like elastase family member 1 (Cela1), Rho-related GTP-binding protein RhoB (Rhob), carboxypeptidase A1 (Cpa1), electron carrier protein (LOC690675), histone H3 (Hist2h3c2), GDH/6PGL endoplasmic bifunctional protein precursor (H6pb), calcium/calmodulin-dependent protein kinase type 1 (Camk1), protein LOC298795 (Sfn), actin (Acta1) and solute carrier family 2 (Slc2a5). A number of proteins were associated with pancreatic secretion, protein digestion and absorption: Cela2a, Cela3b, Cpa1, Ctrl, Pnlip, Prss1, Try4; with metabolic pathways: Alpi, Asah2, Asns, Atp5g1, Cpox, Cth, Ctps, Gatm, Gmppb, H6pd, LOC690675, Mri1, Nsdhl, Pc, Pnlip, Psat1, Pycr1, Tst; with proteasome: psma1, psma2, psma3, psma4, psmb7; and with structural molecule activity: Rplp1, Rps27l, RGD1564095, Rps3a, Rps11, Rps15, Rps16, Csnk1d; which were differentially expressed through Mn exposure. Mn is a cofactor in numerous enzymatic processes, so it is essential for brain biology and physiology. But excessive levels of Mn in two neurodegenerative syndromes (Huntington and Parkinson's disease) result in neuronal loss in the basal ganglia,51,52 pathological changes in the striatum,53 as well as having effects on mitochondrial dysfunction and disruption of cellular energy metabolism.7,54 Specifically, we found that four proteins were involved in Huntington's disease, Alzheimer's disease and Parkinson's disease: Gapdh, Atp5g1, OC690675, and Creb1. S100 proteins are calcium-binding proteins that are involved in controlling diverse extracellular and intracellular processes such as differentiation, cell growth, and antimicrobial function.55,56 S100 protein family members exhibit variable affinities for oligomerization properties, post-translational modifications, divalent metal ions, and unique spatial/temporal expression patterns.57 In our study, the mineral absorption associated protein (S100G) was also differentially expressed. Results of the current study indicated that Mn exposure altered the small intestinal proteomes in rats.4. Conclusions
All organisms require mechanisms for sensing small fluctuations to maintain a controlled balance of efflux, uptake, and sequestration in metal levels and to ensure that metal availability is physiologically controlled.2 In conclusion, chronic exposure to a high concentration of Mn significantly changed the behavioral patterns in the rat producing motor disturbances and altering the protein expression profile in rats' intestinal mucosa cells. A total of 175 intestinal mucosa proteins that may regulate Mn levels were identified using iTRAQ-based proteomic technology under biologically relevant Mn exposures. These proteins are involved in pancreatic secretion, protein digestion and absorption, fat digestion and absorption, biosynthesis of amino acids, Alzheimer's and Parkinson's disease, etc. The targets of these proteins related to Mn exposure may improve our understanding of the regulation of Mn homeostasis and intracellular and cellular Mn trafficking.Conflict of interest
The authors declare no competing financial interest.Acknowledgements
Financial support from the Gansu Provincial Natural Science Foundation (No. 1606RJYA224), the National Key Research and Development Plan (No. 2016YFD0501200) and the Central Public-interest Scientific Institution Basal Research Fund (No. 1610322013003) is greatly appreciated. We would also like to thank Shanghai Applied Protein Technology Co. Ltd for their technical assistance.Notes and references
- Z. Ma, F. E. Jacobsen and D. P. Giedroc, Chem. Rev., 2009, 109, 4644–4681 CrossRef CAS PubMed.
- M. I. Hood and E. P. Skaar, Nat. Rev. Microbiol., 2012, 10, 525–537 CrossRef CAS PubMed.
- S. Tottey, K. J. Waldron, S. J. Firbank, B. Reale, C. Bessant, K. Sato, T. R. Cheek, J. Gray, M. J. Banfield, C. Dennison and N. J. Robinson, Nature, 2008, 455, 1138–1142 CrossRef CAS PubMed.
- K. J. Waldron and N. J. Robinson, Nat. Rev. Microbiol., 2009, 7, 25–35 CrossRef CAS PubMed.
- D. P. Giedroc and A. I. Arunkumar, Dalton Trans., 2007, 29, 3107–3120 RSC.
- D. W. Domaille, E. L. Que and C. J. Chang, Nat. Chem. Biol., 2008, 4, 168–175 CrossRef CAS PubMed.
- K. J. Horning, S. W. Caito, K. G. Tipps, A. B. Bowman and M. Aschner, Annu. Rev. Nutr., 2015, 35, 71–108 CrossRef CAS PubMed.
- B. Michalke, J. Trace Elem. Med. Biol., 2016, 31, 50–61 Search PubMed.
