Open Access Article
Open Access ArticleBiofilm formation and control strategies of foodborne pathogens: food safety perspectives
Xihong Zhao *a,
Fenghuan Zhaoa,
Jun Wangb and
Nanjing Zhongc
*a,
Fenghuan Zhaoa,
Jun Wangb and
Nanjing Zhongc
aResearch Center for Environmental Ecology and Engineering, Key Laboratory for Green Chemical Process of Ministry of Education, Key Laboratory for Hubei Novel Reactor & Green Chemical Technology, School of Chemical Engineering and Pharmacy, Wuhan Institute of Technology, Wuhan 430073, P. R. China. E-mail: xhzhao2006@gmail.com; Fax: +86-27-89474980; Tel: +86-27-89474980
bCollege of Food Science and Engineering, Qingdao Agricultural University, Qingdao, Shandong 266109, P. R. China
cSchool of Food Science, Guangdong Pharmaceutical University, Zhongshan 528458, P. R. China
First published on 25th July 2017
Abstract
Foodborne pathogens are the main factors behind foodborne diseases and food poisoning and thus pose a great threat to food safety. Many outbreaks have been found to be associated with biofilms. It is well documented that biofilms have become an urgent problem in the current food industry as biofilms can renders inhabitants on such films resistant to antimicrobial agents and cleaning. In this paper, recent literature related to the biofilm formation and on the factors that influence foodborne pathogens are reviewed. In addition, the control and removal strategies of foodborne pathogens used to efficiently maintain food safety in the food industry are also discussed.
1. Introduction
Foodborne pathogens are a major cause of foodborne diseases and food poisoning and thus pose a serious threat to food safety.1 There are many kinds of foodborne pathogens, which commonly contaminate fruits, vegetables, meat, and seafood, such as Escherichia coli O157:H7, Salmonella, Vibrio parahaemolyticus, Staphylococcus aureus, Clostridium perfringens, and Listeria monocytogenes.2 In recent years, diseases caused by foodborne pathogens have become an important public health problem in many parts of the world, producing a significant rate of morbidity and mortality.3,4 According to a World Health Organization (WHO) report, foodborne diseases are considered as an emergent public health problem in both developed and developing countries.5 In addition, many other outbreaks of pathogens have been found to be associated with biofilms.6Food is rich in nutrients and suitable for the growth and reproduction of pathogens. Bacteria exist in food in two ways: bacteria can be suspended in liquid food, usually living planktonically; in solid or viscous food, bacteria can easy adhere to the surface of food materials, food processing equipment, and the surface of pipelines and can eventually form a bacterial biofilm.7 The theory of biofilms was developed from the study of bacterial plaque, which was first discovered and reported by Costerton in 1978, who subsequently found that there was a relationship between biofilms and pathogenicity,8 while later knowledge on the formation and influence of biofilm was greatly advanced. In the natural environment, a small part of the bacteria exists in a floating way, while most of the bacteria live by the way of a biofilm in order to adapt to the adverse environment. Once a biofilm is formed, its susceptibility to various chemical fungicides and environmental changes is greatly reduced, and thus it is difficult to be removed completely. Thus, high dosages of antibiotics cannot remove infectious biofilms since bacteria are well protected by the biofilm matrix. Biofilms are more resistant to antimicrobials compared to planktonic cells, which make their elimination from food processing facilities a great challenge.9 Therefore, the emergence of bacteria resistant to conventional antimicrobials clearly shows that new biofilm control strategies are required. The present review summarizes the mechanisms involved in biofilm formation and the factors influencing such a formation as well as the current and emergent control strategies, providing new insights for developing biofilm-free food processing systems.
2. Biofilm formation
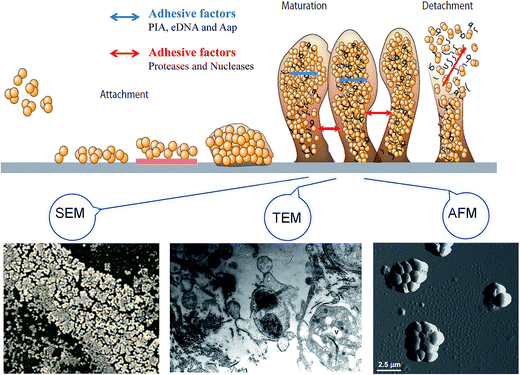
Bacterial biofilms are associated with a self-produced hydrated matrix of extracellular polymeric substances (EPSs) and are comprised of polysaccharides, proteins, lipids, and extracellular DNA (eDNA), which adhere to each other and adhere to the surface of an object. There are some features of biofilms different from planktonic bacteria: (i) the intercellular signaling system,10 such as quorum sensing (produces signaling molecules and regulates the development of biofilm), (ii) cyclic nucleotide second messengers,11 such as: cyclic-diguanosine-GMP (regulates biofilm formation and dispersal by influencing flagellar motility, and the attachment and production of extracellular polysaccharides), (iii) bap protein12 (assembles matrix scaffold proteins and builds the biofilm matrix). Most of the bacteria live in a biofilm to adapt to special environments. Biofilms are found extensively on moist surfaces, such as food, food processing equipment, water pipelines, industrial piping, ventilation, medical devices, pathological human tissues and organs and so on. A biofilm is a growth mode involving adhesion to a surface, and is usually made up of many a variety of bacteria.13 Biofilm formation is also influenced by environmental factors, such as temperature and pH, since these factors may facilitate bacterial adhesion. The biofilm formation mainly includes four stages: bacterial attachment, microcolony formation, bacterial biofilm maturation, and dispersion. Initial attachment may occur to moist food and pipeline surfaces, such as the surfaces of fruit products and food processing utensils. The initial step of biofilm formation depends on the interaction between the environmental changes and bacteria signals regulation. The attachment is influenced mostly by the surface's properties, such as roughness and hydrophobic interactions. The process of biofilm maturation and the architecture of the biofilm formation are mainly regulated by signals communicated by different bacteria. Additionally, a biofilm will be more stable with certain adhesive factors, including accumulation-associated protein (Aap), eDNA, polysaccharide intercellular adhesion. Otherwise, biofilm may tend to disperse with the addition of disruptive factors, such as proteases and nucleases. The last step of detachment is driven commonly by external environmental changes, such as various enzymes that can react with the EPSs (Fig. 1).2.1 Initial attachment
Bacteria's initial attachment consists of reversible attachment and irreversible attachment. In the case of reversible attachment, bacteria casually stick to the carrier surface by some extracellular organelles, such as flagellum, pili, and a small amount of EPSs.14 Since the attachments are not yet committed to the differentiation series of morphological changes, this stage of attachment is easy to return to the original planktonic lifestyle. The main components of the EPSs include PIA, eDNA, protein, lipids, and so on. When these environmental factors change, it promotes the attachment to the surface, driving attachment toward irreversible attachment (Fig. 1).15 In the stage of reversible attachment, the level of EPSs secreted by bacteria reaches a certain degree, which generates a strong interaction between the bacteria and the surface, and then the biofilm enters the stage of irreversible attachment. From the stage of reversible attachment to the irreversible attachment, the time is as short as several minutes.16 EPSs secreted by cells in the stage of irreversible attachment enhance the cell-to-surface adhesion, which requires a strong shear or disinfectant to remove such a bacterial attachment.2.2 Microcolony formation
After irreversible attachment, with the accumulation of a certain number of bacteria and their secretion of extracellular polymers, the binding between bacteria and the surface becomes close under appropriate growth conditions. In the meantime, the process of micro-colonies being formed increases significantly, and gradually small colonies are formed (Fig. 1). For Staphylococcus epidermidis, in the irreversible phase, the polysaccharide intercellular adhesin (PIA), encoded by the icaADBC locus, is the main component mediating intercellular adhesion.17 Among biofilm formation in Pseudomonas aeruginosa, for irreversible attachment, the SadB protein level appears to have a positive correlation, but RpoN and FleR appear to negatively affect the SadB levels.18 Research has found that lap mutants were unable to progress from the reversible attachment to the irreversible attachment stage of biofilm development, since the lap mutant strains are impaired in an early step in biofilm formation and are unable to develop the mature biofilm, so a lapEBC-encoded ABC transporter is required for biofilm formation by Pseudomonas fluorescens WCS365 in the irreversible attachment stage.19 Quorum sensing occurs mainly in the microcolony formation stage. Quorum sensing is a bacterial intercommunication system for when the population density of bacteria reach a certain level. The result of quorum sensing is the secretion of signal molecules regulating the expression of the corresponding gene, and secreting EPSs.202.3 Biofilm maturation
The attachment of small colonies grows into the mature biofilm with the characteristic three-dimensional (3D) biofilm structure. The attachment between cells and carriers and cells-to-cells mainly relies on the EPSs so that the colony can withstand a certain degree of mechanical pressure to prevent shedding from the carrier surface. Cell lysis and released extracellular DNA are critical for the initial biofilm attachment, and released eDNA remains an important matrix component in biofilm maturation. The regulation of quorum sensing and surfactants has extensive importance for biofilm maturation processes. The appearance of a mature biofilm can be justified obviously by using a microscope, such as scanning electron microscopy (SEM), atomic force microscopy (AFM), and transmission electron microscopy (TEM) (Fig. 1). The mature biofilm is composed of three layers, where the inner layer is a regulating film, which forms a network structure instead of covering completely the surface of the living or non-living body; the middle layer is a compact microbial basement membrane; the outermost surface film is where the plankton lives. The mature biofilm, with pathogenicity, which increases resistance to antibacterial agents, is more difficult to contact and remove bacteria, and the detachment of the biofilm often leads to human infection because of S. epidermidis and other foodborne pathogens.21 However, biofilm maturation can be influenced with disruptive factors, since enzymes can react with the biofilm matrix and cause the biofilm to change into free bacteria finally.2.4 Dispersion
The outermost film is inhabited by plankton, which are free to float to other areas and form a new biofilm in some special conditions. It has been shown that biofilm bacteria can be detached by disruptive factors, such as catabolite repression, nutrient limitation, and secretory proteins (Fig. 1). More biofilm formation can occur though, since the maturation biofilm will change into planktonic bacteria in an appropriate environment. The reasons for biofilm separation include external environmental effects, such as increased shear stress and a lack of nutrient supply, and internal biochemical changes in bacteria, such as endogenous enzyme degradation, EPS or surface-binding protein releasing.22 P. aeruginosa biofilm was found to be regulated by the gene and signal molecules, and the biofilm dispersion locus bdlA gene is mutated under the management of biocide H2O2, suggesting that BdlA protein might be the signal molecules leading to biofilm dispersal.23 Furthermore, P. aeruginosa is regulated by two quorum-sensing systems, LasI/LasR and RhlI/RhlR, and quorum sensing promotes biofilm dispersion at least by reducing the synthesis of rhamnolipids.24 The detachment from the biofilm is thought to be a key reason for the spread of pathogens, so it is important to study the mechanism of biofilm detachment and its inhibition for preventing foodborne pathogens infection.3. Factors behind biofilm formation
3.1 Bacterial factors
The initial attachment of bacteria involves casually sticking to the carrier surface by flagellum, pili, and fimbriae. Flagella and type IV pili were found not to be necessary for P. aeruginosa initial attachment, as wild type, flagella, and type IV pili mutants formed biofilms with different structures, but they also contributed to the biofilm architecture and allowed for the formation of voluminous biofilms.25 Bacteria fimbriae integrate with the surface of biological materials by non-specific electrical attraction or hydrophobic interaction, which is one of the important means of biofilm formation.26 In the process of Klebsiella pneumoniae colonization, type 3 fimbriae promote the attachment of bacteria to non-biological surfaces and the formation of K. pneumoniae C3091 biofilm, compare to type 1.27 Fluorescence detection of the adherent fimbriae SEF17 in Salmonella enteritidis confirmed the association of the fimbriae with aggregated cells of the biofilm.28 The microbial colonization of surfaces is a very complex process and depends on the generation of extracellular molecules. EPSs are the major metabolites of bacteria, which stick the cell to other substances.29 EPSs account for a large proportion of the dry weight of the biofilm. Protein and polysaccharides are the main components of the EPSs and play a major role in the biofilm formation. The production of EPSs occurs in the whole stage of biofilm formation.30 EPS molecules enhance the interaction among microorganisms, which determines the process of the cell aggregate formation on a solid surface. Studies have shown that modification of the EPS components can affect the formation of virulence and pathogens related to biofilm-associated infections.31 For the Pseudomonas fluorescens strain B52, the polysaccharides associated with the planktonic forms are different from biofilm polysaccharides, and one study reported that the polysaccharides in the biofilm had glucuronic and guluronic acids as the main components, showing that biofilm formation produces more polysaccharides than free bacteria.32 In conclusion, it finds that flagellum, pili, fimbriae, and the EPSs of bacteria secretion play a role in promoting biofilm formation.3.2 Surface characteristics
While biofilm forms in a certain material carrier, the character of the surface, such as the carrier interface, electrostatic, hydrophobic, interface roughness, and morphological characteristics, will affect the cell attachment and biofilm formation, with the roughness of the interface in particular playing a major role. Furthermore, the effect of surface roughness on the biofilm is conditional. It was found that L. monocytogenes formed a strong biofilm, which differed in surface roughness at 30 °C. In contrast, the ability of L. monocytogenes to form a biofilm at 15 °C decreased with the decreasing roughness of the surface.33 In addition, there are different consequences for the roughness of a nanoscale surface that affect bacterial attachment. E. coli, P. aeruginosa, and S. aureus show a greater tendency to adhere to “nanosize” surfaces.34 Furthermore, the chemical composition, critical surface tension, surface energy, hydrophilicity or hydrophobicity, and surface charge on the surface of the adherent material also have a great influence on bacterial attachment, leading to a different ability to adhere to a surface. It was found that the attachment of the hydrophobic matrix of the strains was greater than the hydrophilic surface, and due to the hydrophilic surface, i.e., polyurethane, bacterial attachment decreased.35 Proteins and high concentrations of other nutrients promote the attachment of a biofilm. A study showed that nutrient-rich media containing glucose, vitamin K, and other nutrients enhanced the formation of Enterococcus faecalis biofilms, which reached maturity at 72 h.36 In summary, the characteristics of a surface, such as roughness, hydrophilicity or hydrophobicity, and nutrients, can contribute to biofilm formation as the environmental conditions change.3.3 Gene regulation
Generally, biofilm formation is mediated by the production of the polysaccharide intercellular adhesin (PIA), which is synthesized with the enzyme encoded by the ica operon. However, some bacteria may form a protein biofilm, regulated by fibronectin-binding proteins (FnBPs),37 which forms PIA-dependent and PIA-independent biofilms. Among these, the FnBPs-mediated protein biofilm does not exist in fibrous, net-like structures, and ethanol can induce protein biofilm formation.38 Another study of the S. epidermidis icaC IS256 insertion mutant found that the transcription of aap encoding the accumulation-associated protein Aap was enhanced in a producing a proteinaceous biofilm, while transcription of the Bap-homologous protein gene bhp was down-regulated. The results showed that PIA-independent biofilms differed from the wild type in gene regulation.39 PIA is the main biofilm component, which is also known as poly-N-acetylglucosamine (PNAG).39 PIA with its positive charge can be easier to combine with a negative charge surface. The icaADBC locus includes four genes (A, D, B, C), which are affected by environmental conditions. IcaA encodes the N-acetylglucosamyl transferase, being responsible for the synthesis of PIA, and the co-expression of the icaD in vitro increases the activity. IcaB encodes deacetylase, which is responsible for the maturation of the PIA, and the trans-membrane protein of icaC appears to be involved in the polysaccharides involved in externalization and extension.40 The transcriptional level of ica, which encodes the essential enzyme for PIA biosynthesis, was reduced after treatment with thiols, resulting in the inhibition of PIA biosynthesis and an obvious inhibition of biofilm formation in S. aureus.41 Real-time PCR showed that the mutation in the sarA gene led to down-regulation of the ica operon transcription, and that sarA can activate the S. aureus development of biofilm by both enhancing the ica operon transcription and by suppressing the transcription of either the protein involved in the turnover of PIA/PNAG or a repressor of its synthesis.42 In addition, the pel genes encoded enzymes were involved in the production of polysaccharides. P. aeruginosa PAK strains, which construct pel gene mutants, and there are serious flaws in mutants in the solid attachment process of the initial biofilm formation, proposed pel gene cluster regulate biofilm formation of Gram-negative bacteria. However, pel gene clusters may be conserved in other Gram-negative bacteria, since the pel mutants in P. aeruginosa PAK had little influence on biofilm initiation.43 Here, the pel operon associated with quorum-sensing regulation may regulate the chemical complementation of 3-oxo-dodecanoyl homoserine lactone to restore the biofilm formation ability, since transcription of the pel operon was greatly reduced in lasI and rhlI mutants.44 To sum up, except for the common gene regulation of the ica operon, like the pel operon, other gene regulation will likely be discovered in the future.3.4 Quorum sensing
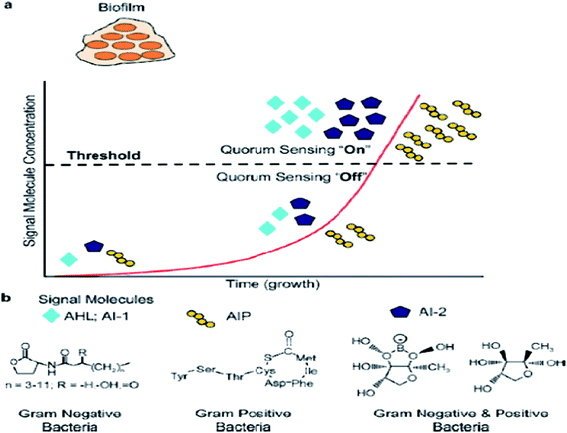
In previous studies, quorum sensing has been demonstrated to play an important role in biofilm formation. Quorum sensing is a cell-to-cell communication process that enables bacteria to secrete special signals in response to changes in the cell density of the surrounding microbial community. Bacteria sense the quorum sensing density through these molecules. The quorum sensing can be turned “off” with a few bacteria in the environment. When the number of bacteria reaches a certain threshold, the activation of the corresponding genes are expressed in cells, and as more signal molecules release, it can lead to bacteria secreting toxins, biofilm formation, and bioluminesense with the quorum sensing “on”45 (Fig. 2a). Quorum sensing regulates the whole stages of biofilm formation, which activates certain genes in the bacteria to secrete the extracellular matrix, such as EPS and protein, and gradually a full biofilm structure is formed, in which the bacteria have drug resistance. The normal operation of a quorum-sensing system requires the participation of signal molecules, and different types of bacteria secrete different signal molecules, such as AHL secreted by Gram-negative bacteria, AIP secreted by Gram-positive bacteria, and AI-2 secreted both by Gram-negative and Gram-positive bacteria (Fig. 2b). However, the regulation of biofilm formation is complex and involves several regulatory mechanisms.46 AI-2 and LuxS seem to act as a regulator of biofilm formation and LPS synthesis in sessile K. pneumoniae cells, since the mutations in LuxS and the AI-2 transport systems induced an increase in the expression of two lipopolysaccharide-synthesis-related genes.47 P. aeruginosa employs acyl homoserine lactone signals, with two AHL signaling systems, to express genes for the production of polysaccharides, rhamnolipid, and other virulence factors.46The signal molecules of the Gram-negative bacteria quorum-sensing system are mainly N-acyl homoserine lactones (AHLs) (Fig. 2b), which are synthesized by LuxI-type enzymes (AHLs synthetase) under certain environments. AHLs, which are able to diffuse freely through the bacterial membrane, can reach the extracellular cells through specific channels and can accumulate in the environment. When the AHLs concentration in the environment reaches a certain threshold, AHLs automatically enter the cell and combine with intracellular LuxR (regulatory protein), which causes an expression of the bacterial cell-specific functional gene.48 4-Coumaroyl-homoserine lactone synthase is another AHL synthetase, which produces p-coumaroyl-homoserine lactone (HSL). Many Gram-negative bacteria produce and detect several autoinducers, such as C4-HSL, 3OH-C4-HSL, and lso valery-HSL, and different kinds of HSL bind to the receptor through different structures.49 Aeromonas hydrophila, the Gram-negative bacteria, possesses an N-acylhomoserine lactone (AHL)-dependent quorum-sensing system based on the ahyRI locus. Research showed that the ahyI mutant cannot produce C4-HSL or form a mature biofilm, but the addition of exogenous C4-HSL partly restores the biomembrane differentiation defects, and it has been supposed that the A. hydrophila LuxR homolog is responsible for facilitating biofilm maturation via C4-HSL/C6-HSL.50
The signal molecules of the Gram-positive quorum-sensing system are mainly autoinducing peptides (AIPs) (Fig. 2b). Bacteria sense the environment through two-component proteins and transmit the signal to the cell through the mechanism of phosphorylation and dephosphorylation, and regulate the expression of related genes in a cell.51 Since AIP cannot pass through the cytoderm by itself, quorum sensing will play a role in the ATP-binding cassette transporter delivery system or in other membrane channel protein transport to the extracellular systems.52 The auxiliary gene regulator (agr) operon in S. aureus possesses the agr quorum-sensing system in response to AIPs.53 When reaching the threshold level, AIP will bind AgrC and encode the necessary attachment molecules for biofilm formation. In the agr quorum-sensing system of S. aureus, the repression of agr is necessary to form a biofilm, and the addition of AIP or the depletion of glucose can lead to agr activation in the biofilm initial detachment.54
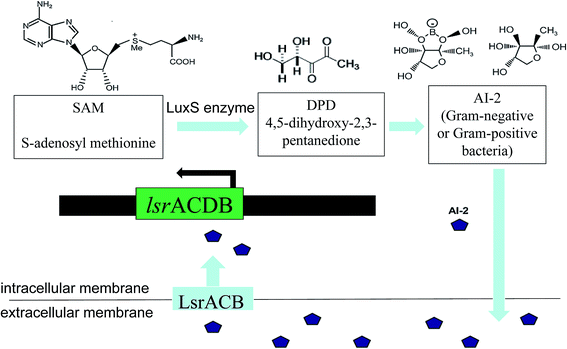
The LuxS/2 autoinducer2 (AI-2) quorum-sensing system is widely present in Gram-positive and Gram-negative bacteria, known as the interspecies quorum-sensing system (Fig. 2b). Its signal molecule, AI-2, is considered to be a universal signal molecule and involved in regulating the biofilm of many bacteria as the number of cells increases.55 It has been reported that the synthesis, secretion, and signal transfer of AI-2 is regulated by the LuxS-regulated operon in E. coli. The precursor to AI-2 is 4,5-dihydroxy-2,3-pentanedione (DPD), a product of the LuxS enzyme in the catabolism of S-ribosylhomocysteine, which is a byproduct of the S-adenosyl methionine (SAM) metabolism, and SAM is widely present in various microorganisms. Since the intra-molecular cyclization of DPD in organisms may generate different AI-2 molecules, different AI-2 signals are produced by different kinds of bacteria. When AI-2 has been formed in the AI-2 biosynthesis pathway, it is exported into the extracellular medium directly by the lipid bilayer. With more bacteria growth, AI-2 increasingly accumulates in the extracellular membrane. Finally, through the Lsr transporter, which is synthesized in the Lsr operon, AI-2 is imported into the intracellular membrane. When the accumulation of AI-2 reaches a certain threshold, the extracellular membrane AI-2 will enter into cells by LsrACB, resulting in the expression of the lsrACDB gene relevant to biofilm formation (Fig. 3). Researchers found that the LuxS mutant, without a lack of AI-2, enhanced biofilm formation and reduced adhesion ability.56 AI-2 could increase the biofilm formation and adhesion of A. pleuropneumoniae independent of LuxS, and growth under iron-restricted conditions could be controlled by LuxS. AI-2 can decrease biofilm formation in S. aureus via icaR activation.57 AI-2 may play different roles in different bacteria, since several AI-2 analogs were found to function as synergistic quorum-sensing agonists in Vibrio harveyi, while two of these analogs inhibited quorum sensing in P. aeruginosa.58 However, other researchers have shown that the exogenous addition of in vitro synthesized AI-2 has an inhibitory effect on biofilm formation by B. cereus and promotes dispersion of the biofilm.59 The study of the Streptococcus oralis LuxS mutant found that AI-2 acts as an interspecies signal and its concentration is critical for biofilm formation, and also an optimal concentration of DPD was determined, above and below which biofilm formation was suppressed.60 Many studies have shown that the inhibition of quorum sensing can depress the formation of biofilms, and have also demonstrated that baicalin, flavonoids, emodin, and other quorum-sensing inhibitors play an important role in the inhibition of biofilm formation.61
3.5 Environmental factors
Environmental factors regulate bacterial biofilm formation through several pathways. Mature biofilms change with the environmental conditions in order to obtain the best nutrients to survive and reproduce. At either extreme, nutrient-rich, or very nutrient-poor conditions, biofilms will be inclined to be planktonic.62 Researchers have found that the biofilm formation of Lactobacillus rhamnosus GG in vitro is associated with a gastrointestinal environment, including a low pH, high osmolarity, bile, mucin, and nondigestible polysaccharides.63 The adhesion stage of the E. coli O157:H7 biofilm, in addition to the functioning of quorum sensing and hypertonic pressure promotes bacterial adhesion. Bacteria formed a biofilm in LB broth containing 1 M sucrose, while E. coli K-12 IAM1264 did not form a biofilm in LB broth containing 1 M NaCl, indicating that there were different concentrations of hyperosmotic environment in different solutions during the biofilm formation.64 The biofilm formation of V. parahaemolyticus varies depending on the temperature, where a higher temperature (15 °C to 37 °C) induces stronger biofilm formation, but a lower temperature (4 °C or 10 °C) leads to the attachment of bacterial cells as monolayers.65 At a temperature of 20 °C or 37 °C, the stx coding gene of biofilm could be transferred to potentially pathogenic E. coli O157:H7, where the infection rate at 37 °C is higher than at 20 °C.66 The biofilm toxicity mechanism of Vibrio cholerae may be the adaptive mechanism for the environmental reactions.67 However, another study showed that V. cholerae and P. aeruginosa produced less biofilm at the higher temperature of 37 °C compared with at 30 °C, indicating that the temperature of biofilm formation was different according to different strains of bacteria. In addition, at pHs 5.5, 7.5, and 8.5, pathogenic bacteria, such as P. aeruginosa, produced more biofilm at higher pH.68 In summary, the temperature, pH, and surface conditions also may play a vital role in biofilm formation under quorum-sensing regulation.4. Control and removal strategies
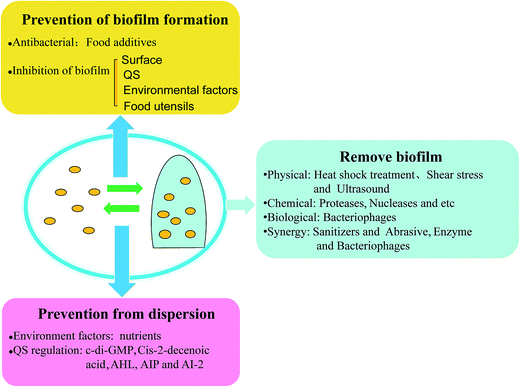
Foodborne pathogenic bacteria cause bacterial food poisoning, which seriously endangers human health and can cause great economic losses, and have become one of the most prominent food hygiene problems. Foodborne pathogens and their biofilm widely exist in the natural environment and in many areas in life, and they can form in a variety of biological materials, such as, surfaces and mucosa in vivo.69 Biofilms can not only protect the bacteria resistance against clinical antibiotics and fungicides in industrial facilities, but can also help the bacteria fight against the body's immune clearance effect. Thus, foodborne pathogens and their biofilms can cause great harm to human health, and make food hygiene increasingly difficult to remove such harmful materials, so effective methods need to be taken to prevent and remove biofilm. It's well known that bacteria widely exist on food surfaces, pipelines, air, and other food contact surfaces. Furthermore, it's easy to remove bacteria by using disinfectant compared with the removal of biofilm. However, in certain environments, bacteria can develop into biofilm. Many studies have also come up with countermeasures, and the prevention of biofilm initial attachment by disinfection of food contact surfaces is one of the most important strategies, such as with essential oils and bacteriophages. In a study on biofilm control, vanillin addition showed the most substantial reduction of biofilm formation, since vanillin may interfere with quorum-sensing molecules.70 There are different strategies for different parts of the cycle of biofilm development: (i) disinfection and cleaning, before a biofilm develops, (ii) removal of mature biofilm using both mechanical treatment and chemical reagents, and the (iii) prevention of biofilm dispersion, to hinder other biofilm formation (Fig. 4).4.1 Prevention of biofilm formation
(i) Measures aimed at the contact surface. In a study, it is easy to form the biofilm in outside moisture and on nutrient-rich surfaces, so it vital to maintain a dry and clean environment. A good cleaning process can remove food residues and other compounds, preventing the accumulation of nutrients, which decreases the conditions for biofilm formation. Cleaning-In-Place (CIP) is a process that allows the entire system to be cleaned without manual disassembly of the entire system or manual cleaning by an operator, which involves spraying the entire plant on the surface or inside. The equipment can be effectively cleaned in time with increased turbulence and flow. The addition of caustic and nitric acid additives can enhance the removal of bacterial biofilm of CIP.74 Also, with regard to an S. aureus thick biofilm, CIP cleaning agents, such as strong alkaline CIP cleaning agents, have been demonstrated to function effectively for the removal of S. aureus biofilm, so CIP cleaning with strong alkaline acidic CIP cleaning agents is recommend to remove biofilm completely in the pipelines in the food environment.75 Studies have shown that different food matrices have a strong impact on the efficiency of L. monocytogenes in cleaning and disinfection. Cleaning and disinfection also lead to the inhibition of biofilm formation of the initial stage without sterilization.33 Many different disinfectants can be used for cleaning, including surfactants and alkaline products, which can suspend and dissolve food residues. Many studies have been performed that show that sodium hypochlorite (NaClO), hydrogen peroxide (H2O2), and ozone (O3) can be used as disinfectants. Rinsing with 3% NaClO on the root canal has a strong bactericidal effect on biofilm. Another study showed that E. faecalis cells in the starvation phase are responsive to 5.25% NaClO during biofilm formation, but the impact on biofilm decreased as the biofilm matured.76 H2O2, at a concentration of 3% and 5%, rapidly eradicates S. epidermidis biofilms, as hydrogen peroxide at 3% and 5% led to a log reduction of the biofilm after incubation for 5 min, while povidone–iodine was less effective.77 With high concentrations of gaseous and aqueous O3 applied dose-dependently, biofilm cells can be almost eliminated by highly concentrated aqueous O3 (20 μg ml−1) compared with invalid H2O2.78 Essential oils are capable of inhibiting or slowing the growth of bacteria since they contain a wide variety of secondary metabolites. Natural antimicrobial agents, such as the essential oil of Satureja thymbra, can be used as food biofilm disinfectants, and have been shown to remove the biofilm of Staphylococcus simulans and Lactobacillus formed on stainless steel.79 Kavanaugh and Ribbeck demonstrated that cassia, peru balsam, and red thyme essential oils are more effective in disc diffusion assay in eradicating Pseudomonas and S. aureus, compared with selective important antibiotics.80 However, a biofilm cannot be removed completely without a disinfectant. Furthermore, nutrient limitation is one way to control biofilm development without increasing the use of disinfectant. For instance, biofilm development can be limited by a combination of organic carbon removal and enhanced disinfectant persistence.81
(ii) Measures aimed at quorum sensing. Biofilm formation is regulated by quorum-sensing molecules, which in many Gram-positive bacteria are small peptides, whereas in Gram-negative bacteria, these are N-acyl-l-homoserine lactones. Therefore, the use of something that can disturb the production or transportation of quorum-sensing molecules can decrease biofilm formation. In addition to traditional disinfectants, such as chlorine, hydrogen peroxide, now there are also a large number of studies on quorum-sensing inhibitors or quorum quenchers. Last but not least, the selection of quorum-sensing inhibitors depends on the types of quorum-sensing system. Studies have shown that a variety of traditional Chinese medicine components inhibit the formation of biofilm by inhibiting quorum sensing. Emodin and rographolide have a strong permeability in S. epidermidis biofilm.82 Extracts of radix, radix scutellariae, and rhubarb can improve the antibacterial activity with penicillin, gentamicin, and other four kinds of antibiotic.83 Rosa rugosa tea polyphenol extract exhibited inhibition in the swarming motility (84.90% and 78.03%) and biofilm formation (67.02% and 72.90%) of E. coli K-12 and P. aeruginosa PAO1, showing that polyphenols and flavonoid may block bacterial quorum-sensing systems and biofilm formation.84 In addition, besides synergistic effects on the sterilization of in vitro biofilms, when treating mice with both quorum-sensing inhibitors and tobramycin, there could be achieved synergistic antimicrobial efficacy, showing synergistic effects on the killing of P. aeruginosa biofilms in vivo.85 Moreover, quorum quenchers are of interest since they can weaken the virulence factors of bacteria and promote the removal of biofilms. In this study, those bioactive agents from coral associated bacteria could inhibit the production of quorum-sensing-dependent virulence factors, such as swarming motility and biofilm formation in Serratia marcescens, which is promising to treat drug resistant bacteria pathogens.86
(iii) Measures aimed at environmental factors. Temperature, pH, and water hardness are important factors affecting the effectiveness of disinfectants. Generally, high temperature and low pH could be in favor for biofilm removal.68 Regarding environmental stress factors, a study of S. aureus reported that most bacteria formed more biofilm at 37 °C, and had a higher biofilm production with the addition of glucose. Thus, to choose the safe environmental condition is essential to reduce the chances of biofilm formation on food contacts surfaces and to insure food safety.87
(iv) Measures aimed at food utensils. It is widely known that the antibacterial ability of silver nanoparticles (AgNPs) have been used in the rapid detection field, while the anti-biofilm activities of AgNPs are still evolving. A P. aeruginosa biofilm was inhibited by the addition of 20 μg ml−1 of AgNPs with an inhibition rate of 67%. However, the anti-biofilm activities of AgNPs decreased as the bacterial concentration was enhanced.88 The anti-biofilm ability of AgNPs depends on the form of application, 0.1% AgNP could not disrupt the E. faecalis biofilm structure, while 0.02% AgNP gel caused the disruption of the structural integrity of the biofilm, which showed that AgNPs can be used as a medicament to eliminate biofilms.89 With regard to the anti-biofilm activities of AgNPs, the application of AgNPs on food utensils can prevent bacteria growth and biofilm formation. Moreover, the usage of AgNPs on utensils, such as coating on surfaces and vessels, is promising to greatly decrease the application of antiseptics.
4.2 Removal of biofilms
It is difficult to remove a biofilm once formed, since mature biofilm have been shown to be highly resistant to a lot of antimicrobials, let alone the bacteria protected in biofilm. The removal of mature biofilm has thus aroused growing concern. It is obvious that a single method cannot disrupt biofilm efficiently. Here, treatments are classified as physical, chemical, biological, and synergetic treatment to improve the efficiency of biofilm disruption.4.3 Prevention of dispersion
The biofilm dispersion-spontaneous regulation of biofilm, the last step in the biofilm developmental cycle, not only produces another new biofilm, but also causes human infection by free bacteria, since dispersed bacteria may carry different phenotypic properties and transcriptional profiles compared to both biofilms and planktonic bacteria, leading to a significantly increased virulence. Thus, it is necessary to control the biofilm dispersion to prevent serious bacterial infection. Does this mean that biofilms cannot be disturbed? In fact, biofilms cannot be dispersed by the regulation of bacteria themselves, because the dispersed single bacteria will be easier to form another biofilm, and it may occur that the bacteria has a new toxin.24 As mentioned above, it is the signal molecule of quorum sensing that triggers biofilm dispersion. Biofilm dispersion is both affected by the environment factors and via regulation by quorum sensing, so its prevention mainly depends on environmental changes. Therefore, prevention from dispersion depends on control of the environment factors and quorum-sensing signals. Chemotaxis protein BdlA (polypeptide chains) activate in response to environmental cues provides biofilm dispersion signal and elevates c-di-GMP levels and protease ClpP, which is required for P. aeruginosa biofilms to disperse. It is a cyclic di-GMP that could degrade the protein BdlA and phosphodiesterases, resulting in biofilm dispersion.23 One study demonstrated that the dispersion deficient phenotype of the bdlA mutant was confirmed as an obstacle to biofilm development since the microscopic observations of bdlA mutant was similar to the treatment with the biocide H2O2. The usage of biocide H2O2 may obstruct biofilm dispersion.111 When the dispersed gene is inhibited, or the presence of substances in the environment impede dispersion from occurring, the mature biofilm cannot enter the planktonic state, which prevents pathogens from new attachment and prevents human infection to a certain extent. Nutrients increase, such as succinate, glutamate, glucose, and induce cell dispersion of the biofilm, while a nutrient reduction can inhibit dispersion.23 Cis-2-decenoic acid, an organic compound produced in P. aeruginosa, which is functionally and structurally similar to the short chain fatty acid (cell-to-cell communication molecules), has shown the ability to induce the dispersion of P. aeruginosa PAO1 biofilm,112 and so preventing the production of cis-2-decenoic acid is an obstacle to biofilm dispersion. Studies have also shown that mimicking the quorum-sensing signals would induce dispersion of the biofilm, which also means hindering the regulation of the quorum-sensing signals, such as AHL, AIP, and AI-2, to prevent bacteria formation and detachment.24 Signals (febrile-range temperatures, norepinephrine, extracytoplasmic ATP, and nutrient increase availability) induce the release of bacteria from biofilms into free bacteria in a newly developed biofilm model, causing the infection of Streptococcus pneumoniae. The study showed that interkingdom signals were recognized by bacteria to stop detachment.113 Hence, to search for chemicals for preventing biofilm dispersion is meaningful for biofilm disruption and bacteria removal.5. Conclusions and future perspectives
Foodborne pathogens can attach and produce EPS on many food matrices and environmental surfaces. Pathogenic bacteria can also coexist within biofilms with other environmental microorganisms. The formation of pathogenic biofilms depends not only on genetic bases and regulation, but also on properties of the substratum and bacterial cells as well as environmental factors, including temperature, pH, surface characteristic, and nutrient components. Pathogenic biofilms have attracted great interest from many research groups involved in food safety. Foodborne pathogens in biofilms formed in different food industry settings are a source of food contamination. With the increased demand for fresh, ready-to-eat, processed foods and for better food safety, many studies are needed to address biofilm control and removal in the food industries.The control of bacterial biofilms in food processing facilities is vital in food processing environments. An early control strategy mainly employs chemical cleaning, sanitation, and equipment to prevent the biofilm formation, such as CIP.114 The use of a control strategy in food processing facilities is promising to enhance the prevention of bacteria and biofilms. Cleaning and disinfection and the use of effective cleaning procedures with chemical disinfectants are usually mentioned to remove more microorganisms and prevent microbial growth on surfaces. Recently studies have similarly focused on the emerging strategies to control biofilms in the food industry, including cleaning and disinfection, enzymatic treatment, and phages treatment. Simões et al.115 regarded the use of enzymes, phages, and bioregulation as a green strategy for biofilms control. It is suggested that the metabolites of some microorganisms can interfere with biofilm formation and development. Surfactin from B. subtilis caused biofilm dispersion without affecting cell growth, preventing biofilm formation in S. enterica and E. coli O157:H7. In addition to biological and chemical alternatives, new physical processes were suggested in this review, such as laser decontamination devices and the conception of an antimicrobial coating on food surfaces. The application of these physical treatments for bacteria attachment prevention still needs to be further investigated. It is surprising to propose protective biofilms as barrier microflora against the potential foodborne pathogens.116 Many previous studies have been sure that disruption of the biofilm could kill bacteria, but this study holds different views. It finds that resident microflora caused the inhibition of the pathogen L. monocytogenes, guessing that other resident microflora may form a biofilm, and these protective biofilms could then fight against the formation of foodborne pathogenic biofilms by the activity of competition, exclusion, and displacement. The feasibility of the protective biofilms still needs to be taken into deep consideration, since protective biofilms pose a potential threat. Once a biofilm is destroyed by disruptive factors, foodborne pathogens can rapidly form a biofilm.
Combination technology is a promising new approach, which involves integration with two or more different control techniques to control biofilm. The combination treatments of chemical agents with UV irradiation were shown to remove a P. aeruginosa biofilm effectively. In a recent review, conventional and emergent control strategies of biofilms were discussed abundantly, since such control strategies according to different food areas are worth recommending.117 The cycle of biofilm development may exert a great influence on the chosen biofilm control strategy, since different periods of biofilm should be treated with different methods, resulting in a high efficiency of biofilm removal, which provides a new and targeted solution as a biofilm control strategy.
Nowadays, the conventional control strategies, including good production hygiene, a rational running of the process line, and the effective use of cleaning and disinfectant products, are still used and being developed further. However, due to the increased resistance of biofilms to conventional disinfection processes, a more efficient and environmental friendly control strategy is indispensable to satisfy the needs of food safety in food processing systems. It should be considered that disinfection methods should provide ideal, cost-effective results without any adverse effects on human health and the environment. Therefore, we strongly recommend combination technology because it provides synergistic effects and reduces the material and energy consumption. In addition, the inhibition of biofilms and quorum sensing by natural biological agents will also be helpful for solving biofilm problems.
Acknowledgements
This work has been supported by the National Natural Science Foundation of China (Grant No. 31501582).References
- S. P. Oliver, B. M. Jayarao and R. A. Almeida, Foodborne Pathog. Dis., 2005, 2, 115–129 CrossRef CAS PubMed.
- X. H. Zhao, J. L. Zhong, C. J. Wei, C. W. Lin and T. Ding, Front. Microbiol., 2017, 8, 580 Search PubMed.
- X. Zhao, C. W. Lin, J. Wang and D. H. Oh, J. Microbiol. Biotechnol., 2014, 24, 297–312 CrossRef CAS PubMed.
- X. Zhao, C. Wei, J. Zhong and S. Jin, Biotechnol. Biotechnol. Equip., 2016, 30, 1–7 CrossRef.
- W. H. Organization, WHO, Food safety and foodborne illness, http://www.who.int/mediacentre/factsheets/fs237/en/print.html, Januray 2002.
- K. Aarnisalo, J. Lundén, H. Korkeala and G. Wirtanen, LWT--Food Sci. Technol., 2007, 40, 1041–1048 CrossRef CAS.
- M. Nitschke, L. V. Araújo, S. G. Costa, R. C. Pires, A. E. Zeraik, A. C. Fernandes, D. M. Freire and J. Contiero, Lett. Appl. Microbiol., 2009, 49, 241–247 CrossRef CAS PubMed.
- J. W. Costerton, K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta and T. J. Marrie, Annu. Rev. Microbiol., 1987, 41, 435–464 CrossRef CAS PubMed.
- M. Simões, L. C. Simões, I. Machado, M. O. Pereira and M. J. Vieira, Food Bioprod. Process., 2007, 84, 338–345 CrossRef.
- A. K. Bhardwaj, K. Vinothkumar and N. Rajpara, Recent Pat. Anti-Infect. Drug Discovery, 2013, 8, 68–83 CrossRef CAS.
- R. Hengge, A. Gründling, U. Jenal, R. Ryan and F. Yildiz, J. Bacteriol., 2015, 198, 15–26 CrossRef PubMed.
- A. Taglialegna, S. Navarro, S. Ventura, J. A. Garnett, S. Matthews, J. R. Penades, I. Lasa and J. Valle, PLoS Pathog., 2016, 12(6), e1005711 Search PubMed.
- L. Hallstoodley and P. Stoodley, Trends Microbiol., 2005, 13, 7–10 CrossRef CAS PubMed.
- M. Klausen, A. Heydorn, P. Ragas, L. Lambertsen, A. Aaesjørgensen, S. Molin and T. Tolkernielsen, Mol. Microbiol., 2003, 48, 1511–1524 CrossRef CAS PubMed.
- H. C. Flemming, T. R. Neu and D. J. Wozniak, J. Bacteriol., 2007, 189, 7945–7947 CrossRef CAS PubMed.
- J. Palmer, S. Flint and J. Brooks, J. Ind. Microbiol. Biotechnol., 2007, 34, 577–588 CrossRef CAS PubMed.
- Z. Qin, X. Yang, L. Yang, J. Jiang, Y. Ou, S. Molin and D. Qu, J. Med. Microbiol., 2007, 56, 83–93 CrossRef CAS PubMed.
- N. C. Caiazza and G. A. O'Toole, J. Bacteriol., 2004, 186, 4476–4485 CrossRef CAS PubMed.
- S. M. Hinsa, M. Espinosa-Urgel, J. L. Ramos and G. A. O'Toole, Mol. Microbiol., 2003, 49, 905–918 CrossRef CAS PubMed.
- B. K. Hammer and B. L. Bassler, Mol. Microbiol., 2003, 50, 101–114 CrossRef CAS PubMed.
- M. Otto, Annu. Rev. Med., 2013, 64, 175–188 CrossRef CAS PubMed.
- K. Sauer, M. C. Cullen, A. H. Rickard, L. A. H. Zeef, D. G. Davies and P. Gilbert, J. Bacteriol., 2004, 186, 7312–7326 CrossRef CAS PubMed.
- R. Morgan, S. Kohn, S. H. Hwang, D. J. Hassett and K. Sauer, J. Bacteriol., 2006, 188, 7335–7343 CrossRef CAS PubMed.
- C. Solano, M. Echeverz and I. Lasa, Curr. Opin. Microbiol., 2014, 18, 96–104 CrossRef CAS PubMed.
- C. Prigent-Combaret, G. Prensier, T. T. L. Thi, O. Vidal, P. Lejeune and C. Dorel, Environ. Microbiol., 2000, 2, 450–464 CrossRef CAS PubMed.
- C. N. Murphy and S. Clegg, Future Microbiol., 2015, 7, 991–1002 CrossRef PubMed.
- C. Schroll, K. B. Barken, K. A. Krogfelt and C. Struve, BMC Microbiol., 2010, 10, 1–10 CrossRef PubMed.
- J. W. Austin, G. Sanders, W. W. Kay and S. K. Collinson, FEMS Microbiol. Lett., 1998, 162, 295–301 CrossRef CAS PubMed.
- Y. Chai, P. B. Beauregard, H. Vlamakis, R. Losick and R. Kolter, mBio, 2012, 3, e00184 CrossRef CAS PubMed.
- K. Czaczyk and K. Myszka, Pol. J. Environ. Stud., 2007, 16, 799–806 CAS.
- C. Vuong, S. Kocianova, J. M. Voyich, Y. Yao, E. R. Fischer, F. R. Deleo and M. Otto, J. Biol. Chem., 2004, 279, 54881–54886 CrossRef CAS PubMed.
- J. Kives, B. Orgaz and C. Sanjosé, Colloids Surf., B, 2006, 52, 123–127 CrossRef CAS PubMed.
- Y. Chaturongkasumrit, H. Takahashi, S. Keeratipibul, T. Kuda and B. Kimura, Food Control, 2011, 22, 1893–1899 CrossRef CAS.
- N. Mitik-Dineva, J. Wang, V. K. Truong and P. Stoddart, Curr. Microbiol., 2009, 58, 268–273 CrossRef CAS PubMed.
- N. Cerca, G. B. Pier, M. Vilanova, R. Oliveira and J. Azeredo, Res. Microbiol., 2005, 156, 506–514 CrossRef CAS PubMed.
- C. J. Seneviratne, J. W. Yip, J. W. Chang, C. F. Zhang and L. P. Samaranayake, Arch. Oral Biol., 2013, 58, 1327–1334 CrossRef CAS PubMed.
- E. O'Neill, C. Pozzi, P. Houston, H. Humphreys, D. A. Robinson, A. Loughman, T. J. Foster and J. P. O'Gara, J. Bacteriol., 2008, 190, 3835–3850 CrossRef PubMed.
- M. Vergara-Irigaray, J. Valle, N. Merino, C. Latasa, B. García, d. L. M. I. Ruiz, C. Solano, A. Toledo-Arana, J. R. Penadés and I. Lasa, Infect. Immun., 2009, 77, 3978–3991 CrossRef CAS PubMed.
- S. Hennig, N. W. Sun and W. Ziebuhr, Int. J. Med. Microbiol., 2007, 297, 117–122 CrossRef CAS PubMed.
- B. Diemondhernández, F. Solórzanosantos, B. Leañosmiranda, L. Peregrinobejarano and G. Mirandanovales, BMC Infect. Dis., 2010, 10, 30–33 CrossRef PubMed.
- X. Wu, Y. Wang and L. Tao, FEMS Microbiol. Lett., 2011, 316, 44–50 CrossRef CAS PubMed.
- J. Valle, A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penadés and I. Lasa, Mol. Microbiol., 2003, 48, 1075–1087 CrossRef CAS PubMed.
- P. Vasseur, I. Vallet-Gely, C. Soscia, S. Genin and A. Filloux, Microbiology, 2005, 151, 985–997 CrossRef CAS PubMed.
- Y. Sakuragi and R. Kolter, J. Bacteriol., 2007, 189, 5383–5386 CrossRef CAS PubMed.
- J. M. Yarwood and P. M. Schlievert, J. Clin. Invest., 2003, 112, 1620–1625 CrossRef CAS PubMed.
- W. R. J. D. Galloway, J. T. Hodgkinson, S. D. Bowden, M. Welch and D. R. Spring, Chem. Rev., 2011, 111, 28–67 CrossRef CAS PubMed.
- D. Balestrino, J. A. J. Haagensen, C. Rich and C. Forestier, J. Bacteriol., 2005, 187, 2870–2880 CrossRef CAS PubMed.
- J. S. Dickschat, Nat. Prod. Rep., 2010, 27, 343–369 RSC.
- M. R. Parsek and E. P. Greenberg, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 8789–8793 CrossRef CAS.
- M. J. Lynch, S. Swift, D. F. Kirke, C. W. Keevil, C. E. R. Dodd and P. Williams, Environ. Microbiol., 2002, 4, 18–28 CrossRef CAS PubMed.
- T. Abee, A. T. Kovács, O. P. Kuipers and d. V. S. Van, Curr. Opin. Biotechnol., 2011, 22, 172–179 CrossRef CAS PubMed.
- V. G. Lewis, M. P. Ween and C. A. Mcdevitt, Protoplasma, 2012, 249, 919–942 CrossRef CAS PubMed.
- K. E. Beenken, P. M. Dunman, F. Mcaleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins and M. S. Smeltzer, J. Bacteriol., 2004, 186, 4665–4684 CrossRef CAS PubMed.
- B. R. Boles and A. R. Horswill, PLoS Pathog., 2008, 4, 4191 Search PubMed.
- R. J. Worthington, J. J. Richards and C. Melander, Org. Biomol. Chem., 2012, 10, 7457–7474 CAS.
- S. Guo, Y. Li, J. Chu, L. Wang, X. Zhao, N. Zhong, S. Qiu, J. Chen and Y. Li, Afr. J. Biotechnol., 2011, 10, 12847–12851 CrossRef CAS.
- X. H. Zhao, Y. M. Li, M. Park, J. Wang, Y. H. Zhang, X. W. He, F. Forghani, L. Wang, G. C. Yu and D. H. Oh, J. Microbiol. Biotechnol., 2013, 23, 246–250 CrossRef CAS PubMed.
- H. Ganin, X. Tang and M. M. Meijler, Bioorg. Med. Chem. Lett., 2009, 19, 3941–3944 CrossRef CAS PubMed.
- S. Auger, E. Krin, S. Aymerich and M. Gohar, Appl. Environ. Microbiol., 2006, 72, 937–941 CrossRef CAS PubMed.
- P. E. Kolenbrander, R. J. Palmer Jr, S. Periasamy and N. S. Jakubovics, Nat. Rev. Microbiol., 2010, 8, 471–480 CrossRef CAS PubMed.
- J. Y. Liu, R. Zhou, L. Li, B. M. Peters, B. Li, C. W. Lin, T. L. Chuang, D. Q. Chen, X. H. Zhao, Z. Y. Xiong, Z. B. Xu and M. E. Shirtliff, Res. Microbiol., 2017, 168, 188–193 CrossRef CAS PubMed.
- M. Janczarek, Int. J. Mol. Sci., 2011, 12, 7898–7933 CrossRef CAS PubMed.
- S. Lebeer, T. L. A. Verhoeven, M. P. Vlez, J. Vanderleyden and S. C. J. D. Keersmaecker, Appl. Environ. Microbiol., 2007, 73, 6768–6775 CrossRef CAS PubMed.
- T. Kawarai, S. Furukawa, N. Narisawa, C. Hagiwara, H. Ogihara and M. Yamasaki, J. Biosci. Bioeng., 2009, 107, 630–635 CrossRef CAS PubMed.
- N. Han, M. F. R. Mizan, I. K. Jahid and S. D. Ha, Food Control, 2016, 70, 161–166 CrossRef CAS.
- H. T. Solheim, C. Sekse, A. M. Urdahl, Y. Wasteson and L. L. Nesse, Appl. Environ. Microbiol., 2013, 79, 896–900 CrossRef CAS PubMed.
- G. Reguera and R. Kolter, J. Bacteriol., 2005, 187, 3551–3555 CrossRef CAS PubMed.
- A. Hoštacká, I. Čižnár and M. Štefkovičová, Folia Microbiol., 2010, 55, 75–78 CrossRef PubMed.
- H. D. Kusumaningrum, G. Riboldi, W. C. Hazeleger and R. R. Beumer, Int. J. Food Microbiol., 2003, 85, 227–236 CrossRef CAS PubMed.
- K. Ponnusamy, S. Kappachery, M. Thekeettle, J. H. Song and J. H. Kweon, World J. Microbiol. Biotechnol., 2013, 29, 1695–1703 CrossRef CAS PubMed.
- G. P. Zhao, Y. Q. Li, J. YanG and K. Y. Cui, Czech J. Food Sci., 2016, 34, 197–203 CrossRef.
- Z. Lei, Z. Heyan, H. Tianyang and L. Siran, Food Chem., 2015, 187, 370–377 CrossRef PubMed.
- M. K. Smith, L. A. Draper, P. J. Hazelhoff, P. D. Cotter, R. P. Ross and C. Hill, Front. Microbiol., 2016, 7, 1939 Search PubMed.
- P. J. Bremer, S. Fillery and A. J. Mcquillan, Int. J. Food Microbiol., 2006, 106, 254–262 CrossRef CAS PubMed.
- S. Furukawa, Y. Akiyoshi, M. Komoriya, H. Ogihara and Y. Morinaga, Food Control, 2010, 21, 669–672 CrossRef CAS.
- H. Liu, X. Wei, J. Ling, W. Wang and X. Huang, J. Endod., 2010, 36, 630–635 CrossRef PubMed.
- E. Presterl, M. Suchomel, M. Eder, S. Reichmann, A. Lassnigg, W. Graninger and M. Rotter, J. Antimicrob. Chemother., 2007, 60, 417–420 CrossRef CAS PubMed.
- K. C. Huth, M. Quirling, S. Maier, K. Kamereck, M. Alkhayer, E. Paschos, U. Welsch, T. Miethke, K. Brand and R. Hickel, Int. Endod. J., 2009, 42, 3–13 CrossRef CAS PubMed.
- N. G. Chorianopoulos, E. D. Giaouris, P. N. Skandamis, S. A. Haroutounian and G. J. E. Nychas, J. Appl. Microbiol., 2008, 104, 1586–1596 CrossRef CAS PubMed.
- N. L. Kavanaugh and K. Ribbeck, Appl. Environ. Microbiol., 2012, 78, 4057–4061 CrossRef CAS PubMed.
- X. H. Zhao, L. Wang, J. Chu, Y. M. Li, Y. Y. Li, Z. B. Xu, L. Li, M. E. Shirtliff, X. W. He, Y. Liu, J. H. Wang and L. S. Yang, Food Sci. Biotechnol., 2010, 19, 1655–1659 CrossRef.
- Y. Chen, T. Liu, K. Wang, C. Hou, S. Cai, Y. Huang, Z. Du, H. Huang, J. Kong and Y. Chen, PLoS One, 2016, 11, e0153468 Search PubMed.
- Z. C. Yang, B. C. Wang, X. S. Yang, Q. Wang and L. Ran, Colloids Surf., B, 2005, 41, 79–81 CrossRef CAS PubMed.
- J. Zhang, X. Rui, L. Wang, Y. Guan, X. Sun and M. Dong, Food Control, 2014, 42, 125–131 CrossRef CAS.
- L. D. Christensen, M. V. Gennip, T. H. Jakobsen, M. Alhede, H. P. Hougen, N. Høiby, T. Bjarnsholt and M. Givskov, J. Antimicrob. Chemother., 2012, 67, 1198–1206 CrossRef CAS PubMed.
- D. Bakkiyaraj, C. Sivasankar and S. K. Pandian, Bioorg. Med. Chem. Lett., 2012, 22, 3089–3094 CrossRef CAS PubMed.
- D. Vázquez-Sánchez, O. Habimana and A. Holck, Curr. Microbiol., 2013, 66, 110–121 CrossRef PubMed.
- N. K. Palanisamy, N. Ferina, A. N. Amirulhusni, Z. Mohdzain, J. Hussaini, L. J. Ping and R. Durairaj, J. Nanobiotechnol., 2014, 12, 2–7 CrossRef PubMed.
- D. Wu, W. Fan, A. Kishen, J. L. Gutmann and B. Fan, J. Endod., 2014, 40, 285–290 CrossRef PubMed.
- M. Farhat, M. C. Trouilhé, E. Briand, M. Molettadenat, E. Robine and J. Frère, J. Appl. Microbiol., 2010, 108, 1073–1082 CrossRef CAS PubMed.
- X. H. Zhao, J. Wang, F. Forghani, J. H. Park, M. S. Park, K. H. Seo and D. H. Oh, J. Microbiol. Biotechnol., 2013, 23, 1708–1716 CrossRef CAS PubMed.
- T. Karosi, I. Sziklai and P. Csomor, Laryngoscope, 2013, 123, 17–23 CrossRef PubMed.
- N. He, J. Hu, H. Liu, T. Zhu, B. Huang, X. Wang, Y. Wu, W. Wang and D. Qu, Antimicrob. Agents Chemother., 2011, 55, 5331–5337 CrossRef CAS PubMed.
- R. A. N. Chmielewski and J. F. Frank, Compr. Rev. Food Sci. Food Saf., 2003, 2, 22–32 CrossRef CAS.
- J. B. Xavier, C. Picioreanu, S. A. Rani, M. C. van Loosdrecht and P. S. Stewart, Microbiology, 2005, 151, 3817–3832 CrossRef CAS PubMed.
- J. S. Son, S. J. Lee, S. Y. Jun, S. J. Yoon, S. H. Kang, H. R. Paik, J. O. Kang and Y. J. Choi, Appl. Microbiol. Biotechnol., 2010, 86, 1439–1449 CrossRef CAS PubMed.
- X. H. Zhao, L. Wang, J. Chu, Y. Y. Li, Y. M. Li, Z. B. Xu, L. Li, M. E. Shirtliff, X. W. He, Y. Liu, J. H. Wang and L. S. Yang, Food Sci. Biotechnol., 2010, 19, 1191–1197 CrossRef.
- I. Gilan and A. Sivan, FEMS Microbiol. Lett., 2013, 342, 18–23 CrossRef CAS PubMed.
- Z. B. Xu, L. Li, X. H. Zhao, J. Chu, B. Li, L. Shi, J. Y. Su and M. E. Shirtliff, Afr. J. Biotechnol., 2011, 5, 1869–1873 CAS.
- M. R. Kiedrowski, J. S. Kavanaugh, C. L. Malone, J. M. Mootz, J. M. Voyich, M. S. Smeltzer, K. W. Bayles and A. R. Horswill, PLoS One, 2011, 6, e26714 CAS.
- A. M. Robinson, M. Bannister, J. E. Creeth and M. N. Jones, Colloids Surf., A, 2001, 186, 43–53 CrossRef CAS.
- R. D. Harper, M. R. T. H. Parracho, J. Walker, R. Sharp and G. Hughes, Antibiotics, 2014, 3, 270–284 CrossRef.
- D. Kelly, O. Mcauliffe, R. P. Ross and A. Coffey, Lett. Appl. Microbiol., 2012, 54, 286–291 CrossRef CAS PubMed.
- C. Anneleen, C. Pieter-Jan, T. S. Jeroen, V. P. Helena, N. Jean-Paul, O. V. Shaburova, V. N. Krylov, V. Guido and L. Rob, PLoS One, 2011, 6, e18597 Search PubMed.
- W. Fu, T. Forster, O. Mayer, J. J. Curtin, S. M. Lehman and R. M. Donlan, Antimicrob. Agents Chemother., 2010, 54, 397–404 CrossRef CAS PubMed.
- L. Carson, S. P. Gorman, B. F. Gilmore, G. Donelli, R. Bayston, W. B. Costerton and M. E. Shirtliff, FEMS Immunol. Med. Microbiol., 2010, 59, 447–455 CrossRef CAS PubMed.
- M. Corcoran, D. Morris, N. De Lappe, J. O'Connor, P. Lalor, P. Dockery and M. Cormican, Appl. Environ. Microbiol., 2014, 80, 1507–1514 CrossRef CAS PubMed.
- U. T. Nguyen and L. L. Burrows, Int. J. Food Microbiol., 2014, 187, 26–32 CrossRef CAS PubMed.
- L. B. Coulter, R. J. C. Mclean, R. E. Rohde and G. M. Aron, Viruses, 2014, 6, 3778–3786 CrossRef CAS PubMed.
- Y. Zhang and Z. Hu, Biotechnol. Bioeng., 2013, 110, 286–295 CrossRef CAS PubMed.
- O. E. Petrova and K. Sauer, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16690–16695 CrossRef CAS PubMed.
- D. G. Davies and C. N. Marques, J. Bacteriol., 2009, 191, 1393–1403 CrossRef CAS PubMed.
- L. R. Marks, B. A. Davidson, P. R. Knight and A. P. Hakansson, mBio, 2013, 4, e00438 CrossRef PubMed.
- R. A. N. Chmielewski and J. F. Frank, Compr. Rev. Food Sci. Food Saf., 2010, 2, 22–32 CrossRef.
- M. Simões, L. C. Simões and M. J. Vieira, LWT--Food Sci. Technol., 2010, 43, 573–583 CrossRef.
- A. Bridier, P. Sanchez-Vizuete, M. Guilbaud, J. C. Piard, M. Naïtali and R. Briandet, Food Microbiol., 2015, 45, 167–178 CrossRef CAS PubMed.
- S. Srey, I. K. Jahid and S. D. Ha, Food Control, 2013, 31, 572–585 CrossRef.
| This journal is © The Royal Society of Chemistry 2017 |