Open Access Article
Open Access ArticlepH-Controlled recognition of amino acids by urea derivatives of β-cyclodextrin†
Pawel
Stepniak
 a,
Bruno
Lainer
ab,
Kazimierz
Chmurski
b and
Janusz
Jurczak
a,
Bruno
Lainer
ab,
Kazimierz
Chmurski
b and
Janusz
Jurczak
 *a
*a
aInstitute of Organic Chemistry, Polish Academy of Sciences, Kasprzaka 44/52, 01-224 Warsaw, Poland. E-mail: jurczak_group@icho.edu.pl
bFaculty of Chemistry, University of Warsaw, Pasteura 1, 02-093 Warsaw, Poland
First published on 9th March 2017
Abstract
Water soluble amphiphilic urea-substituted β-cyclodextrins were synthesized and applied as amino acid receptors. A great affinity towards nonpolar amino acids was observed, ranging from 2300 M−1 for alanine to 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 M−1 for tryptophan in a highly competitive environment (pH 8 phosphate-buffered water solution). Significant selectivity was observed, since carboxylates without the amino group (or having hydroxyl instead) remained not bound. Endoenthalpic effects were recorded for amino acids during titrations, pointing to a different mode of complex formation as compared to native β-cyclodextrin. We found a dramatic change in binding strength within the physiological pH range: with pH change from 8 to 6, affinity towards amino acids dropped from 54
800 M−1 for tryptophan in a highly competitive environment (pH 8 phosphate-buffered water solution). Significant selectivity was observed, since carboxylates without the amino group (or having hydroxyl instead) remained not bound. Endoenthalpic effects were recorded for amino acids during titrations, pointing to a different mode of complex formation as compared to native β-cyclodextrin. We found a dramatic change in binding strength within the physiological pH range: with pH change from 8 to 6, affinity towards amino acids dropped from 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 500 M−1, making the receptors potentially appropriate for biotechnological purposes.
000 to 500 M−1, making the receptors potentially appropriate for biotechnological purposes.
Introduction
The ubiquity of amino acids is one of the main factors propelling various research efforts to develop more sophisticated tools to identify, bind, and control them. They are present in all living organisms, playing key roles in many biochemical processes, e.g. signal transmission and cellular structure building.1 Therefore, identifying objects that interact with amino acids strongly and selectively is an ongoing objective of supramolecular chemistry. Analytical quantitative detection of amino acids is usually based on chromatographical or electrochemical techniques; their main drawback is that analytes often need to be derivatized prior to analysis.2,3 The supramolecular approach to amino acid identification and binding appears to offer a solution, based on strong and selective interactions between amino acids and electrically neutral molecular receptors.4–6 One very demanding issue in this field is that of pH-controlled binding-release systems, allowing for specific and targeted control of processes that involve amino acids.7 An analogous approach is investigated in the field of specialized drug delivery systems and signal transduction regulation.8,9The most common strategy towards the formation of complexes of receptors with amino acids utilizes structures with cavities available for the inclusion of guest species. Usually, a type of hydrophobic effect is employed to situate the amino acid inside the cavity.10,11 This allows for the usage of such receptors under aqueous conditions, but, on the other hand, limits the range of guests to less polar ones. In this manner, cucurbiturils,12–14 calixarenes,15 pilarenes,16 and cyclodextrins17–19 are exploited for amino acid binding. Another, widely applied approach involves the utilization of hydrogen bond formation, whereby various nucelophilic acceptors are usually bound by receptors.20–22 This strategy is limited by the competing environment, e.g. polar solvents.23–25 Therefore, efficient binding of nonderivatized amino acids in aqueous solutions still remains a challenge.1
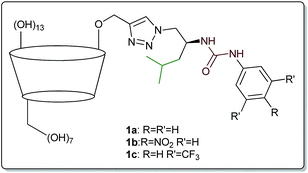
In our recent studies we have shown that combining these two modes of interaction results in substantially improved recognition of amino acids in water.26 The introduction of an amphiphilic substituent containing urea moiety to β-cyclodextrin (β-CD) resulted in receptors 1a–c (Fig. 1), which appeared to be 1000-fold more efficient than native (underivatized) β-CD. Binding constants of up to 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 M−1 (for L-tryptophane) were obtained, and the affinity was correlated with both the acidity of the urea protons and the lipophilicity of the amino acid. Unexpectedly, exclusion of isobutyl moiety from the side-chain structure caused a dramatic decrease in binding constants.
400 M−1 (for L-tryptophane) were obtained, and the affinity was correlated with both the acidity of the urea protons and the lipophilicity of the amino acid. Unexpectedly, exclusion of isobutyl moiety from the side-chain structure caused a dramatic decrease in binding constants.
Inspired by these regularities, we decided to investigate how changes in the amphiphilic side-chain or pH of the environment would affect binding of various amino acids and carboxylates. We assumed that introduction of an additional aromatic ring (instead of isobutyl moiety) in the side-chain would enhance the interaction with nonpolar species, in particular with hydrophobic amino acids. We additionally assumed that receptors having such a structure would not undergo deprotonation, which would make them useful in a wide range of pH. In this report, we present such new β-CD derivatives with side-chains based on phenylalanine and leucine and describe their utilization in amino acid binding.
Results and discussion
Synthesis
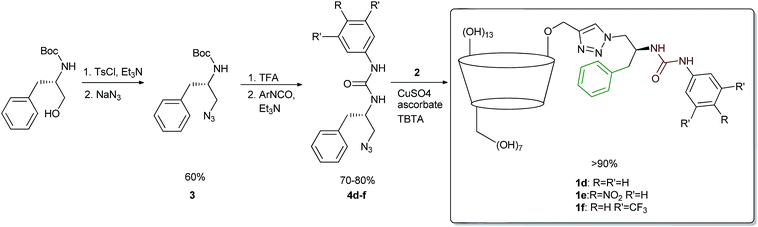
The structure of the target receptors was designed based on our previous findings concerning the behaviour of cyclodextrins and various platforms exploiting hydrogen bonding in carboxylate binding.27,28 We showed that in the case of C-6 substituted β-CDs, intramolecular inclusion complex formation may occur, making the CD cavity much less available for the guest.29 Therefore, we have developed a synthetic approach to efficient preparation of mono-2-functionalized β-CD, which can be then further modified via the azide–alkyne click chemistry methodology.30 In such a manner we used mono-2-propargyl-β-CD (2) to synthesize compounds 1d–f, being phenyl analogues of the previously reported receptors 1a–c. In this case N-Boc-L-phenylalaninol was tosylated and subsequently transformed into azide 3. After Boc deprotection the amino group was functionalized with the corresponding commercially available aromatic isocyanate, yielding desired compound 4. Next, azide–alkyne coupling with monopropargylated CD 2 was carried out (according to the known procedure), to yield receptors 1d–f almost quantitatively (>90%). Consequently, we had in hand six receptors of type 1, with varying side-chains originating from leucine and phenylalanine, having different acidity of the group. None of the compounds had any hydrogen atoms that would be deprotonated under pH < 10 (Scheme 1).Binding measurements
We were able to assess the role of the isobutyl/phenyl substituent in the complexation process, comparing it with previously obtained reference compound having just the ethylene group between azide and urea functionalities.26 We envisioned that the presence of the additional aromatic moiety should enhance the guest binding process without causing any additional self-inclusion complex formation (if the latter had been the case, leucine-derived compounds 1a–c would have suffered from this as well). We decided to investigate the amino acid binding properties of receptors of type 1 at pH 8, being in the upper part of the physiological condition range. We used 50 mM phosphate buffer with ionic strength of about 140 mM (which corresponds well to the intracellular environment). We utilized isothermal titration calorimetry to gain fuller insight into the thermodynamics of the complexation process as well as to compare with previously reported data. In this case, titrations of L-amino acid into 2 mM solutions of receptors provided satisfactory sigmoidal curves allowing for the determination of enthalpies, binding constants, and interaction stoichiometries (Fig. 2). Similarly to our previous findings, receptors 1d–f also exhibited tendencies to form higher-ordered superstructures, which was confirmed with dynamic light scattering (with aggregates of 164, 122, and 255 nm identified for respective receptors 1d–f). However, during titration a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (host
1 (host![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
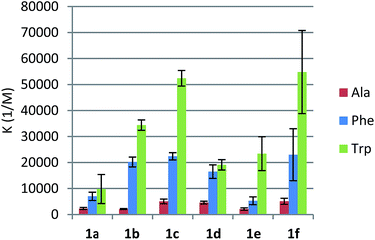
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) guest) complexation model was usually identified, which confirmed the same mode of operation for receptors 1d–f as compared to leucine-derived 1a–c. Consequently, we applied the typical model of n-independent binding sites in the fitting procedure. First, we tested how these receptors complex nonpolar amino acids. The apparent binding constants are listed in Table 1.
guest) complexation model was usually identified, which confirmed the same mode of operation for receptors 1d–f as compared to leucine-derived 1a–c. Consequently, we applied the typical model of n-independent binding sites in the fitting procedure. First, we tested how these receptors complex nonpolar amino acids. The apparent binding constants are listed in Table 1.
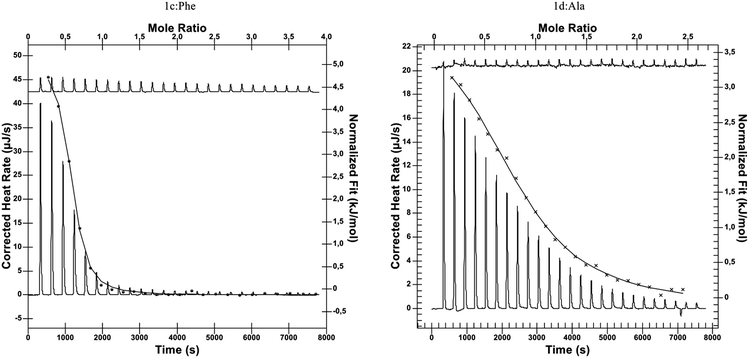 | ||
| Fig. 2 Representative thermograms of ITC experiments: L-Phe titrated to 1c (left) and L-Ala titrated to 1d (right). | ||
We found that phenylalanine-derived receptors 1d and 1f bound alanine, phenylalanine, and tryptophan substantially more strongly than their analogues 1a and 1c. In the case of p-nitrourea derivatives 1b and 1e, this tendency was reversed. Slightly lower binding constants for compound 1e could be explained by π-stacking leading to the rearrangement of the binding site. As we have already shown, the presence and proper positions of two lipophilic substituents of the side-chain is essential for the binding, since an analogue with no substituent at the ethylene linker between urea and triazole groups showed no affinity towards amino acids at all. In the case of both series of receptors, an increase in the urea hydrogens' acidity resulted in higher Ka's (Fig. 3). This is consistent with our assumption that the urea moiety provides hydrogen bonding for the carboxylate group of an amino acid.
All the reactions proved to be endothermic, strongly entropy-driven, with enthalpies varying between 1.5 and 5.5 kJ mol−1 (Table 2). This would confirm that the mode of interaction with amino acids is similar for all the receptors. We observed that for corresponding pairs of leucine- and phenylalanine-derived receptors the enthalpies were lower, probably indicating better conformational preorganization of compounds 1d–f for guest binding.
Next, we investigated the scope of guests that could interact with receptors of type 1. We analysed the previously thoroughly examined model receptor 1c in binding of tyrosine, phenylglycine, and serine as well as 2-phenylbutyric and mandelic acids (Table 3). Tyrosine exhibited a binding constant two times lower than phenylalanine. The conformationally more constrained phenylglycine appeared to be even less effectively bound by receptor 1c. It could be inferred that, for tight interaction, both the carboxylic group and the pendant arm of the guest have to be in a proper position, which would make these two structural elements essential for complex formation. On the other hand, serine was bound with similar strength to phenylalanine, even though it is far more polar and does not contain aromatic moiety. Replacing the amino group with hydroxyl or aliphatic substituent led to dramatic decreases in affinity constants, which proved that the presence of the amino group in the guest structure is necessary for efficient binding. In fact, the same phenomenon was observed in the case of ibuprofen binding by β-CD and its derivatives (Table 4). Addition of the side-chain into the β-CD structure provided weaker interaction between ibuprofen and receptors 1a, 1c, and 1d, compared to native β-CD. In all these cases the complexation reactions were exothermic, showing that the mode of interaction is different than observed for amino acid binding. These results were consistent with complexation enthalpies of 2-phenylbutyrate and mandelate (Table 3). This proves that when typical inclusion complex formation occurs, β-CD functions better than receptors of type 1, since the latter are impaired by competing self-inclusion complex formation and aggregation. However, for amino acids specific interactions appear, leading to a different mechanism of complexation (positive enthalpies) and much stronger affinities.
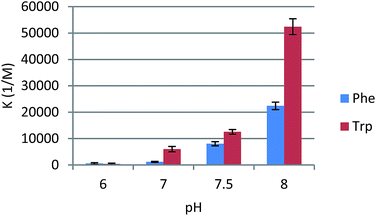
We next investigated how pH affects the effectiveness of receptors of type 1. Using model compound 1c, we measured binding constants in the pH range of 6–8 (Fig. 4). The more basic the conditions, the more efficient the receptor proved to be. Phenylalanine and tryptophan were bound about 4 times more weakly when pH dropped from 8 to 7.5. Under neutral conditions the receptor was still 10-fold better than native β-CD, but affinities were substantially lower. Under slightly acidic conditions (comparable to the pH of fast-proliferating cells) binding constants dropped to 500 M−1. These results were in agreement with the abundance of anionic form of amino acids. At pH 6 almost all phenylalanine and tryptophan molecules are in their zwitterionic forms, in which they apparently form weak complexes with receptors of type 1. At pH 8 anionic forms are generated, which are strongly bound by urea-derived β-CD receptors. We observed approximately linear correlation between effective concentration of anionic form of amino acid and Ka values. This shows that this type of receptor is especially effective in amino acid binding in the narrow physiological pH range. A change of two pH units induces a 40- and 140-fold increase in binding of phenylalanine and tryptophan, respectively.
Conclusions
We have presented a spectrum of synthetic opportunities and supramolecular applications of amphiphilic urea derivatives of β-CD. We again demonstrated that our approach allows for obtaining pure, well-defined mono-2-substituted β-CDs with excellent yields. A repertoire of various receptors were tested for selectivity towards amino acids, finding that receptors 1a–f interact strongly only with amino acids among which nonpolar ones are bound with Ka's up to 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 M−1 in phosphate buffered water solution. Presence of the amino group is essential for the binding occurrence and is an origin of its pH-dependency. At pH 8 receptors present great affinities, while at pH 6 there is almost no binding at all. These results make urea derived β-CDs an interesting prospective tool for bioengineering and drug delivery, since dramatic changes occur within the physiological pH range.
800 M−1 in phosphate buffered water solution. Presence of the amino group is essential for the binding occurrence and is an origin of its pH-dependency. At pH 8 receptors present great affinities, while at pH 6 there is almost no binding at all. These results make urea derived β-CDs an interesting prospective tool for bioengineering and drug delivery, since dramatic changes occur within the physiological pH range.
Acknowledgements
This research was financed by the European Union within the European Regional Development Fund, Project POIG.01.01.02-14-102/09.Notes and references
- H.-J. Schneider, Angew. Chem., Int. Ed., 2009, 48, 3924–3977 CrossRef CAS.
- M. C. Waldhier, M. A. Gruber, K. Dettmer and P. J. Oefner, Anal. Bioanal. Chem., 2009, 394, 695–706 CrossRef CAS.
- T. J. Ward and K. D. Ward, Anal. Chem., 2012, 84, 626–635 CrossRef CAS.
- A. M. Costero, G. M. Rodríguez-Muñiz, S. Gil, S. Peransi and P. Gaviña, Tetrahedron, 2008, 64, 110–116 CrossRef CAS.
- D. Lichosyt, S. Wasiłek and J. Jurczak, J. Org. Chem., 2016, 81, 7342–7348 CrossRef CAS.
- M. M. Stone, A. H. Franz and C. B. Lebrilla, J. Am. Soc. Mass Spectrom., 2002, 13, 964–974 CrossRef CAS.