- D. L. Stredrick, A. H. Stokes, T. J. Worst, W. M. Freeman, E. A. Johnson, L. H. Lash, M. Aschner and K. E. Vrana, Neurotoxicology, 2004, 25, 543–553 CrossRef CAS PubMed.
- A. Takeda, Brain Res. Rev., 2003, 41, 79–87 CrossRef CAS PubMed.
- H. Wang, Z. Liu, S. Wang, D. Cui, X. Zhang, Y. Liu and Y. Zhang, Metallomics, 2017, 9, 192–203 RSC.
- S. C. Pearce, S. M. Lonergan, E. Huff-Lonergan, L. H. Baumgard and N. K. Gabler, PLoS One, 2015, 10, e0143099 Search PubMed.
- K. K. Kumar, E. W. Lowe Jr, A. A. Aboud, M. D. Neely, R. Redha, J. A. Bauer, M. Odak, C. D. Weaver, J. Meiler, M. Aschner and A. B. Bowman, Sci. Rep., 2014, 4, 6801 CrossRef CAS PubMed.
- A. Nel, T. Xia, H. Meng, X. Wang, S. Lin, Z. Ji and H. Zhang, Acc. Chem. Res., 2012, 46, 607–621 CrossRef PubMed.
- L. Su, L. Cao, R. Zhou, Z. Jiang, K. Xiao, W. Kong, H. Wang, J. Deng, B. Wen, F. Tan, Y. Zhang and L. Xie, PLoS One, 2013, 8, e54237 CAS.
- A. Glen, C. A. Evans, C. S. Gan, S. S. Cross, F. C. Hamdy, J. Gibbins, J. Lippitt, C. L. Eaton, J. Noirel, P. C. Wright and I. Rehman, Prostate, 2010, 70, 1313–1332 CAS.
- J. H. Zhang, Y. Li, X. B. Song, X. H. Ji, H. N. Sun, H. Wang, S. B. Fu, L. J. Zhao and D. J. Sun, Toxicol. Res., 2016, 5, 651–659 RSC.
- T. Monsinjon and T. Knigge, Proteomics, 2007, 7, 2997–3009 CrossRef CAS PubMed.
- M. F. Lemos, A. M. Soares, A. C. Correia and A. C. Esteves, Proteomics, 2010, 10, 873–887 CrossRef CAS PubMed.
- E. F. Petricoin, A. M. Ardekani, B. A. Hitt, P. J. Levine, V. A. Fusaro, S. M. Steinberg, G. B. Mills, C. Simone, D. A. Fishman, E. C. Kohn and L. A. Liotta, Lancet, 2002, 359, 572–577 CrossRef CAS.
- P. G. Reeves, F. H. Nielsen and G. C. Fahey, J. Nutr., 1993, 123, 1939–1951 CAS.
- WHO, Guidelines for drinking-water quality, Health criteria and other supporting information, World Health Organization, Geneva, 2nd edn, 1996, vol. 2 Search PubMed.
- X. Wang, S. Qiao, Y. Yin, L. Yue, Z. Wang and G. Wu, J. Nutr., 2007, 137, 1442–1446 CAS.
- H. Wang, Z. Liu, M. Huang, S. Wang, D. Cui, S. Dong, S. Li, Z. Qi and Y. Liu, Biol. Trace Elem. Res., 2016, 172, 326–335 CrossRef CAS PubMed.
- J. R. Wiśniewski, A. Zougman, N. Nagaraj and M. Mann, Nat. Methods, 2009, 6, 359–362 CrossRef PubMed.
- A. Sandberg, G. Lindell, B. N. Källström, R. M. Branca, K. G. Danielsson, M. Dahlberg, B. Larson, J. Forshed and J. Lehtiö, Mol. Cell. Proteomics, 2012, 11 CAS , M112.016998.
- E. Choleris, A. W. Thomas, M. Kavaliers and F. S. Prato, Neurosci. Biobehav. Rev., 2001, 25, 235–260 CrossRef CAS PubMed.
- Q. Ye, J. E. Park, K. Gugnani, S. Betharia, A. Pino-Figueroa and J. Kim, Metallomics, 2017 10.1039/c7mt00079k.
- A. Uchino, T. Noguchi, K. Nomiyama, Y. Takase, T. Nakazono, J. Nojiri and S. Kudo, Neuroradiology, 2007, 49, 715–720 CrossRef CAS PubMed.
- J. R. Wiśniewski, A. Friedrich, T. Keller, M. Mann and H. Koepsell, J. Proteome Res., 2014, 14, 353–365 CrossRef PubMed.
- H. M. Zeron, M. R. Rodriguez, S. Montes and C. R. Castaneda, J. Trace Elem. Med. Biol., 2011, 25, 225–229 Search PubMed.
- L. S. Kaminsky and Q. Y. Zhang, Drug Metab. Dispos., 2003, 31, 1520–1525 CrossRef CAS PubMed.
- T. Nilsson, M. Mann, R. Aebersold, J. R. Yates, A. Bairoch and J. J. Bergeron, Nat. Methods, 2010, 7, 681–685 CrossRef CAS PubMed.
- J. Storch and L. McDermott, J. Lipid Res., 2009, 50, S126–S131 CrossRef PubMed.
- J. Wang, Y. Y. Wang, L. Lin, Y. Gao, H. S. Hong and D. Z. Wang, J. Proteomics, 2012, 75, 2038–2052 CrossRef CAS PubMed.
- T. Jursa and D. R. Smith, Toxicol. Sci., 2009, 107, 182–193 CrossRef CAS PubMed.
- K. A. Brenneman, R. C. Cattley, S. F. Ali and D. C. Dorman, Neurotoxicology, 1999, 20, 477–487 CAS.
- X. M. Shen and G. Dryhurst, Chem. Res. Toxicol., 1998, 11, 824–837 CrossRef CAS PubMed.
- A. W. Dobson, K. M. Erikson and M. Aschner, Ann. N. Y. Acad. Sci., 2004, 1012, 115–128 CrossRef CAS PubMed.
- D. HaMai and S. C. Bondy, Ann. N. Y. Acad. Sci., 2004, 1012, 129–141 CrossRef CAS PubMed.
- A. K. Chandra, R. Ghosh, A. Chatterjee and M. Sarkar, J. Inorg. Biochem., 2007, 101, 944–956 CrossRef CAS PubMed.
- F. Di Domenico, R. Sultana, G. F. Tiu, N. N. Scheff, M. Perluigi, C. Cini and D. A. Butterfield, Brain Res., 2010, 1333, 72–81 CrossRef CAS PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Nucleic Acids Res., 2009, 37, 1–13 CrossRef CAS PubMed.
- M. Lechpammer, M. S. Clegg, Z. Muzar, P. A. Huebner, L. W. Jin and S. M. Gospe Jr, Ann. Neurol., 2014, 75, 608–612 CrossRef CAS PubMed.
- A. Franceschini, D. Szklarczyk, S. Frankild, M. Kuhn, M. Simonovic, A. Roth, J. Lin, P. Minguez, P. Bork, C. von Mering and L. J. Jensen, Nucleic Acids Res., 2013, 41, D808–D815 CrossRef CAS PubMed.
- M. Nagata, T. Kagawa, D. Koutou, T. Matsushita, Y. Yamazaki and K. Murase, Radiological Physics and Technology, 2011, 4, 7–12 CrossRef PubMed.
- A. Meyer, K. Stolz, W. Dreher, J. Bergemann, V. H. Thimmashetty, N. Lueschen, Z. Azizi, V. Khobragade, K. Maedle and E. Kuestermann, Diabetes, 2015, 64, 2138–2147 CrossRef CAS PubMed.
- J. J. Cao and F. H. Nielsen, Curr. Opin. Clin. Nutr. Metab. Care, 2010, 13, 698–702 CrossRef CAS PubMed.
- A. B. Bowman, G. F. Kwakye, E. H. Hernandez and M. Aschner, J. Trace Elem. Med. Biol., 2011, 25, 191–203 CAS.
- C. W. Olanow, Ann. N. Y. Acad. Sci., 2004, 1012, 209–223 CrossRef CAS PubMed.
- Q. Yu, Y. Xiong, H. Gao, J. Liu, Z. Chen, Q. Wang and D. Wen, Virol. J., 2015, 12, 115 CrossRef PubMed.
- B. B. Williams, G. F. Kwakye, M. Wegrzynowicz, D. Li, M. Aschner, K. M. Erikson and A. B. Bowman, Toxicol. Sci., 2010, 117, 169–179 CrossRef CAS PubMed.
- J. L. Madison, M. Wegrzynowicz, M. Aschner and A. B. Bowman, PLoS One, 2012, 7, e31024 CAS.
- M. Aschner, K. M. Erikson, E. Herrero Hernández and R. Tjalkens, NeuroMol. Med., 2009, 11, 252–266 CrossRef CAS PubMed.
- R. Donato, B. R. Cannon, G. Sorci, F. Riuzzi, K. Hsu, D. J. Weber and C. L. Geczy, Curr. Mol. Med., 2013, 13, 24–57 CrossRef CAS PubMed.
- J. H. Kwek, A. Wynne, C. Lefèvre, M. Familari, K. R. Nicholas and J. A. Sharp, Mol. Phylogenet. Evol., 2013, 69, 4–16 CrossRef CAS PubMed.
- D. B. Zimmer, J. O. Eubanks, D. Ramakrishnan and M. F. Criscitiello, Cell Calcium, 2013, 53, 170–179 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |