Open Access Article
Open Access ArticleMetabolomic studies on the systemic responses of mice with oxidative stress induced by short-term oxidized tyrosine administration
Yuhui Yang a,
Biao Yana,
Xiangrong Chenga,
Yinyi Dinga,
Xu Tiana,
Yonghui Shiab and
Guowei Le
a,
Biao Yana,
Xiangrong Chenga,
Yinyi Dinga,
Xu Tiana,
Yonghui Shiab and
Guowei Le *ab
*ab
aThe Laboratory of Food Nutrition and Functional Factors, School of Food Science and Technology, Jiangnan University, 1800 Lihu Road, Wuxi 214122, China. E-mail: lgw@jiangnan.edu.cn; Fax: +86 510 85869236; Tel: +86 510 85917789
bThe State Key Laboratory of Food Science and Technology, School of Food Science and Technology, Jiangnan University, Wuxi 214122, China
First published on 31st May 2017
Abstract
Oxidized tyrosine (O-Tyr) has attracted more interest in recent years because many researchers have discovered that it and its product (dityrosine) are associated with pathological conditions and metabolic disorders, especially various age-related disorders in biological systems. However, biochemical responses of an organism to short-term O-Tyr and dityrosine (Dityr) administration are unclear. Therefore, our objective is to provide insight into the effects of O-Tyr and Dityr administration on internal metabolic processes. In this study, three groups of Kunming mice were respectively given O-Tyr (320 μg kg−1 body weight), Dityr (320 μg kg−1 body weight) and sterile saline (control group) via gavage once daily for 7 consecutive days. We systematically analysed the O-Tyr and Dityr-induced metabonomic changes in mice serum and urine using proton nuclear magnetic resonance-based metabonomics approaches in conjunction with body weight, indices of oxidative damage, antioxidant capacity assessments, and antioxidant enzymes mRNA expressions. Compared with mice in the control group, O-Tyr and Dityr administration elevated oxidative damage to proteins and lipids, reduced antioxidant capacity, and suppressed antioxidant enzymes mRNA expression in mice. What's more, O-Tyr and DT administration can alter certain systemic metabolic processes in common, including enhanced fatty acid oxidation, glycolysis, glucose–alanine cycle, tricarboxylic acid (TCA) cycle metabolism, induced oxidative stress responses, elevated metabolism of vitamin-B3, and altered gut microbiota functions. Our work provides a comprehensive view of the effects of O-Tyr and Dityr administration, implies an excess intake of oxidative proteins may result in deficiency of vitamin-B3 in body, and reveals it is absolutely essential to avoid overly processed foods. These findings are very important for animal and human food safety.
Introduction
Food proteins are vulnerable to being oxidized during the handling, processing, cooking and storage of food materials.1,2 The oxidation of proteins leads to a variety of physicochemical modifications including protein unfolding and denaturation, cleavage of peptide bonds, formation of cross-links, and loss of functionality.3 Particularly, specific amino acid side chains are susceptible to oxidation, leading to various chemical modifications such as loss of sulfhydryl groups and amino groups, the formation of carbonyl compounds and other oxidation derivatives.4,5 Formation of oxidized protein products in the food system has been proven to cause a significant decrease in protein nutritional value in terms of availability of essential amino acids and digestibility of oxidized proteins.2,5,6 Furthermore, consumption of oxidized protein products may have adverse health effects.7–10Tyrosine (Tyr) is sensitive to various reactive oxygen species (ROS) and other free radicals, leading to formation of oxidized tyrosine (O-Tyr) derivatives (dityrosine, 3-nitrotyrosine, AOPPs, 3-chlorotyrosine, and 3,4-dihydroxyphenylalanine).11,12 These derivatives have been widely detected in food systems, and suggested as promising biomarkers for oxidative damage to proteins.13–17 Our past findings have shown that dietary O-Tyr could induce oxidative stress, inflammation, hepatic fibrosis, and renal fibrosis in rats.18,19 Moreover, our previous experiments also provide proof that Dityr (accounting for 22% of the total O-Tyr material) may be responsible for O-Tyr-induced injury.19 What's more, our study also found that dityrosine exposure impairs hippocampus-dependent non-spatial memory accompanied by modulation of NMDA receptor subunits and expression of brain-derived neurotrophic factor (Bdnf).20 Traditional studies on O-Tyr and Dityr administration have been performed by measuring and comparing a single or several biochemical markers. However, these studies did not sufficiently reflect the overall metabolic status of animals or humans exposed to O-Tyr and Dityr. Therefore, a holistic investigation of the systemic metabolic effects of O-Tyr and Dityr on a whole living bio-system is required to better understand the relationship between O-Tyr administration, Dityr administration, and human health.
Metabonomics, a powerful top-down systems biological tool based on proton nuclear magnetic resonance (1H NMR) spectroscopy or mass spectrometry techniques, together with multivariate statistical analysis methods, provides an effective method for evaluating metabolic responses of living organisms to nutritional intervention.21,22 1H NMR spectroscopy is one of the major techniques used in metabolomic studies as the spectra of biofluids or tissues contain a wealth of metabolic information and provide novel insight into the intervention effect or perturbation of diets with regard to nutrient metabolism and health.23 On the basis of 1H NMR analysis, Stella et al.24 applied an NMR-based metabonomic approach for characterization of the metabolic effects of vegetarian, low meat, and high meat diets in humans. This study illustrates the efficacy of the metabonomic approach for measuring influence of dietary modulations on short-term human metabolism. He et al.25 reported that dietary arginine administration alters the catabolism of fat and amino acids in the whole body, enhances protein synthesis in skeletal muscle, and modulates intestinal microbial metabolism in growing pigs using a 1H NMR technique. A 1H NMR-based metabonomic approach was also applied to evaluate global metabolic responses to L-tryptophan, cysteamine, spermine, chlorogenic acid, green tea and black tea, pea fiber and wheat bran fiber, and drinking water administration in animals.26–32 Thus, 1H NMR-based metabolomics was shown to be very useful for exploring the complex relationship between nutritional intervention and metabolism in order to clarify the role of dietary components in maintaining health and developing disease.33
Therefore, we hypothesized that O-Tyr and Dityr administration causes systemic metabolic alterations in mouse urine and serum. To test this hypothesis, 1H NMR spectroscopy coupled with appropriate multivariate data analysis techniques were used to examine the effects of O-Tyr and Dityr administration on biochemical profiles of urine and serum from mice. Our objective was to provide insight into the effects of O-Tyr and Dityr administration on internal metabolic processes. Metabolic profiles of O-Tyr and Dityr administration in mice can provide clues on the relationship between metabolites and nutritionally influenced biochemical O-Tyr and Dityr mechanisms, contribute baseline data for future metabolomic experiments on O-Tyr and Dityr administration, and also search for further associations between O-Tyr and Dityr administration and health or disease risk.
Materials and methods
Materials
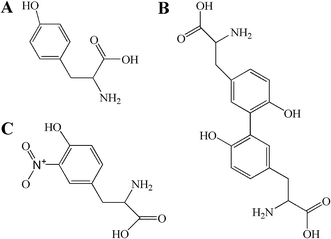
Tyrosine was obtained from Sigma Chemical Co. (St. Louis, MO, USA). Dityr was obtained from Xiamen Huijia Biotechnology Co., Ltd. (Xiamen, China). NaH2PO4·2H2O, and K2HPO4·3H2O were purchased from Guoyao Chemical Co. Ltd. (Shanghai, China). TMSP [3-(trimethylsilyl) propionic-(2,2,3,3-d4) acid sodium salt], and D2O (99.9% in D) were bought from Cambridge Isotope Laboratories (MA, USA). Phosphate buffer was prepared with K2HPO4 and NaH2PO4 for their good solubility and low-temperature stability, as previously reported.34 O-Tyr was prepared by our laboratory according to previously reported methods.18,19,35 Chemical structures of tyrosine, dityrosine and 3-nitrotyrosine are shown in Fig. 1. ELISA kits of advanced oxidized protein products (AOPPs), Dityr and 3-nitrotyrosine (3-NT) were purchased from Xiamen Huijia Bioengineering Institute (Xiamen, China). Detection kits for total antioxidant capacity (T-AOC), malondialdehyde (MDA), catalase (CAT), superoxide dismutase (SOD), glutathione peroxidase (GSH-PX), reduced glutathione (GSH) and oxidized glutathione (GSSG) were purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). All chemicals used in the experiments were analytical grade.Animal experiments and sample collections
All animal experimental procedures were performed according to the National Guidelines for Experimental Animal Welfare (MOST of PR China, 2006), and approved by the Jiangnan University Institutional Animal Care and Use Committee. Female Kunming mice weighing approximately 25 g (4 weeks old) were purchased from Suzhou University (Suzhou, China). A total of 30 mice were housed in a SPF animal laboratory, and kept at a constant temperature of 22 ± 2 °C and relative humidity of 50 ± 10% with a 12/12 h light/dark cycle. After acclimatization for one week, mice were randomly divided into three groups (n = 10): the control group (gavage administration of physiological saline solution once daily for 7 days), the O-Tyr group (gavage administration of O-Tyr once daily for 7 days), and the Dityr group (gavage administration of Dityr once daily for 7 days). O-Tyr and Dityr were dissolved in physiological saline solution and gavage administered at a dose of 320 μg kg−1 body weight, respectively. The dosages of O-Tyr and Dityr for this study were selected based on previous reports from our laboratory.18–20 All mice were given free access to the same standard diet and drinking water throughout the study. The mice diet was conducted according to the general quality standard for formula feeds of laboratory animals in China (GB14924.1, 2001).Urine samples were collected from each mouse into ice-cooled vessels from day 6 to day 7 of the treatment period (24 h). At the end of the experimental period, all mice were sacrificed after an overnight fast. Blood samples were collected (10:00–11:00 a.m.) from eyeballs under anesthesia (intraperitoneal injection of sodium pentobarbital), placed into Eppendorf tubes, kept at 4 °C for 30 min, and centrifuged at 3500g for 15 min at 4 °C to obtain serum. All urine and serum samples were stored at −80 °C until ready for NMR spectroscopy analysis. Liver and kidney tissues from each group were immediately collected for subsequent analysis.
Analysis of oxidative damage and total antioxidant capacity
The concentrations of AOPPs, Dityr, and 3-NT in liver and kidney were assayed using kits and following manufacturer's instructions. The activity of T-AOC, CAT, SOD, and GSH-PX, and the concentration of GSH, GSSG, and MDA in serum, liver, and kidney also were assayed using kits and following manufacturer's instructions.Assay of mRNA expression related in antioxidant enzymes
For determining mRNA expression, total RNA was first extracted from frozen tissues with Trizol reagent. The amount and purity of the RNA were verified by measuring the A260/A280 ratio and gel electrophoresis, respectively. Total RNA was reverse-transcribed to cDNA according to manufacturer's instructions (MultiScribe Reverse Transcriptase; Applied Biosystems). The mRNA expression was quantified using Real-Time Polymerase Chain Reaction (RT-PCR). Primers were designed and synthesized by Generay Biotech Co., Ltd. (Shanghai, China). The primer sequences of CAT, SOD1, SOD2, and GSH-PX are listed in Table 1.| Gene symbol | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| CAT | CCTTCAAGTTGGTTAATGCAGA | CAAGTTTTTGATGCCCTGGT |
| SOD1 | ATGGCGATGAAAGCGGTGT | CCTTGTGTATTGTCCCCATACTG |
| SOD2 | CAGACCTGCCTTACGACTATGG | CTCGGTGGCGTTGAGATTGTT |
| GSH-PX | TTTCCCGTGCAATCAGTTC | TCGGACGTACTTGAGGGAAT |
Sample preparation and 1H NMR spectroscopy
Serum and urine samples were thawed to room temperature prior to NMR analysis, and prepared according to the method of Zhu et al.36 with a little modification. Briefly, a total of 200 μL of urine was mixed with 350 μL of D2O and 50 μL of phosphate buffer (1.5 M Na+/K+ buffer, pH = 7.4) containing 0.1% 3-(trimethylsilyl) propionic-(2,2,3,3-d4) acid sodium salt (TSP) as a chemical shift reference. After vortexing and centrifugation at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C for 10 min, 550 μL of supernatant was transferred into a 5 mm NMR tube for analysis. Serum samples were prepared by mixing 100 μL of serum with 500 μL of saline solution containing 75% D2O as a field frequency lock. These mixtures were vortexed and centrifuged at 11
000g, 4 °C for 10 min, 550 μL of supernatant was transferred into a 5 mm NMR tube for analysis. Serum samples were prepared by mixing 100 μL of serum with 500 μL of saline solution containing 75% D2O as a field frequency lock. These mixtures were vortexed and centrifuged at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C for 10 min, and then 550 μL of supernatant from each sample was transferred into a 5 mm NMR tube for analysis.
000g, 4 °C for 10 min, and then 550 μL of supernatant from each sample was transferred into a 5 mm NMR tube for analysis.
In this study, NMR detection of all samples was conducted by Wuhan Zhongke Metaboss Technology CO., LTD. All one-dimensional 1H NMR spectra of urine and serum samples were recorded at 298 K on a Bruker Avance II 600 MHz NMR spectrometer (Bruker Biospin, Germany) operating at 600.58 MHz for proton frequency and equipped with a 5 mm broadband observe probe. The 1H NMR spectra of urine samples were recorded using water-suppressed standard one-dimensional nuclear overhauser effect spectroscopy (NOESYPR1D) pulse sequence (recycle delay–90°–t1–90°–tm–90°–acquisition) for representation of the total metabolite composition with a recycle delay of 2 s, t1 of 3 μs, mixing time (tm) of 100 ms, and 90° pulse length of 12.00 ms. A total of 64 transients were collected in 32k data points using a spectral width of 8417.5 Hz and an acquisition time of 3.89 s. For serum, a water-presaturated Carr–Purcell–Meiboom–Gill (CPMGPR1D) pulse sequence [recycle delay–90°–(τ–180°–τ)n–acquisition] was employed to attenuate the NMR signals from macromolecules. These spectra were measured using a spin–spin relaxation delay (2nτ) of 80 ms and a spin-echo delay τ of 400 μs. A total 32 transients were collected into 32k data points using a spectral width of 9615.4 Hz with a relaxation delay of 3 s. Metabolite assignments were usually made by considering chemical shifts, coupling constants, and relative intensities, as in previous reports.37,38
Data processing for metabonomic analysis
Free induction decays were multiplied by an exponential window function with a 1 Hz line-broadening factor prior to Fourier transformation. All 1H NMR spectra from urine and serum samples were then phase-adjusted and baseline-corrected manually employing Mestrenova 6.1.1 software (Mestrelab Research S.L., Spain). A urinary spectral region δ 0.5 to δ 9.5 was bucketed into regions with 0.01 ppm widths, and a serum spectral region δ 0.5 to δ 8.5 was integrated into regions with equal 0.01 ppm widths. TSP with a chemical shift at δ 0.00 was used as a spectral reference for urine, whereas NMR spectra of the serum were referenced to the methyl proton of L-lactate at δ 1.33. Regions containing water (δ 4.68 to δ 5.20) were removed to avoid the effects of imperfect water suppression. The urea signal was excluded to avoid any contributions of urea to intergroup differentiations in order to eliminate the effects of variation in urea signals caused by partial cross-solvent saturation via solvent-exchanging protons. Discarded regions in the urine spectra included δ 4.68 to δ 5.00 for H2O and δ 5.70 to δ 6.00. Subsequently, all remaining regions of the spectra were normalized to the total integrated area of the spectra to reduce any significant concentration differences before multivariate data analysis.Normalized NMR datasets were exported to TXT files using Mestrenova 6.1.1 software, imported into Microsoft Office Excel 2007 (Microsoft Corporation, Redmond, Washington, USA) for integration, and then imported into the software package SIMCA-P 13.0 (Umetrics, Umea, Sweden) for multivariate data analysis. Initially, in a unsupervised manner, principal component analysis (PCA) was performed on the dataset to generate an overview and identify possible outliers within the dataset.39 Each point in the score plots represented an individual spectrum of a sample. If one point position was outside the Hotelling's T2 ellipse on the score plot, it was removed from the dataset. Subsequently, partial latent structure-discriminate analysis (PLS-DA) was conducted to distinguish experimental group mice and control group mice in a supervised manner. The quality of the model was evaluated by the parameters: R2X (representing the total explained variation) and Q2 (standing for the model predictability). R2 > 0.50 and Q2 > 0.50 indicates that the model is robust and has good fitness and prediction. Leave-one-out cross validation and the response of the permutation test (200 cycles) should be used to evaluate the reliability of the model when a small number of samples is adopted. Furthermore, a supervised pattern recognition approach known as an orthogonal projection to latent structures discriminant analysis (OPLS-DA) was used to improve classification of the different groups while screening biomarkers. With an aim to discover potential variables contributing to the differentiation, we generated an S-plot for the OPLS-DA model used to define metabolites significantly contributing to the separation of the two groups.40 Metabolites with VIP (variable importance in the projection) values ≥1.0 were regarded as significant in this study. VIP is a computation of the influence of every X term in the model on the Y variable. Larger VIP values indicate a greater influence of a term X on the Y variable.41 Meanwhile, an independent sample t-test was further used to validate those major contributing variables from the PLS-DA and OPLS-DA models using the software of SPSS 17.0 (SPSS Inc., Chicago, USA). Values of P < 0.05 were regarded as significant statistical significance. Only those metabolites that meet the two criteria are eventually considered as potential biomarkers.42
Statistical analysis
All other experimental results were also statistically analyzed using the software of SPSS 17.0. Statistical significance was determined by a one-way analysis of variance (ANOVA) followed by the Tuckey post hoc test.43 Data were presented as mean ± standard error of the mean (SEM). P values less than 0.05 were accepted as statistically significant.Results
Effects of O-Tyr and DT administration on body weights in mice
Effects of O-Tyr and DT administration on the body weight of mice are listed in Table 2. At the beginning of the study, the body weights of mice in the O-Tyr group and DT group were the same as in the control group. At the end of the experiments, O-Tyr and DT administration had a tendency to decrease the body weights of mice, but there were no statistically significant differences between the three groups (P > 0.05).| Parameters | Control group | O-Tyr group | Pa value | Dityr group | Pb value |
|---|---|---|---|---|---|
| a Data from each group (n = 10) were averaged and presented as means ± SEM. P values <0.05 were regarded as significant statistical significance. | |||||
| Initial weight (g) | 25.53 ± 0.52 | 25.60 ± 0.37 | 0.906 | 25.49 ± 0.25 | 0.949 |
| Finish weight (g) | 26.99 ± 0.35 | 26.86 ± 0.58 | 0.852 | 26.67 ± 0.42 | 0.567 |
Effects of O-Tyr and DT administration on protein oxidation of mice liver and kidney
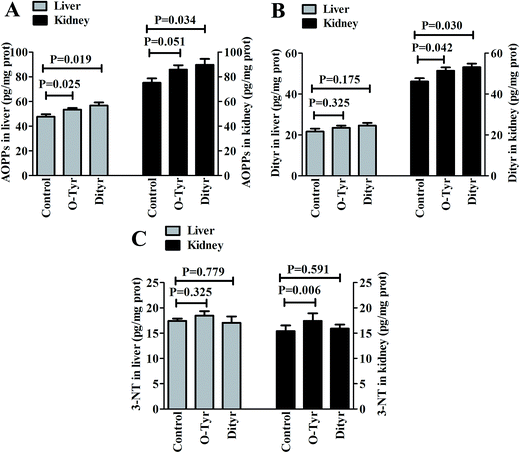
Effects of O-Tyr and DT administration on the concentrations of AOPPs, Dityr and 3-NT in mice liver and kidney are shown in Fig. 2. Compared with control group, O-Tyr administration significantly increased the concentration of AOPPs in liver, and the concentrations of Dityr and 3-NT in kidney (P < 0.05). DT administration significantly increased the concentration of AOPPs in liver, and the concentrations of AOPPs and Dityr in kidney (P < 0.05).Effects of O-Tyr and DT administration on the antioxidant capacity of mice serum, liver, and kidney
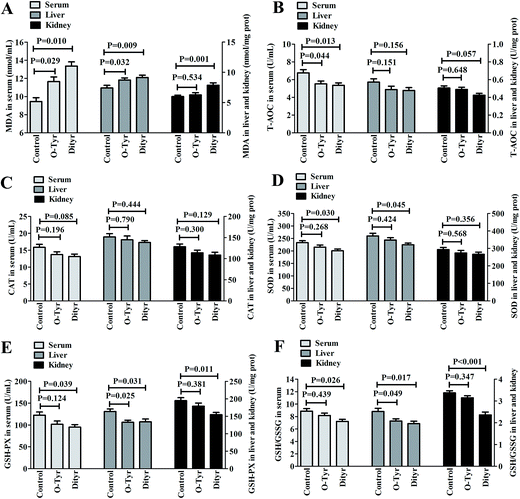
Effects of O-Tyr and DT administration on T-AOC, CAT, SOD, and GSH-PX activity, and GSH/GSSG and MDA content in mice serum, liver, and kidney are shown in Fig. 3. O-Tyr administration significantly decreased the activity of T-AOC in serum and GSH-PX in liver, and reduced the value of GSH/GSSG in liver, and increased the content of MDA in serum and liver, compared to the control group (P < 0.05). DT administration significantly decreased the activity of T-AOC in serum, and declined the activity of SOD in serum and liver, decreased the activity of GSH-PX in serum, liver, and kidney and reduced the value of GSH/GSSG in serum, liver, and kidney, and increased the content of MDA in serum, liver, and kidney (P < 0.05).Effects of O-Tyr and DT administration on the antioxidant enzymes mRNA expression of mice liver and kidney
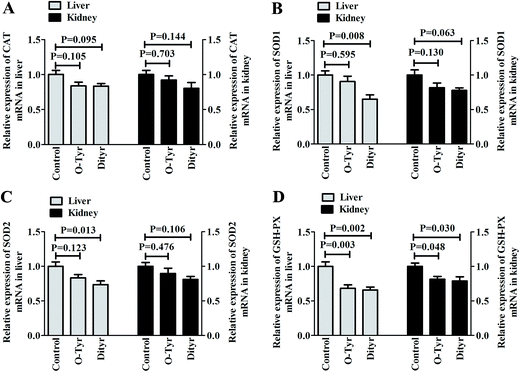
Effects of O-Tyr and DT administration on the antioxidant enzymes mRNA expression of mice liver and kidney are shown in Fig. 4. Quantitative RT-PCR analysis showed that O-Tyr administration significantly reduced the antioxidant enzyme of GSH-PX expression in liver and kidney tissues (P < 0.05). DT administration significantly decreased the antioxidant enzymes of SOD1, SOD2 and GSH-PX expressions in liver tissue, and declined the antioxidant enzyme of GSH-PX expression in kidney tissue (P < 0.05).1H NMR spectra of urine and serum samples
Fig. 5 and 6, respectively, show representative 1H NMR spectra of serum and urine samples taken from randomly selected mice in the control group, O-Tyr group, and DT group. A total of 47 metabolites in serum and 61 metabolites in urine were unambiguously assigned from the 1H NMR spectra. Assignment of metabolites was made on the basis of comparisons with the published literature44–47 and existing databases (such as the Human Metabolome Data Base).48 In addition, for exact assignment purposes, five two-dimensional (2D) NMR spectra including 1H–1H J-resolved spectroscopy (J-Res), 1H–1H correlation spectroscopy (COSY), 1H–1H total correlation spectroscopy (TOCSY), 1H–13C heteronuclear single quantum coherence spectroscopy (HSQC), and 1H–13C heteronuclear multiple bond correlation spectroscopy (HMBC) were obtained from previous studies as ref. 49–51. Their chemical shifts, peak multiplicity, and the corresponding 1H NMR signal multiplicities are listed in Tables 3 and 4, respectively. The spectra of serum samples contained resonances from several amino acids, short-chain fatty acids, nitrogenous compounds, acetyl glycoproteins, lipoproteins, glucose, ketones, TCA cycle intermediates, and choline metabolites. The urine samples mainly contained several amino acids, glucose, organic acids, allantoin, TCA cycle intermediates, short-chain fatty acids, nitrogenous compounds, and metabolites of amino acids, choline, and vitamin B3. In order to obtain more details about metabonomic changes in O-Tyr and DT group mice, multivariate data analyses were further conducted on 1H NMR data for serum and urine.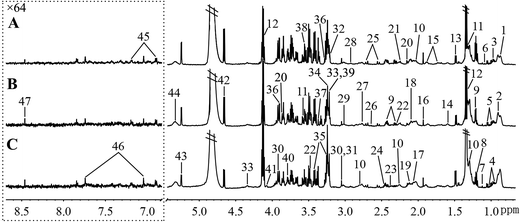 | ||
| Fig. 5 Typical 600 MHz 1H NMR spectra of serum taken from mice with the control group (A), O-Tyr group (B) and DT group (C). The region of δ 6.83–8.67 was magnified 64 times compared with corresponding region of δ 0.71–5.44 for the purpose of clarity. Metabolite keys are given in Table 3. | ||
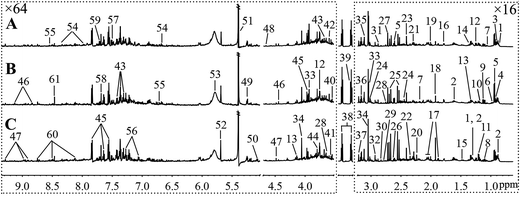 | ||
| Fig. 6 Typical 600 MHz 1H NMR spectra of urine taken from mice with the control group (A), O-Tyr group (B) and DT group (C). The regions of δ 0.64–3.21 and δ 3.53–9.33 were magnified 16 and 64 times compared with corresponding region of δ 3.24–3.49 for the purpose of clarity, respectively. Metabolite keys are given in Table 4. | ||
| Keys | Metabolites | Moieties | δ1H (ppm) and multiplicity |
|---|---|---|---|
| a LDL, low density lipoprotein; VLDL, low density lipoprotein; TMAO, trimethylamine-N-oxide; GPC, glycerophosphorylcholine; s, singlet; d, doublet; t, triplet; q, quartet; dd, doublet of doublets; m, multiplet. | |||
| 1 | LDL | CH3(CH2)n | 0.88(m) |
| 2 | VLDL | CH3CH2CH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) |
0.90(t) |
| 3 | Leucine | αCH, βCH2, γCH, δCH3 | 3.73(t), 1.72(m), 0.96(d), 0.91(d) |
| 4 | Isoleucine | αCH, βCH, βCH3, γCH2, δCH3 | 3.68(d), 1.99(m), 1.01(d), 1.26(m), 1.47(m), 0.94(t) |
| 5 | Valine | αCH3, βCH, γCH3 | 3.62(d), 2.28(m), 0.99(d), 1.04(d) |
| 6 | Propionate | CH3, CH2 | 1.08(t), 2.18(q) |
| 7 | Isobutyrate | CH3 | 1.12(d) |
| 8 | Ethanol | CH3, CH2 | 1.19(t), 3.66(q) |
| 9 | 3-Hydroxybutyrate | αCH2, βCH, γCH3 | 2.31(dd), 2.42(dd), 4.16(m), 1.21(d) |
| 10 | Lipids | 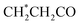 , CH2–C , CH2–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
1.29(m), 1.58(m), 2.02(m) |
CH2–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, CH–O–CO O, CH–O–CO |
2.25(m), 2.77(m) | ||
| 11 | Threonine | αCH, βCH, γCH3 | 1.32(d), 4.25(m), 3.58(d) |
| 12 | Lactate | αCH, βCH3 | 1.33(d), 4.11(q) |
| 13 | Alanine | αCH, βCH3 | 3.77(q), 1.48(d) |
| 14 | Citrulline | αCH2, βCH2, γCH2 | 3.70(m), 1.59(m), 3.15(t) |
| 15 | Lysine | αCH, βCH2, γCH2, δCH2 | 3.77(t), 1.89(m), 1.73(m) |
| 16 | Acetate | CH2–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
1.93(s) |
| 17 | N-Acetyl glycoprotein | CH3 | 2.05(s) |
| 18 | O-Acetyl glycoprotein | CH3 | 2.09(s) |
| 19 | Glutamate | αCH, βCH2, γCH2 | 3.75(m), 2.12(m), 2.35(m) |
| 20 | Methionine | αCH, βCH2, γCH2, S–CH3 | 3.87(t), 2.16(m), 2.65(t), 2.14(s) |
| 21 | Acetone | CH3 | 2.24(s) |
| 22 | Acetoacetate | CH3, CH2 | 2.29(s), 3.49(s) |
| 23 | Pyruvate | CH3 | 2.38(s) |
| 24 | Glutamine | αCH, βCH2, γCH2 | 3.68(t), 2.15(m), 2.45(m) |
| 25 | Citrate | CH2 | 2.54(d), 2.69(d) |
| 26 | Methylamine | CH3 | 2.64(s) |
| 27 | Dimethylamine | CH3 | 2.73(s) |
| 28 | Trimethylamine | CH3 | 2.92(s) |
| 29 | Albumin | Lysyl–CH2 | 3.02(s) |
| 30 | Creatine | N–CH3, CH2 | 3.05(s), 3.93(s) |
| 31 | Creatinine | CH3, CH2 | 3.05(s), 4.05(s) |
| 32 | Choline | N–(CH3)3, αCH2, βCH2 | 3.20(s), 4.05(t), 3.51(t) |
| 33 | GPC | N–(CH3)3, OCH2, NCH2 | 3.22(s), 4.33(t), 3.51(t) |
| 34 | TMAO | CH3 | 3.26(s) |
| 35 | Taurine | N–CH2, S–CH2 | 3.27(t), 3.43(t) |
| 36 | Betaine | CH3, CH2 | 3.28(s), 3.90(s) |
| 37 | Proline | βCH2, γCH2, δCH2 | 2.02–2.33(m), 2.00(m), 3.35(t) |
| 38 | Glycine | CH2 | 3.56(s) |
| 39 | Phosphorylcholine | N(CH3)3, OCH2, NCH2 | 3.22(s), 4.21(t), 3.61(t) |
| 40 | Ornithine | CH2, αCH | 3.80(s), 3.79(t) |
| 41 | Myo-inositol | 5-CH, 4,6-CH, 2-CH | 3.30(t), 3.63(t), 4.06(t) |
| 42 | β-Glucose | 2-CH, 1-CH | 3.25(dd), 4.65(d) |
| 43 | α-Glucose | 1-CH | 5.26(d) |
| 44 | Unsaturated lipids | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH2–C C–CH2–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) , –CH , –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH– CH– |
5.19(m), 5.31(m) |
| 45 | Tyrosine | 2,6-CH, 3,5-CH | 7.19(dd), 6.90(d) |
| 46 | 1-Methylhistidine | 4-CH, 2-CH | 7.04(s), 7.75(s) |
| 47 | Formate | CH | 8.46(s) |
| Keys | Metabolites | Moieties | δ1H (ppm) and multiplicity |
|---|---|---|---|
| a TMAO, trimethylamine-N-oxide; 4-PY, N1-methyl-4-pyridone-5-carboxamide; 2-PY, N1-methyl-2-pyridone-5-carboxamide; s, singlet; d, doublet; t, triplet; q, quartet; dd, doublet of doublets; m, multiplet. | |||
| 1 | α-Hydroxy-n-valerate | CH3, γCH2 | 0.86(t), 1.31(m) |
| 2 | Valerate | δCH3, γCH2, βCH2, αCH2 | 0.88(t), 1.31(m), 1.61(m), 2.28(t) |
| 3 | α-Hydroxybutyrate | CH3 | 0.89(t) |
| 4 | Butyrate | CH3 | 0.92(t) |
| 5 | 2-Keto-iso-valerate | CH3, CH | 0.94(d), 3.03(m) |
| 6 | α-Hydroxy-iso-valerate | δCH3 | 0.97(d) |
| 7 | Propionate | CH3, CH2 | 1.06(t), 2.18(q) |
| 8 | 2-Keto-isovalerate | CH3, CH2 | 1.11(d), 3.03(m) |
| 9 | Isobutyrate | CH3 | 1.14(d) |
| 10 | Ethanol | CH3, CH2 | 1.19(t), 3.66(q) |
| 11 | 3-Hydroxybutyrate | γCH3, αCH2, βCH | 1.20(d), 2.28(dd), 2.42(dd), 4.16(m) |
| 12 | Methylmalonate | CH3, CH | 1.25(d), 3.75(m) |
| 13 | Lactate | βCH3, αCH | 1.33(d), 4.13(q) |
| 14 | 2-Hydroxyisobutyrate | CH3 | 1.36(s) |
| 15 | Alanine | βCH3, αCH | 1.48(d), 3.77(q) |
| 16 | Putrescine | CH2, CH2–NH2 | 1.78(m), 3.06(t) |
| 17 | N-Acetylglutamate | γCH2, CH3, βCH2 | 1.88(m), 2.04(s), 2.07(m) |
| 18 | Acetate | CH3 | 1.92(s) |
| 19 | Acetamide | CH3 | 2.00(s) |
| 20 | Acetone | CH3 | 2.24(s) |
| 21 | Acetoacetate | CH3 | 2.3(s) |
| 22 | Pyruvate | CH3 | 2.35(s) |
| 23 | Succinate | CH2 | 2.41(s) |
| 24 | α-Ketoglutarate | βCH2, γCH2 | 2.45(t), 3.01(t) |
| 25 | Citrate | CH2 | 2.55(d), 2.68(d) |
| 26 | Methylamine | CH3 | 2.62(s) |
| 27 | Dimethylamine | CH3 | 2.73(s) |
| 28 | Sarcosine | CH3, CH2 | 2.76(s), 3.65(s) |
| 29 | Succinimide | CH2 | 2.78(s) |
| 30 | Methylguanidine | CH3 | 2.83(s) |
| 31 | Trimethylamine | CH3 | 2.88(s) |
| 32 | Dimethylglycine | CH3 | 2.93(s) |
| 33 | Creatine | CH3, CH2 | 3.04(s), 3.93(s) |
| 34 | Creatinine | CH3, CH2 | 3.05(s), 4.05(s) |
| 35 | Ethanolamine | CH2 | 3.13(t) |
| 36 | Malonate | CH2 | 3.16(s) |
| 37 | Choline | OCH2, NCH2, N(CH3)3 | 4.07(t), 3.53(t), 3.20(s) |
| 38 | Taurine | –CH2–S, –CH2–NH2 | 3.27(t), 3.43(t) |
| 39 | TMAO | CH3 | 3.28(s) |
| 40 | Phenylacetate | CH2, 2,6-CH, 4-CH, 3,5-CH | 3.55(s), 7.28(m), 7.29(m), 7.32(m) |
| 41 | Glycine | CH2 | 3.57(s) |
| 42 | p-Hydroxyphenylacetate | 6-CH, 2-CH, 3,5-CH | 3.6(s), 6.87(d), 7.15(d) |
| 43 | Phenylacetylglycine | 2,6-CH, 3,5-CH, 7-CH, 10-CH | 7.31(t), 7.37(m), 7.42(m), 3.68(s) |
| 44 | Guanidoacetate | CH2 | 3.80(s) |
| 45 | Hippurate | CH2, 3,5-CH, 4-CH, 2,6-CH | 3.97(d), 7.57(t), 7.65(t), 7.84(d) |
| 46 | Trigonelline | 2-CH, 4-CH, 6-CH, 5-CH, CH3 | 9.12(s), 8.85(m), 8.83(dd), 8.19(m), 4.44(s) |
| 47 | N-Methylnicotinamide | CH3, 5-CH, 4-CH, 6-CH, 2-CH | 4.48(s), 8.19(m), 8.91(m), 8.97(d), 9.28(s) |
| 48 | β-Glucose | 1-CH, 2-CH, 3-CH, 4-CH, 5-CH, 6-CH | 4.65(d), 3.25(dd), 3.49(t), 3.41(dd), 3.46(m), 3.73(dd), 3.90(dd) |
| 49 | α-Glucose | 1-CH, 2-CH, 3-CH, 4-CH, 5-CH, 6-CH | 5.24(d), 3.54(dd), 3.71(dd), 3.42(dd), 3.84(m), 3.78(m) |
| 50 | 4-Cresol glucuronide | CH3, 2,6-CH, 3,5-CH, 5-CH, 4-CH, 3-CH | 2.30(s), 7.05(d), 7.23(d), 5.08(d), 3.61(m), 3.89(m) |
| 51 | Allantoin | CH | 5.40(s) |
| 52 | cis-Aconitate | CH2, CH | 3.12(s), 5.69(s) |
| 53 | Urea | NH2 | 5.80(s) |
| 54 | 4-PY | N–CH3, 3-CH, 2-CH, 6-CH | 3.89(s), 6.71(d), 7.83(dd), 8.55(d) |
| 55 | 2-PY | N–CH3, 3-CH, 4-CH, 6-CH | 3.64(s), 6.67(d), 7.96(dd), 8.33(d) |
| 56 | m-Hydroxyphenylacetate | 6-CH, 4-CH, 3-CH | 6.92(m), 7.05(d), 7.27(t) |
| 57 | Nicotinate | 2,6-CH, 4-CH, 5-CH | 8.62(d), 8.25(d), 7.50(dd) |
| 58 | Guanine | CH | 7.68(s) |
| 59 | 4-Aminohippurate | CH2 | 7.71(d) |
| 60 | Nicotinamide N-oxide | 5-CH, 6-CH, 2-CH, 4-CH | 7.74(m), 8.12(m), 8.75(m), 8.49(m) |
| 61 | Formate | CH | 8.46(s) |
Multivariate data analysis of 1H NMR data
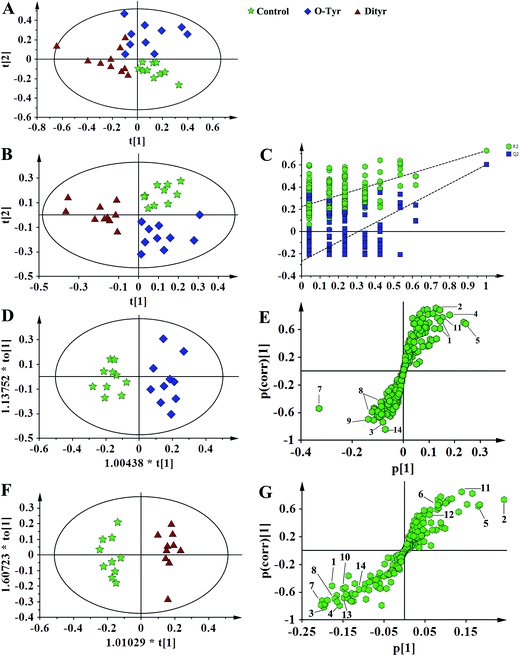
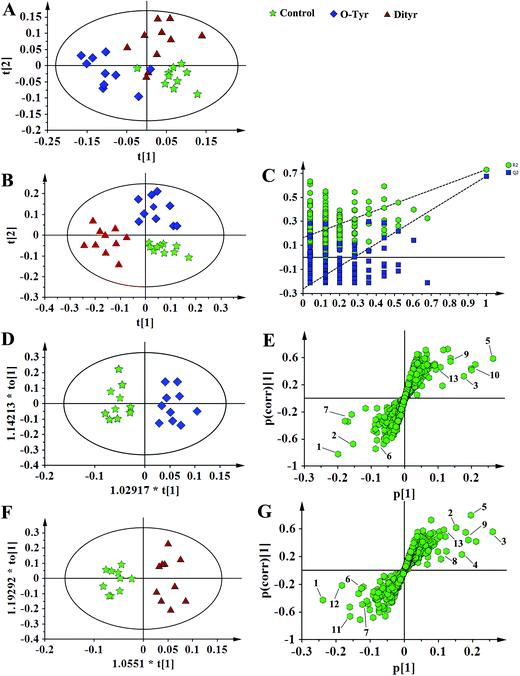
1H NMR spectral data sets from serum and urine were initially analyzed by PCA. The PCA score plots of serum and urine (Fig. 7A and 8A, respectively) showed there were no outliers within the dataset, and separations were absent in the metabolic serum and urine profiles of mice from the control group, O-Tyr group, and DT group. Therefore, PLS-DA was applied to explore the intrinsic differences between these three groups. Samples from different groups were separated and classified into three distinct clusters presented in the PLS-DA score plot (Fig. 7B and 8B, respectively). The model parameters (serum: R2X = 0.428, R2Y = 0.734, Q2 = 0.571. And urine: R2X = 0.591, R2Y = 0.753, Q2 = 0.704) and the validated models (permutation number: 200) indicated no over fitting (Fig. 7C and 8C), respectively. All the results indicated the existence of differences between the three groups.Furthermore, spectral data sets from serum and urine were analyzed by OPLS-DA to maximize discrimination between the three groups (Fig. 7D and F and 8D and F), respectively. The supervised model of OPLS-DA could develop a better separation into two clusters and contribute to the discovery of biomarkers. The O-Tyr group and DT group were well separated from the control group in the OPLS-DA scores plot of spectral data sets from serum and urine, as well as in permutation tests and variance analysis of the cross-validated residuals (CV-ANOVA) (P < 0.05), respectively. Metabolites responsible for a significant contribution to separation of the two groups are indicated in the corresponding S-plots (Fig. 7E and G and 8E and G) and marked with a number. The VIP statistics of the first principal component of OPLS-DA model (threshold ≥ 1), together with the P-value of the independent sample t-test (threshold < 0.05) were used for selecting significant variables responsible for group separation.52 The identified potential markers in serum and urine are listed in Tables 5 and 6, respectively. O-Tyr administration significantly increased the serum levels of propionate, isobutyrate, 3-hydroxybutyrate, acetoacetate, citrate, methylamine and choline, and decreased the serum levels of isoleucine, valine and α-glucose compared with the control group (P < 0.05, Table 5). Moreover, O-Tyr administration significantly increased the urine levels of acetoacetate, pyruvate, succinate, allantoin, N1-methyl-4-pyridone-5-carboxamide (4-PY), N1-methyl-2-pyridone-5-carboxamide (2-PY), m-hydroxyphenylacetate, N-methylnicotinamide and trigonelline compared with the control group (P < 0.05, Table 6). Dityr administration significantly increased the serum levels of propionate, isobutyrate, 3-hydroxybutyrate, acetoacetate, citrate, trimethylamine, choline and GPC, and decreased the serum levels of isoleucine, valine, alanine, betaine and α-glucose compared with the control group (P < 0.05, Table 5). Also, DT administration significantly increased the urine levels of acetoacetate, pyruvate, succinate, citrate, allantoin, 4-PY, 2-PY, m-hydroxyphenylacetate, nicotinamide N-oxide and trigonelline, and decreased the urine levels of p-hydroxyphenylacetate and hippurate compared with the control group (P < 0.05, Table 6).
| Metabolites | NMR chemical shift (δ) | O-Tyr vs. control | Dityr vs. control | ||||
|---|---|---|---|---|---|---|---|
| VIP | p value | Change | VIP | p value | Change | ||
| a O-Tyr vs. control, O-Tyr group compared with the control group; Dityr vs. control, Dityr group compared with the control group; VIP was obtained from OPLS-DA model. Compared with the control group, ↑ indicates a relative increase, and ↓ indicates a relative decrease in the integral value for the region containing the identified metabolite. Significant differences are set at P < 0.05. | |||||||
| Isoleucine | 1.01(d) | 1.34 | 0.041 | ↓ | 1.52 | 0.036 | ↓ |
| Valine | 1.04(d) | 1.97 | 0.049 | ↓ | 2.64 | 0.008 | ↓ |
| Propionate | 1.08(t) | 1.39 | 0.045 | ↑ | 1.76 | 0.033 | ↑ |
| Isobutyrate | 1.12(d) | 1.54 | 0.023 | ↑ | 1.87 | 0.046 | ↑ |
| 3-Hydroxybutyrate | 1.21(d) | 2.97 | 0.020 | ↑ | 2.40 | 0.012 | ↑ |
| Alanine | 1.48(d) | — | — | — | 1.27 | 0.023 | ↓ |
| Acetoacetate | 2.29(s) | 2.33 | 0.021 | ↑ | 3.58 | 0.003 | ↑ |
| Citrate | 2.54(d) | 1.99 | 0.023 | ↑ | 1.84 | 0.033 | ↑ |
| Methylamine | 2.63(s) | 1.30 | 0.011 | ↑ | — | — | — |
| Trimethylamine | 2.92(s) | — | — | — | 1.62 | 0.027 | ↑ |
| Choline | 3.20(s) | 1.66 | 0.035 | ↑ | 1.70 | 0.044 | ↑ |
| GPC | 3.22(s) | — | — | — | 1.84 | 0.031 | ↑ |
| Betaine | 3.28(s) | — | — | — | 1.06 | 0.047 | ↓ |
| α-Glucose | 5.26(d) | 1.12 | 0.047 | ↓ | 1.19 | 0.024 | ↓ |
| Metabolites | NMR chemical shift (δ) | O-Tyr vs. control | Dityr vs. control | ||||
|---|---|---|---|---|---|---|---|
| VIP | p value | Change | VIP | p value | Change | ||
| a O-Tyr vs. control, O-Tyr group compared with the control group; Dityr vs. control, Dityr group compared with the control group; VIP was obtained from OPLS-DA model. Compared with the control group, ↑ indicates a relative increase, and ↓ indicates a relative decrease in the integral value for the region containing the identified metabolite. Significant differences are set at P < 0.05. | |||||||
| Acetoacetate | 2.30(s) | 2.64 | 0.044 | ↑ | 3.57 | 0.049 | ↑ |
| Pyruvate | 2.35(s) | 2.13 | 0.037 | ↑ | 1.78 | 0.024 | ↑ |
| Succinate | 2.41(s) | 1.55 | 0.49 | ↑ | 2.01 | 0.041 | ↑ |
| Citrate | 2.68(d) | — | — | — | 1.46 | 0.032 | ↑ |
| Allantoin | 5.40(s) | 1.86 | 0.045 | ↑ | 2.49 | 0.037 | ↑ |
| 4-PY | 6.71(d) | 1.20 | 0.036 | ↑ | 1.88 | 0.020 | ↑ |
| 2-PY | 8.33(d) | 1.62 | 0.049 | ↑ | 1.54 | 0.048 | ↑ |
| p-Hydroxyphenylacetate | 6.87(d) | — | — | — | 1.27 | 0.024 | ↓ |
| m-Hydroxyphenylacetate | 7.05(d) | 1.31 | 0.041 | ↑ | 1.62 | 0.049 | ↑ |
| N-Methylnicotinamide | 8.91(m) | 1.59 | 0.027 | ↑ | — | — | — |
| Hippurate | 7.84(d) | — | — | — | 1.13 | 0.010 | ↓ |
| Nicotinamide N-oxide | 8.12(m) | — | — | — | 1.82 | 0.028 | ↑ |
| Trigonelline | 9.12(s) | 1.15 | 0.038 | ↑ | 1.22 | 0.046 | ↑ |
Discussion
Tyrosine (Tyr) is one of the major targets of protein oxidation; oxidation products of Tyr (such as Dityr and 3-NT) are universal biomarkers of protein oxidation and have been demonstrated to be associated with metabolic disorders in biological systems.18 In this study, 1H nuclear magnetic resonance spectroscopy was used to demonstrate the effects of O-Tyr and Dityr administration on metabolomes in the serum and urine of mice. Our results support the hypothesis that O-Tyr and Dityr administration can cause systemic metabolic alterations in serum and urine (Tables 5 and 6, respectively).O-Tyr and Dityr administration induced oxidative stress responses in mice. Ketone bodies, such as acetoacetate and 3-hydroxybutyrate, are the products of β-oxidation of fatty acids in mitochondria.53,54 Increased serum acetoacetate and 3-hydroxybutyrate levels suggested that O-Tyr and Dityr administration promoted β-oxidation of fatty acids. The ratio of acetoacetate to 3-hydroxybutyrate is a useful indicator of the mitochondrial redox state.55 O-Tyr and Dityr administration were found to increase the urinary level of acetoacetate, but the concentration of 3-hydroxybutyrate in urine remained unchanged. As a result, acetoacetate/3-hydroxybutyrate ratios were increased, respectively. These results further suggest a highly oxidized cell state, which can be brought about by increased oxidation of fatty acids. Lipid oxidation can generate large amounts of electrons entering a mitochondrial respiratory chain to produce excess reactive oxygen species (ROS),56 thereby causing free radical oxidative damage.57,58 Oxidative stress followed by lipid oxidation is believed to be an important cause of destruction and damage to cell membranes.59 In support of this theory, O-Tyr administration increased the serum level of choline while DT administration increased the serum levels of choline and GPC, and decreased the level of betaine compared with the control group. Choline and GPC are essential for structural integrity of the cell membrane.60,61 These compounds also have important functions in cell metabolism and signaling processes.62,63 Oxidation of choline in liver mitochondria results in the formation of betaine.64,65 Some studies have shown that betaine functions to spare choline for the formation of phospholipids.66,67 In the present study, serum levels of choline and GPC were increased and betaine was decreased. A possible explanation is that structural integrity of the cell membrane was decreased.
Moreover, O-Tyr and Dityr administration induced elevation of urinary allantoin, which is an end product of the oxidation of uric acid by purine catabolism; this result also supports enhanced oxidative stress in agreement with previous studies that considered the elevation of urinary allantoin as an indicator for oxidative stress.68,69 Furthermore, elevation of urinary 4-PY, and 2-PY in mice administration with O-Tyr, and the elevation of urinary nicotinamide N-oxide, 4-PY, and 2-PY in mice administration with Dityr suggested that metabolisms of nicotinate and nicotinamide (vitamin B3) were probably associated with progression of oxidative stress. Nicotinamide is a component of nicotinamide adenine dinucleotide (NAD) involved in intracellular respiration to oxidize fuel substrates,70 while nicotinamide N-oxide, 4-PY, and 2-PY are oxidation metabolites of nicotinamide in liver.44 Past research found that increased levels of nicotinamide N-oxide, 4-PY, and 2-PY imply elevated oxidative stress response in vivo.71 Therefore, the changes of these nicotinamide metabolites are probably also indications that O-Tyr and Dityr administration can induce oxidative stress responses in mice.
What's more, oxidative stress ultimately triggers an antioxidative response in an organism. Trigonelline and N-methylnicotinamide are the respective methylated metabolites of nicotinate and nicotinamide, which can be generated during the conversion of S-adenosylmethionine to S-adenosylhomocysteine during biosynthesis of cysteine, an essential amino acid of glutathione synthesis.51,72 A previous report suggests that elevated levels of N-methylnicotinamide and decreased levels of nicotinate indicate increased oxidative stress response in vivo.73 In our study, the elevation of urinary trigonelline and N-methylnicotinamide in mice administration with O-Tyr, and the elevation of urinary trigonelline in mice administration with Dityr, implied that mice can utilize an antioxidative vitamin B3 cycle to decrease oxidative stress induced by O-Tyr and Dityr administration. The observation of an increased level of m-hydroxyphenylacetate, also implies antioxidative responses activated by O-Tyr and Dityr administration. This is because m-hydroxyphenylacetate is a rutin metabolite and an antioxidant.74
Taken together, O-Tyr and Dityr administration induced oxidative stress responses in mice. The increased levels of AOPPs, Dityr, 3-NT, and MDA (as markers of protein and lipid oxidative injury), decreased activities of T-AOC, SOD, and GSH-PX, and declined antioxidant enzymes mRNA expression of SOD1, SOD2, and GSH-PX, also supports such a theory (Fig. 2 and 3). Moreover, this is in agreement with results from our previous study that 24 week exposure to O-Tyr induces oxidative damage in rats.18,19
A novel and unexpected finding from this study was that O-Tyr and Dityr administration promoted energy metabolism. O-Tyr and Dityr administration significantly decreased serum α-glucose levels compared with the control group. Glucose is a major substrate for energy metabolism. The decreased levels of α-glucose in serum suggested that O-Tyr and Dityr administration promoted glycolysis. Moreover, Dityr administration decreased serum alanine levels. Alanine is an important participant and regulator in glucose metabolism.75 Taken together, Dityr administration enhanced the glucose–alanine cycle metabolism. What's more, the observation of increased levels of pyruvate in urine suggested that O-Tyr and Dityr administration led to a promotion of the TCA cycle. This is because pyruvate is a key metabolite that is important in both glycolysis and the TCA cycle, and can be converted into acetyl-CoA by decarboxylation and transported into the TCA cycle under aerobic conditions.76 Increased pyruvate levels revealed that generation of acetyl-coenzymeA (acetyl-CoA) was up-regulated, resulting in promotion of the TCA cycle. In support of this view, O-Tyr significantly increased serum citrate levels, and urine succinate levels, and Dityr administration significantly increased serum citrate levels, and urine citrate and succinate levels. Citrate and succinate are important intermediates of the TCA cycle. A change in the contents of citrate and succinate would reflect a change in the metabolic rate of the TCA cycle.77,78 Increased levels of citrate and succinate in serum and urine suggests that the TCA cycle is promoted by administration with O-Tyr and Dityr. Our results also showed that levels of branched-chain amino acids were decreased by O-Tyr and Dityr administration. A possible reason is that oxidative stress-induction increased energy expenditure and also caused elevated consumption of amino acids, such as isoleucine, alanine, and valine, to provide energy.
Another novel and unexpected finding from this study was that O-Tyr and Dityr administration caused changes in gut microbiota functions. Previous research from our group has shown that oxidized casein administration induced a decrease of intestinal Lactobacillus and increased intestinal Escherichia coli and Enterococcus in mice.79 Intestinal microbes convert dietary non-digestible fibers into short-chain fatty acids (acetate, propionate, butyrate, and isobutyrate) and other nutrients that can be used by the mammalian host as energy sources or as precursors for fatty acid synthesis.80 In this study, we found that O-Tyr and Dityr administration significantly increased serum propionate and isobutyrate levels, implying that O-Tyr and Dityr administration caused changes in gut microbiota functions. In support of this theory, serum levels of methylamine in the O-Tyr group and trimethylamine in the Dityr group were increased compared with the control group. Serum nitrogenous compounds (methylamine, dimethylamine, trimethylamine, and TMAO) are microbial metabolites of carbohydrates and amino acids, which are probably produced in the lumen of the small and large intestines.23,81 Hippurate is one of the co-metabolites between gut microbiota and a host, and is a product of the conjugation of glycine and benzoic acid,82 which takes place in mitochondria of the liver and kidney.83,84 Benzoic acid is a product of the intestinal microbial degradation of aromatic compounds in food.85–87 Therefore, a change in the excretion of hippurate suggests an alteration in the functional metabolism of microbiota.88 In this study, we found a marked elevation in the levels of hippurate in the urine of mice administration with Dityr; this result also supports the view that Dityr administration changes intestinal microbial metabolism. Moreover, p-hydroxyphenylacetate is a metabolite of tyrosine processed by enteric bacteria. Decreased levels of p-hydroxyphenylacetate in urine also confirmed the association of Dityr administration with a gut microbiota disturbance.
In this work, changes in these metabolites of intestinal microflora may be explained by an increased number and/or altered activity of intestinal microflora. Mammalian metabolism is significantly influenced by interactions with the complex gut microbiota. An introduction of O-Tyr and Dityr ingestion into the mammalian system may displace baseline mammalian-to-microbial behavior, thereby disrupting microbial populations and eventually affecting metabolism. Gut microbiota significantly affect development and structure of the intestinal epithelium, digestive and absorptive properties of the intestine, and the host immune system.89 Possible disturbances of gut microbiota by O-Tyr and Dityr administration can affect health; thus, microbiological identification of specific changes in the microbiota community can help address metabolic implications of O-Tyr and Dityr administration.
Conclusions
This study has shown that O-Tyr and Dityr administration affects the urine and serum metabolome of mice. O-Tyr and Dityr administration can cause oxidative stress and some common systemic metabolic changes, including energy metabolism, glucose–alanine cycle metabolism, glycolysis, cell membrane metabolism, vitamin-B3 metabolism, and gut microbiota metabolism. Therefore, we need additional intake of antioxidant and vitamin-B3 when much more oxidative proteins are ingested. These results are very important for animal and human food safety and disease prevention. To the best of our knowledge, this study is the first in vivo report on the response of animal biological systems to O-Tyr and Dityr administration. Future studies may be directed toward a mechanistic understanding of the effects of O-Tyr and Dityr on animal tissue intermediary metabolism, especially in terms of the associations between deficiency of B vitamins and O-Tyr and Dityr administration.Conflict of interest
The authors declare that they have no conflict of interest. All authors listed have contributed to the work, and agreed to submit the manuscript. No part of the work has been published before.Ethical approval
All procedures performed in studies involving animals were in accordance with the ethical standards laid down in the guidelines of Laboratory Animals Care of Jiangsu Province (China) and in the 1964 Declaration of Helsinki and its later amendments.Acknowledgements
This research was supported by program of “Collaborative innovation center of food safety and quality control in Jiangsu Province”, the National Natural Science Foundation of China (No. 31571841), State Key Laboratory of Food Science and Technology of Jiangnan University in China (SKLF-ZZB-201609), and China Postdoctoral Science Foundation (2015M571669).References
- B. Halliwell, M. A. Murcia, S. Chirico and O. I. Aruoma, Crit. Rev. Food Sci. Nutr., 1995, 35, 7–20 CrossRef CAS PubMed.
- M. N. Lund, M. Heinonen, C. P. Baron and M. Estevez, Mol. Nutr. Food Res., 2011, 55, 83–95 CAS.
- W. Zhang, S. Xiao and D. U. Ahn, Crit. Rev. Food Sci. Nutr., 2013, 53, 1191–1201 CrossRef CAS PubMed.
- E. R. Stadtman and R. L. Levine, Ann. N. Y. Acad. Sci., 2000, 899, 191–208 CrossRef CAS PubMed.
- M. Utrera and M. Estevez, J. Agric. Food Chem., 2012, 60, 8002–8011 CrossRef CAS PubMed.
- E. R. Stadtman, Annu. Rev. Biochem., 1993, 62, 797–821 CrossRef CAS PubMed.
- T. Henle, Mol. Nutr. Food Res., 2007, 51, 1075–1078 CAS.
- Z. L. Li, L. Mo, G. Le and Y. Shi, Food Chem. Toxicol., 2014, 64, 86–93 CrossRef CAS PubMed.
- F. Xie, S. Sun, A. Xu, S. Zheng, M. Xue, P. Wu, J. H. Zeng and L. Bai, Cell Death Dis., 2014, 5, e1006 CrossRef CAS PubMed.
- Z. M. Zhong, L. Bai and J. T. Chen, Cell. Physiol. Biochem., 2009, 24, 105–114 CrossRef CAS PubMed.
- M. J. Davies, Biochim. Biophys. Acta, 2005, 1703, 93–109 CrossRef CAS PubMed.
- A. Hohn, J. Konig and T. Grune, J. Proteomics, 2013, 92, 132–159 CrossRef PubMed.
- G. Cohen, S. Yakushin and D. Dembiec-Cohen, Anal. Biochem., 1998, 263, 232–239 CrossRef CAS PubMed.
- J. R. Crowley, K. Yarasheski, C. Leeuwenburgh, J. Turk and J. W. Heinecke, Anal. Biochem., 1998, 259, 127–135 CrossRef CAS PubMed.
- J. W. Heinecke, F. F. Hsu, J. R. Crowley, S. L. Hazen, C. Leeuwenburgh, D. M. Mueller, J. E. Rasmussen and J. Turk, Methods Enzymol., 1999, 300, 124–144 CAS.
- K. Hensley, K. S. Williamson and R. A. Floyd, Free Radical Biol. Med., 2000, 28, 520–528 CrossRef CAS PubMed.
- C. Leeuwenburgh, P. A. Hansen, J. O. Holloszy and J. W. Heinecke, Am. J. Physiol., 1999, 276, R128–R135 CAS.
- Z. L. Li, Y. Shi, G. Le, Y. Ding and Q. Zhao, Oxid. Med. Cell. Longevity, 2016, 2016, 3123294 Search PubMed.
- Z. L. Li, Y. Shi, Y. Ding, Y. Ran and G. Le, Amino Acids, 2017, 49, 241–261 CrossRef CAS PubMed.
- Y. Ran, B. Yan, Z. Li, Y. Ding, Y. Shi and G. Le, Physiol. Behav., 2016, 164, 292–299 CrossRef CAS PubMed.
- D. P. Jones, Y. Park and T. R. Ziegler, Annu. Rev. Nutr., 2012, 32, 183–202 CrossRef CAS PubMed.
- J. K. Nicholson, J. C. Lindon and E. Holmes, Xenobiotica, 1999, 29, 1181–1189 CrossRef CAS PubMed.
- S. Rezzi, Z. Ramadan, L. B. Fay and S. Kochhar, J. Proteome Res., 2007, 6, 513–525 CrossRef CAS PubMed.
- C. Stella, B. Beckwith-Hall, O. Cloarec, E. Holmes, J. C. Lindon, J. Powell, F. van der Ouderaa, S. Bingham, A. J. Cross and J. K. Nicholson, J. Proteome Res., 2006, 5, 2780–2788 CrossRef CAS PubMed.
- Q. He, X. Kong, G. Wu, P. Ren, H. Tang, F. Hao, R. Huang, T. Li, B. Tan, P. Li, Z. Tang, Y. Yin and Y. Wu, Amino Acids, 2009, 37, 199–208 CrossRef CAS PubMed.
- Z. Ruan, Y. Yang, Y. Wen, Y. Zhou, X. Fu, S. Ding, G. Liu, K. Yao, X. Wu, Z. Deng, G. Wu and Y. Yin, Amino Acids, 2014, 46, 2681–2691 CrossRef CAS PubMed.
- G. Liu, Y. Wang, Z. Wang, J. Cai, X. Lv and A. Zhou, J. Agric. Food Chem., 2011, 59, 5572–5578 CrossRef CAS PubMed.
- G. Liu, T. Fang, T. Yan, G. Jia, H. Zhao, Z. Huang, X. Chen, J. Wang and B. Xue, J. Agric. Food Chem., 2014, 62, 9035–9042 CrossRef CAS PubMed.
- G. Liu, L. Xiao, T. Fang, Y. Cai, G. Jia, H. Zhao, J. Wang, X. Chen and C. Wu, PLoS One, 2014, 9, e115561 Search PubMed.
- F. A. Van Dorsten, C. A. Daykin, T. P. Mulder and J. P. Van Duynhoven, J. Agric. Food Chem., 2006, 54, 6929–6938 CrossRef CAS PubMed.
- Z. Ruan, Y. Yang, Y. Zhou, Y. Wen, S. Ding, G. Liu, X. Wu, P. Liao, Z. Deng, H. Assaad, G. Wu and Y. Yin, Amino Acids, 2014, 46, 2219–2229 CrossRef CAS PubMed.
- Y. Zhang, B. Wu, Z. Y. Zhang and S. P. Cheng, J. Hazard. Mater., 2011, 190, 515–519 CrossRef CAS PubMed.
- Q. He, H. Tang, P. Ren, X. Kong, G. Wu, Y. Yin and Y. Wang, J. Proteome Res., 2011, 10, 5214–5221 CrossRef CAS PubMed.
- C. Xiao, F. Hao, X. Qin, Y. Wang and H. Tang, Analyst, 2009, 134, 916–925 RSC.
- T. Kurahashi, A. Miyazaki, S. Suwan and M. Isobe, J. Am. Chem. Soc., 2001, 123, 9268–9278 CrossRef CAS PubMed.
- X. Zhu, H. Lei, J. Wu, J. V. Li, H. Tang and Y. Wang, J. Proteome Res., 2014, 13, 4436–4445 CrossRef CAS PubMed.
- T. W. Fan, R. M. Higashi, A. N. Lane and O. Jardetzky, Biochim. Biophys. Acta, 1986, 882, 154–167 CrossRef CAS.
- M. Kriat, S. Confort-Gouny, J. Vion-Dury, M. Sciaky, P. Viout and P. J. Cozzone, NMR Biomed., 1992, 5, 179–184 CrossRef CAS PubMed.