- G. Yu, K. Jie and F. Huang, Chem. Rev., 2015, 115, 7240–7303 CrossRef CAS.
- O. Swiech, P. Dutkiewicz, K. Wójciuk, K. Chmurski, M. Kruszewski and R. Bilewicz, J. Phys. Chem. B, 2013, 117, 13444–13450 CrossRef CAS.
- O. Swiech, M. Majdecki, A. Debinski, A. Krzak, T. M. Stępkowski, G. Wójciuk, M. Kruszewski and R. Bilewicz, Nanoscale, 2016, 8, 16733–16742 RSC.
- M. V. Rekharsky and Y. Inoue, J. Am. Chem. Soc., 2002, 124, 2144–2154 Search PubMed.
- K. A. Connors, Chem. Rev., 1997, 97, 1325–1357 CrossRef CAS.
- J. Zhang, Y.-Y. Xi, Q. Li, Q. Tang, R. Wang, Y. Huang, Z. Tao, S.-F. Xue, L. F. Lindoy and G. Wei, Chem.–Asian J., 2016, 11, 2250–2254 CrossRef CAS.
- L. a Logsdon, C. L. Schardon, V. Ramalingam, S. K. Kwee and A. R. Urbach, J. Am. Chem. Soc., 2011, 133, 17087–17092 CrossRef CAS.
- P. Rajgariah and A. R. Urbach, J. Inclusion Phenom. Macrocyclic Chem., 2008, 62, 251–254 CrossRef CAS.
- L. Mutihac, J. H. Lee, J. S. Kim and J. Vicens, Chem. Soc. Rev., 2011, 40, 2777 RSC.
- M. Bojtár, A. Paudics, D. Hessz, M. Kubinyi and I. Bitter, RSC Adv., 2016, 6, 86269–86275 RSC.
- C. Liu, J. Lian, Q. Liu, C. Xu and B. Li, Anal. Methods, 2016, 8, 5794–5800 RSC.
- Q. Huang, L. Jiang, W. Liang, J. Gui, D. Xu, W. Wu, Y. Nakai, M. Nishijima, G. Fukuhara, T. Mori, Y. Inoue and C. Yang, J. Org. Chem., 2016, 81, 3430–3434 CrossRef CAS.
- J. Zhou, Y. Liu, Z. Zhang, S. Yang, J. Tang, W. Liu and W. Tang, Nanoscale, 2016, 5621–5626 RSC.
- V. Amendola, L. Fabbrizzi and L. Mosca, Chem. Soc. Rev., 2010, 39, 3889–3915 RSC.
- N. Busschaert, C. Caltagirone, W. Van Rossom and P. a. Gale, Chem. Rev., 2015, 115, 8038–8155 CrossRef CAS.
- K. Choi and A. D. Hamilton, Coord. Chem. Rev., 2003, 240, 101–110 CrossRef CAS.
- G. V Oshovsky, D. N. Reinhoudt and W. Verboom, Angew. Chem., Int. Ed., 2007, 46, 2366–2393 CrossRef PubMed.
- E. A. Kataev and C. Muller, Tetrahedron, 2014, 70, 137–167 CrossRef CAS.
- G. Castronuovo, V. Elia, A. Pierro and F. Velleca, Can. J. Chem., 1999, 77, 1218–1224 CrossRef CAS.
- P. Stepniak, B. Lainer, K. Chmurski and J. Jurczak, Carbohydr. Polym., 2017, 164, 233–241 CrossRef CAS.
- J. Jurczak, P. Dydio, P. Stepniak and T. Zielinski, RSC Adv., 2016, 6, 41568–41571 RSC.
- J. Jurczak, P. Dydio, P. Stepniak and T. Zielinski, Sens. Actuators, B, 2016, 237, 1–14 CrossRef.
- K. Chmurski, P. Stepniak and J. Jurczak, Carbohydr. Polym., 2016, 138, 8–15 CrossRef CAS.
- K. Chmurski, P. Stepniak and J. Jurczak, Synthesis, 2015, 47, 1838–1843 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra02127e |
| This journal is © The Royal Society of Chemistry 2017 |