- G. M. Liu, G. J. Yang, T. T. Fang, Y. M. Cai, C. M. Wu, J. Wang, Z. Q. Huang and X. L. Chen, RSC Adv., 2014, 4, 23749–23758 RSC.
- J. Trygg and S. Wold, J. Chemom., 2002, 16, 119–128 CrossRef CAS.
- J. Jansson, B. Willing, M. Lucio, A. Fekete, J. Dicksved, J. Halfvarson, C. Tysk and P. Schmitt-Kopplin, PLoS One, 2009, 4, e6386 Search PubMed.
- Z. J. Zou, Z. H. Liu, M. J. Gong, B. Han, S. M. Wang and S. W. Liang, Phytomedicine, 2015, 22, 333–343 CrossRef CAS PubMed.
- D. Granato, V. M. D. Calado and B. Jarvis, Food Res. Int., 2014, 55, 137–149 CrossRef.
- D. Li, L. L. Zhang, F. C. Dong, Y. Liu, N. Li, H. H. Li, H. H. Lei, F. H. Hao, Y. L. Wang, Y. Zhu and H. R. Tang, J. Proteome Res., 2015, 14, 2237–2254 CrossRef CAS PubMed.
- G. M. Liu, T. Yan, J. Wang, Z. Q. Huang, X. L. Chen, G. Jia, C. M. Wu, H. Zhao, B. Xue, L. Xiao and J. Y. Tang, J. Agric. Food Chem., 2013, 61, 11212–11221 CrossRef CAS PubMed.
- T. Suna, A. Salminen, P. Soininen, R. Laatikainen, P. Ingman, S. Makela, M. J. Savolainen, M. L. Hannuksela, M. Jauhiainen, M. R. Taskinen, K. Kaski and M. Ala-Korpela, NMR Biomed., 2007, 20, 658–672 CrossRef CAS PubMed.
- A. Vehtari, V. P. Makinen, P. Soininen, P. Ingman, S. M. Makela, M. J. Savolainen, M. L. Hannuksela, K. Kaski and M. Ala-Korpela, BMC Bioinf., 2007, 8, S8 CrossRef PubMed.
- D. S. Wishart, D. Tzur, C. Knox, R. Eisner, A. C. Guo, N. Young, D. Cheng, K. Jewell, D. Arndt, S. Sawhney, C. Fung, L. Nikolai, M. Lewis, M. A. Coutouly, I. Forsythe, P. Tang, S. Shrivastava, K. Jeroncic, P. Stothard, G. Amegbey, D. Block, D. D. Hau, J. Wagner, J. Miniaci, M. Clements, M. Gebremedhin, N. Guo, Y. Zhang, G. E. Duggan, G. D. MacInnis, A. M. Weljie, R. Dowlatabadi, F. Bamforth, D. Clive, R. Greiner, L. Li, T. Marrie, B. D. Sykes, H. J. Vogel and L. Querengesser, Nucleic Acids Res., 2007, 35, D521–D526 CrossRef CAS PubMed.
- Y. Cai, F.-h. Hao and Y.-l. Wang, Chin. J. Magn. Reson., 2013, 1, 007 Search PubMed.
- X. J. Zhao, C. Huang, H. Lei, X. Nie, H. Tang and Y. Wang, J. Proteome Res., 2011, 10, 5183–5190 CrossRef CAS PubMed.
- C. Y. Huang, H. H. Lei, X. J. Zhao, H. R. Tang and Y. L. Wang, J. Proteome Res., 2013, 12, 537–545 CrossRef CAS PubMed.
- W. K. Ren, J. Yin, W. Gao, S. Chen, J. L. Duan, G. Liu, T. J. Li, N. Z. Li, Y. Y. Peng and Y. L. Yin, RSC Adv., 2015, 5, 59550–59555 RSC.
- M. Lopes-Cardozo and S. G. van den Bergh, Biochim. Biophys. Acta, 1974, 357, 53–62 CrossRef CAS.
- A. B. Wojtczak, Biochem. Biophys. Res. Commun., 1968, 31, 634–640 CrossRef CAS PubMed.
- P. Schonfeld, R. Bohnensack, G. Bohme and W. Kunz, Biochim. Biophys. Acta, 1983, 42, 3–13 CAS.
- Y. An, W. Xu, H. Li, H. Lei, L. Zhang, F. Hao, Y. Duan, X. Yan, Y. Zhao, J. Wu, Y. Wang and H. Tang, J. Proteome Res., 2013, 12, 3755–3768 CrossRef CAS PubMed.
- B. Halliwell and S. Chirico, Am. J. Clin. Nutr., 1993, 57, 715S–724S CAS ; discussion 724S-725S.
- D. Wu and A. I. Cederbaum, Alcohol Res. Health, 2003, 27, 277–284 Search PubMed.
- R. Mittler, Trends Plant Sci., 2002, 7, 405–410 CrossRef CAS PubMed.
- N. F. Sheard and S. H. Zeisel, Nutrition, 1989, 5, 1–5 CAS.
- S. H. Zeisel, Nutrition, 2000, 16, 669–671 CrossRef CAS PubMed.
- J. Klein, J. Neural Transm., 2000, 107, 1027–1063 CrossRef CAS PubMed.
- S. H. Zeisel and J. K. Blusztajn, Annu. Rev. Nutr., 1994, 14, 269–296 CrossRef CAS PubMed.
- P. I. Holm, P. M. Ueland, G. Kvalheim and E. A. Lien, Clin. Chem., 2003, 49, 286–294 CAS.
- J. J. De Ridder and K. van Dam, Biochim. Biophys. Acta, 1973, 291, 557–563 CrossRef CAS.
- M. T. Kidd, P. R. Ferket and J. D. Garlich, World's Poult. Sci. J., 1997, 53, 125–139 CrossRef.
- Z. M. Yao and D. E. Vance, J. Biol. Chem., 1989, 264, 11373–11380 CAS.
- M. Grootveld and B. Halliwell, Biochem. J., 1987, 243, 803–808 CrossRef CAS PubMed.
- T. Mikami, K. Kita, S. Tomita, G. J. Qu, Y. Tasaki and A. Ito, Free Radical Res., 2000, 32, 235–244 CrossRef CAS PubMed.
- H. H. Li, Y. P. An, L. L. Zhang, H. H. Lei, L. M. Zhang, Y. L. Wang and H. R. Tang, J. Proteome Res., 2013, 12, 5520–5534 CrossRef CAS PubMed.
- D. Li, L. Zhang, F. Dong, Y. Liu, N. Li, H. Li, H. Lei, F. Hao, Y. Wang, Y. Zhu and H. Tang, J. Proteome Res., 2015, 14, 2237–2254 CrossRef CAS PubMed.
- X. J. Zhao, F. H. Hao, C. Y. Huang, M. Rantalainen, H. H. Lei, H. R. Tang and Y. L. Wang, J. Proteome Res., 2012, 11, 4712–4721 CrossRef CAS PubMed.
- G. Liu, T. Yan, J. Wang, Z. Huang, X. Chen, G. Jia, C. Wu, H. Zhao, B. Xue, L. Xiao and J. Tang, J. Agric. Food Chem., 2013, 61, 11212–11221 CrossRef CAS PubMed.
- S. Baba, T. Furuta, M. Horie and H. Nakagawa, J. Pharm. Sci., 1981, 70, 780–782 CrossRef CAS PubMed.
- P. Felig, Metab., Clin. Exp., 1973, 22, 179–207 CrossRef CAS PubMed.
- P. Liao, L. Wei, X. Zhang, X. Li, H. Wu, Y. Wu, J. Ni and F. Pei, Anal. Biochem., 2007, 364, 112–121 CrossRef CAS PubMed.
- S. A. Barinova, Mikrobiologiia, 1960, 29, 21–27 CAS.
- H. A. Krebs, Adv. Enzyme Regul., 1970, 8, 335–353 CrossRef CAS PubMed.
- W. Fang, J. Sun, Z. L. Li, G. Le and Y. Shi, Chin. J. Microecol., 2012, 4, 193–196 Search PubMed.
- R. E. Ley, P. J. Turnbaugh, S. Klein and J. I. Gordon, Nature, 2006, 444, 1022–1023 CrossRef CAS PubMed.
- G. W. Tannock, Appl. Environ. Microbiol., 2004, 70, 3189–3194 CrossRef CAS PubMed.
- H. B. Lewis, J. Biol. Chem., 1914, 18, 225–231 CAS.
- J. Caldwell, J. R. Moffatt and R. L. Smith, Xenobiotica, 1976, 6, 275–280 CrossRef CAS PubMed.
- S. J. Gatley and H. S. Sherratt, Biochem. J., 1977, 166, 39–47 CrossRef CAS PubMed.
- D. T. Gilbson, Science, 1968, 161, 1093–1097 CAS.
- J. K. Nicholson and I. D. Wilson, Nat. Rev. Drug Discovery, 2003, 2, 668–676 CrossRef CAS PubMed.
- A. R. Rechner, G. Kuhnle, P. Bremner, G. P. Hubbard, K. P. Moore and C. A. Rice-Evans, Free Radical Biol. Med., 2002, 33, 220–235 CrossRef CAS PubMed.
- E. Holmes, J. V. Li, T. Athanasiou, H. Ashrafian and J. K. Nicholson, Trends Microbiol., 2011, 19, 349–359 CrossRef CAS PubMed.
- J. Xu and J. I. Gordon, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 10452–10459 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |