Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Molecular analysis of polysaccharide accumulation in Dendrobium nobile infected with the mycorrhizal fungus Mycena sp.†
Qing Li ,
Bing Li,
Li-Si Zhou,
Gang Ding,
Biao Li* and
Shun-Xing Guo*
,
Bing Li,
Li-Si Zhou,
Gang Ding,
Biao Li* and
Shun-Xing Guo*
Institute of Medicinal Plant Development, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing 100193, People's Republic of China. E-mail: libiao@126.com; sxguo1986@163.com
First published on 16th May 2017
Abstract
Polysaccharides are among the most important active compounds of Dendrobium nobile Lindl., and they improve immunity, strengthen memory, and prevent and fight tumours. Evidence suggests that the mycorrhizal fungus Mycena sp. may increase the polysaccharide content of D. nobile. To understand the regulatory mechanism by which Mycena sp. promotes polysaccharide accumulation in D. nobile, a model including MF23 (Mycena sp.) and D. nobile was developed. Content measurements and electron microscopy showed that the change in polysaccharides was associated with the level of colonization. Large-scale transcriptome sequencing analyses of D. nobile infected with MF23 and of wild D. nobile revealed that 30 unigenes encoding key enzymes were potentially associated with polysaccharide accumulation. A qRT-PCR experiment involving 13 randomly chosen unigenes suggested that the combined effects of the carbohydrate biosynthesis and consumption pathways resulted in changes in polysaccharide levels. MF23 might increase polysaccharide level by inducing genes involved in photosynthesis (petF), starch and sucrose metabolism (SUS, bcsA, glgA) and fructose and mannose metabolism (PMM, GPMM) and by repressing genes involved in glycolysis (PGK, PFK) and the citrate cycle (PDHB, CS, SDHA). This study provides a good example of an endophyte promoting the synthesis of bioactive compounds in its host and paves the way for further investigations of the associated regulatory mechanism using molecular methods.
Introduction
Dendrobium nobile Lindl. is an endangered orchid species that is distributed in India, Burma, Thailand, Laos, Vietnam, and China.1 For thousands of years, its stem has been used as a tonic herb in traditional medicine, and its remarkable curative effects on gastritis, diabetes, aging and immunological disorders have been proven by modern pharmacological research.2–6 Thus, it has been officially listed as “Shi Hu” in different versions of the Pharmacopoeia of the People's Republic of China. To elucidate the pharmacological mechanism of D. nobile, substantial research has been performed on its compounds, and these studies have demonstrated that polysaccharides are among the most important active compounds of D. nobile.7–9 In addition to their effects on strengthening memory10 and inhibiting thrombokinesis,11 many polysaccharides derived from D. nobile exhibit strong immunomodulatory and antitumour activities.12–14 For instance, the polysaccharide DNP4-2 can significantly increase the immune index, strongly promote the secretion of tumour necrosis factor alpha (TNF-α), interferon-gamma (IFN-γ) and interleukin-2 (IL-2), and reduce the concentrations of malondialdehyde (MDA) in the serum.15 At 200 μg mL−1, the polysaccharides DNP-W1 and DNP-W3 show strong antitumour activity (antitumour rate > 80%),16 suggesting that the polysaccharides of D. nobile could be effective natural antitumour agents. However, the content of polysaccharides in D. nobile is not high,17 which restricts their development and utilization.Orchids are typical mycorrhizal plants. Several photosynthetic orchids have recently been confirmed as mixotrophic, with the ability to recover carbon from mycorrhizal fungi associated with their roots.18 Mycorrhizal orchids can acquire more N, P and water than non-mycorrhizal controls.19,20 To date, our laboratory has isolated numerous mycorrhizal fungi with growth-promoting effects from the roots of wild D. officinale Kimura et Migo and D. nobile.21,22 In addition, some mycorrhizal fungi have been reported to promote polysaccharide accumulation in their hosts. In D. nobile infected with Mycena sp., the polysaccharide level increased over a longer period23 and polysaccharides accumulated to much higher levels than those of controls (76.7% increase).24 However, the relationship between mycorrhizal fungi and their hosts is still not clear, which hinders the application of mycorrhizal fungi. Thus, it is of great importance to understand the influence and regulatory mechanism of mycorrhizal fungi on the polysaccharide metabolism of D. nobile.
The accumulation of polysaccharides can be affected by many factors, such as photosynthesis,25 starch and sucrose metabolism,26,27 and respiration.28 The Illumina HiSeq4000 platform was initially used to focus on the key genes in these processes and to reveal the molecular events underlying the changes in the polysaccharides of symbiotic D. nobile. This study provides an overview of the D. nobile stem transcriptome before and after inoculation. In addition, the results in this report provide a good example of an endophyte promoting the synthesis of bioactive compounds in its host and pave the way for further investigations of the associated regulatory mechanism using molecular methods.
Experimental
Biological materials and culture conditions
D. nobile explants of 3–5 cm in height were cultured on improved 1/2 Murashige and Skoog (MS) medium supplemented with potato juice (200 g L−1 potato, juice boiled for 20 min, 3% (w/v) sucrose, and 0.6% (w/v) agar, pH 5.8). Each bottle contained 9 seedlings. Mycelial plugs with a diameter of 9 mm, which were from 20 day-old MF23 (Mycena sp.)29 grown on potato dextrose agar medium, were placed in the centre of a culture bottle, adjacent to the roots of D. nobile. Then, the culture bottles were kept in a conventional greenhouse with a 10 h light/14 h dark photoperiod at (24 ± 1) °C and an illumination intensity of 1500 Lx. Seedlings that were not inoculated with mycelial plugs were maintained as controls.Plant samples were collected at different stages, including 1, 3, 6 and 9 weeks post-inoculation (model group) and at the same stages for the non-inoculated (control) group. All samples were divided into two portions; some were used for morphological and chemical research, and the rest were frozen using liquid nitrogen and stored until RNA extraction.
Determination of the polysaccharide content of D. nobile
Stems of each treatment were dried at 55 °C, thoroughly blended, and ground to a powder. The polysaccharide extraction method was as in Chen et al.30 The polysaccharide content was determined using the phenol-sulfuric acid method described by the Pharmacopoeia Committee of the People's Republic of China31 and Dubois et al.32 with glucose solutions (0, 5, 10, 20, 30, 40, 50, 60, 70, and 80 μg mL−1) as standards. The linear regression equation was y = 7.6021x + 0.0609 (r = 0.9998).Light microscopy
Fresh root segments were fixed in formalin-acetic acid-alcohol (FAA), an approach described by Feder and O'Brien.33 Samples were dehydrated in a graded ethanol series, embedded in paraffin, stained with safranine and fast green, sealed with Gel Damar and then observed and photographed on a light microscope equipped with a camera (ZEISS Axio Imager A1).RNA isolation and sequencing
Total RNA was extracted from 1 week-old and 9 week-old stem samples of D. nobile using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The quality and concentration of RNA were determined using electrophoresis and a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The best RNA samples, which had an RIN above 8.0, were chosen for cDNA library preparation.Library construction and sequencing were performed by the Novogene Bioinformatics Institute (Beijing, China), and the general experimental pipeline is summarized in previously published papers.34,35 Sequencing libraries were generated using an NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer's recommendations, and index codes were added to attribute the sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using a TruSeq PE Cluster Kit v3-cBot-HS (Illumina). After cluster generation, the library preparations were sequenced on an Illumina HiSeq 4000 instrument with 150 bp paired-end reads.
Transcriptome assembly and annotation
Clean reads were obtained from the raw data by removing adapter-containing reads, poly-N-containing reads and low-quality reads. All downstream analyses were based on high-quality clean data.Transcriptome assembly was processed using Trinity36 with min_kmer_cov set to 2. The longest transcript from each gene was designated a unigene. To assign predicted gene descriptions for the assembled unigenes, a BLASTx search was performed against the following databases: Nr (NCBI non-redundant protein sequences), Nt (NCBI non-redundant nucleotide sequences), Pfam (protein family), KOG (eukaryotic orthologous groups of proteins), Swiss-Prot (a manually annotated and reviewed protein sequence database), KO (KEGG orthologue database) and GO (gene ontology), with a cutoff E-value of 10−5.
Analysis of differentially expressed genes (DEGs)
Differential expression analysis of model and control groups was performed using the DESeq R package (1.10.1).37 The P-value was adjusted using the q-value.38 In this study, genes with transcripts with a significant q-value (<0.005) and |log2(fold change)| > 1 between the model and control groups were considered DEGs.GO and KEGG enrichment analyses of the DEGs were performed using the GOseq R packages based on the Wallenius non-central hyper-geometric distribution39 and KOBAS40 software, respectively.
Validation and expression pattern analysis
To experimentally validate the transcriptional abundance results from sequencing and computational analysis, 13 unigenes were selected for qRT-PCR. Primers were designed using Primer 6.0 and are listed in ESI Table 1.† The actin gene of D. nobile was used as a ref. 41. cDNA templates were obtained by reverse-transcribing the total RNA of the model and control groups after different lengths of time in culture. qRT-PCR was performed using SYBR® Premix Ex Taq™ (TaKaRa, Dalian, China) on a LightCycler480 II System (Roche, USA). The reaction was performed at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Three independent biological replicates were analysed for each sample, and three technical replicates were analysed for each biological replicate. Gene expression was evaluated using the 2−ΔΔCt method.42Statistical analysis
SPSS 19.0 (IBM, Chicago, IL, USA) statistical software was used for the statistical evaluation of the results. All results are expressed as the mean ± standard deviation (SD) of the number of experiments. Unpaired t-tests were performed at P < 0.05.Results
Changes in the polysaccharide content of the D. nobile stem
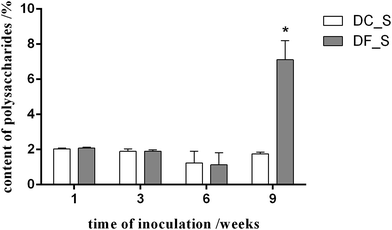
To understand the effect of the mycorrhizal fungus MF23 on the polysaccharide accumulation of D. nobile, the polysaccharide content of D. nobile stems at 1, 3, 6, and 9 weeks after inoculation and in the controls were assessed (Fig. 1). At the first three stages, the stems of both the model and control groups had low polysaccharide contents (less than 2.10%). However, 9 weeks after inoculation, the concentration of polysaccharides increased to 7.11%, which was 4 times greater than that of the controls.Associations between MF23 and D. nobile roots
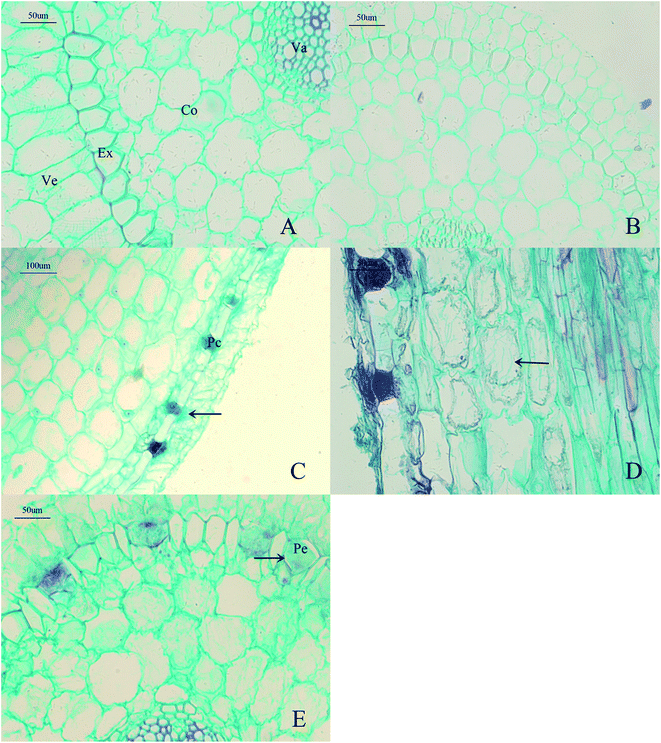
Light microscopy showed no hyphae in 1 week-old and control root tissues (Fig. 2A and B). The hyphae of MF23 first appeared in the velamen cells of the host roots 3 weeks after inoculation and gathered near the passage cells (Fig. 2C). With increasing time, the hyphae penetrated the cell wall of the D. nobile root cortex through the passage cells and spread from cell to cell (Fig. 2D). In the 9th week, when the polysaccharide content of D. nobile had increased significantly, increasing numbers of hyphae appeared at the cortex cells of the host. In addition, pelotons, typical orchid mycorrhizal structures,43 were first observed in the passage cells (Fig. 2E). These findings suggest a possible relationship between polysaccharide accumulation and the formation of pelotons.Transcriptome profiles of inoculated and non-inoculated D. nobile
To determine the D. nobile transcriptome expression profile after MF23 infection, next-generation sequencing was conducted on symbiotic and asymbiotic D. nobile stems 1 and 9 weeks after inoculation. In this study, two biological replicates were sequenced per sample. In total, 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 863
863![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 528–69
528–69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 564
564![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340 raw reads of 150 bp were generated (Table 1). The raw reads are available in the NCBI SRA database under accession number PRJNA338366. After removing adaptor sequences, ambiguous nucleotides and low-quality sequences, 43
340 raw reads of 150 bp were generated (Table 1). The raw reads are available in the NCBI SRA database under accession number PRJNA338366. After removing adaptor sequences, ambiguous nucleotides and low-quality sequences, 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 312
312![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 840–67
840–67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 080
080![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 588 clean reads remained. The assembly of the clean reads resulted in 207
588 clean reads remained. The assembly of the clean reads resulted in 207![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 283 unigenes in the range of 201–17
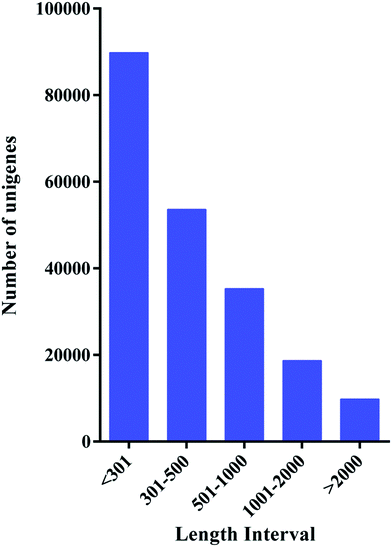
283 unigenes in the range of 201–17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 019 bp, with an N50 length of 857 bp; the length distribution of the unigenes is illustrated in Fig. 3.
019 bp, with an N50 length of 857 bp; the length distribution of the unigenes is illustrated in Fig. 3.
| Sample | Raw reads | Clean reads | Clean bases | Error (%) | Q20 (%) | Q30 (%) | GC content (%) |
|---|---|---|---|---|---|---|---|
| a DC1_S1 & DC1_S2: two biological replicates of the 1 week-old control group.b DF1_S1 and DF1_S2: two biological replicates of the 1 week-old model group.c DC9_S1 & DC9_S2: two biological replicates of the 9 week-old control group.d DF9_S1 & DF9_S2: two biological replicates of the 9 week-old model group. | |||||||
| DC1_S1a | 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 252 252![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 114 114 |
55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 277 277![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 910 910 |
8.29G | 0.01 | 97.53 | 94.05 | 46.16 |
| DC1_S2a | 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 863 863![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 528 528 |
43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 312 312![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 840 840 |
6.5G | 0.01 | 97.56 | 94.09 | 45.75 |
| DF1_S1b | 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 455 455![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 694 694 |
56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340 340![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 260 260 |
8.45G | 0.01 | 97.37 | 93.7 | 46.2 |
| DF1_S2b | 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 912 912![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 596 596 |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 959 959![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 096 |
7.64G | 0.01 | 97.69 | 94.33 | 47.63 |
| DC9_S1c | 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 092 092![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 876 876 |
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 414 414![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 294 294 |
6.81G | 0.01 | 97.25 | 93.47 | 46.48 |
| DC9_S2c | 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 537 537![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 374 374 |
44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 964 964![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 612 612 |
6.74G | 0.01 | 97.49 | 93.96 | 46.59 |
| DF9_S1d | 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 471 471![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 134 134 |
51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 541 541![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 182 182 |
7.73G | 0.01 | 97.54 | 94.09 | 47.3 |
| DF9_S2d | 69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 564 564![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340 340 |
67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 080 080![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 588 588 |
10.06G | 0.01 | 97.55 | 94.12 | 46.94 |
Unigene functional annotation
A total of 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 778 (40.41%) unigenes with significant BLAST hits were returned. Among them, 56
778 (40.41%) unigenes with significant BLAST hits were returned. Among them, 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 378 (27.19%), 34
378 (27.19%), 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 493 (16.64%), 18
493 (16.64%), 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 911 (9.12%), 48
911 (9.12%), 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 431 (23.36%), 46
431 (23.36%), 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 843 (22.59%), 47
843 (22.59%), 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 372 (22.85%), and 27
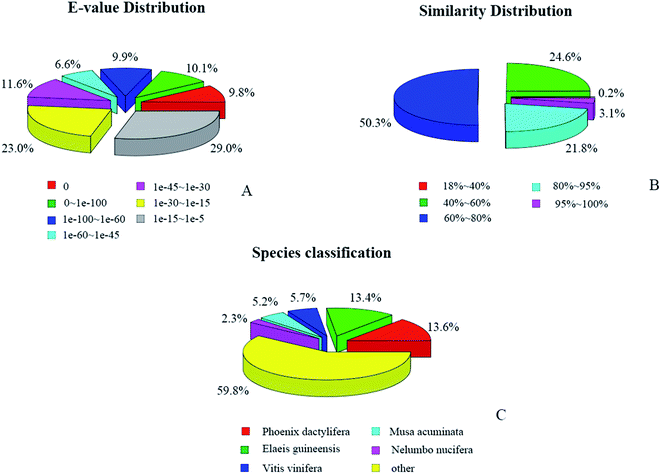
372 (22.85%), and 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 087 (13.06%) unigenes had hits in the Nr, Nt, KO, Swiss-Prot, Pfam, GO, and KOG databases, respectively. The E-values and similarity distributions of the top hits in the Nr database analysis revealed that 36.4% and 24.9% of the genes showed significant homology (E-value < 10−45) or high similarity (greater than 80%), respectively (Fig. 4A and B). Based on Nr annotation, approximately 35.2% of the annotated unigenes were assigned with the best score to sequences from the top five species, Phoenix dactylifera (13.6%), Elaeis guineensis (13.4%), Vitis vinifera (5.7%), Musa acuminate (5.2%), and Nelumbo nucifera (2.3%; Fig. 4C).
087 (13.06%) unigenes had hits in the Nr, Nt, KO, Swiss-Prot, Pfam, GO, and KOG databases, respectively. The E-values and similarity distributions of the top hits in the Nr database analysis revealed that 36.4% and 24.9% of the genes showed significant homology (E-value < 10−45) or high similarity (greater than 80%), respectively (Fig. 4A and B). Based on Nr annotation, approximately 35.2% of the annotated unigenes were assigned with the best score to sequences from the top five species, Phoenix dactylifera (13.6%), Elaeis guineensis (13.4%), Vitis vinifera (5.7%), Musa acuminate (5.2%), and Nelumbo nucifera (2.3%; Fig. 4C).
Analysis of DEGs
To better survey the biological mechanism of polysaccharide biosynthesis induced by mycorrhizal fungi, it is important to identify the DEGs between the two different groups. Differential expression analysis was first performed between the two treatments. DEGs (q-value < 0.005 and |log2(fold change)| > 1) were defined as genes that were significantly enriched or depleted in one treatment relative to the other. In this study, there were a total of 1388 DEGs between DF1_S and DC1_S (1382 up-regulated, 6 down-regulated). Between DF9_S and DC9_S, 2646 genes (1348 up-regulated, 1298 down-regulated) were expressed at significantly different levels. A Venn diagram shows that there were only 28 DEGs in common between DF9_S vs. DC9_S and DF1_S vs. DC1_S, implying that the expression of genes varied significantly with different stages of inoculation (Fig. 5).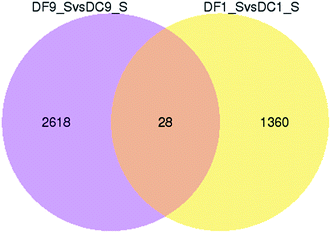 | ||
| Fig. 5 Venn diagram of differentially expressed genes in symbiotic and asymbiotic samples. “X vs. Y” means Y is the control. | ||
Functional distribution of DEGs
To analyse the functions of the DEGs, a GO analysis was performed. The results showed that there were 175 and 5 significantly enriched GO terms related to various biological processes for DF1_S vs. DC1_S and for DF9_S vs. DC9_S, respectively (Fig. 6). For DF1_S vs. DC1_S, ‘metabolic process’ (GO:0008152) was the dominant term, followed by ‘cell’ (GO:0005623), ‘cell part’ (GO:0044464), ‘intracellular’ (GO:0005622), ‘biosynthetic process’ (GO:0009058) and ‘organic substance biosynthetic process’ (GO:0044710). In other words, more DEGs were annotated to enriched GO terms involved in biological processes and cellular components. However, for DF9_S vs. DC9_S, the majority of DEGs were associated with DNA metabolism and substance binding, and there were no significantly enriched GO terms related to cellular components; this result was very different from that of DF1_S vs. DC1_S, implying that MF23 influenced different D. nobile biological processes at different stages.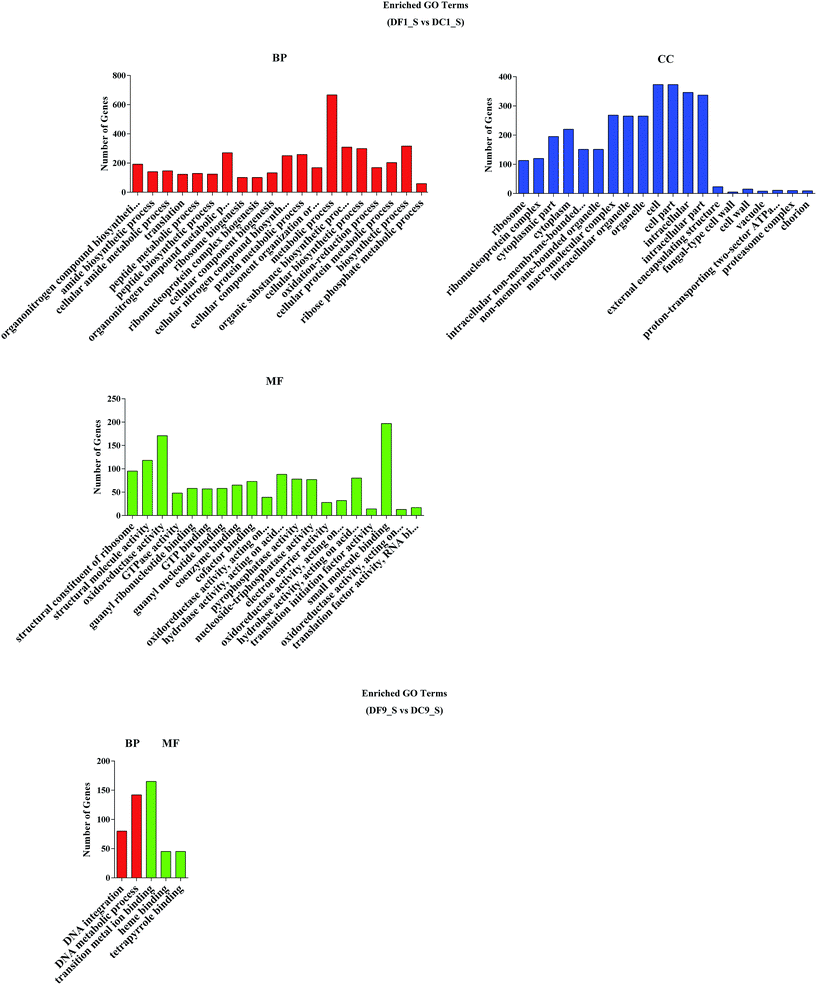 | ||
| Fig. 6 GO functional classification of differentially expressed genes. BP, biological process, CC, cellular component, MF, molecular function. | ||
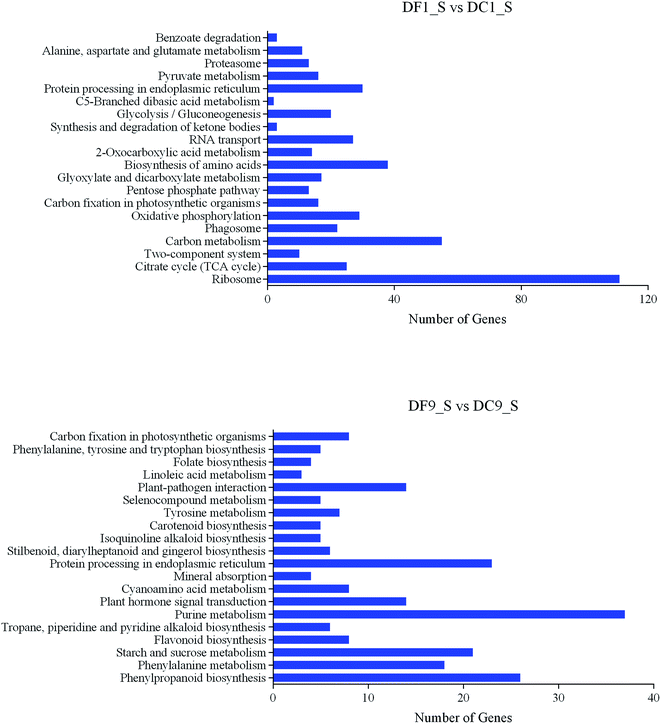
KEGG44,45 is a database resource for understanding high-level functions and utilities of the biological system from molecular-level information. KEGG pathway analyses of the DEGs from DF1_S vs. DC1_S and from DF9_S vs. DC9_S highlighted 113 and 125 pathways, respectively. The top 20 pathways of these two groups are shown in Fig. 7. Notably, among the top 20 KEGG pathways from DF1_S vs. DC1_S, 25% were associated with polysaccharide metabolism even though the changes in polysaccharide content between symbiotic and asymbiotic D. nobile were not significant at this time point; these pathways included the citrate cycle (TCA cycle), carbon metabolism, carbon fixation in photosynthetic organisms, the pentose phosphate pathway and glycolysis/gluconeogenesis. These results suggested that the effect of MF23 on the polysaccharide metabolism of D. nobile began at the early stage of symbiotic culture, even before the hyphae touched the roots.
Unigenes involved in polysaccharide accumulation
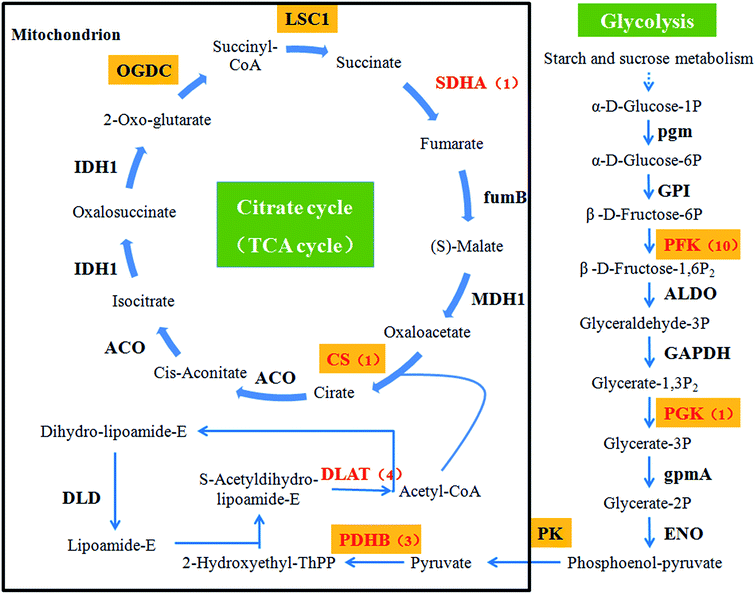
Focusing on the pathways involved in polysaccharide biosynthesis and consumption led to the identification of 6 DEGs from DF9_S vs. DC9_S, including key genes for photosynthesis (ferredoxin (petF), cytochrome c6 (petJ)), starch and sucrose metabolism (sucrose synthase (SUS), cellulose synthase (UDP-forming) (bcsA), starch synthase (glgA)) and glycolysis (phosphoglycerate kinase (PGK)) (Table 2). Furthermore, since apart from glucose, mannose is the main monosaccharide component of D. nobile polysaccharides,46 and because respiration is the major consumer of polysaccharides, the unigenes encoding key enzymes of mannose biosynthesis and respiration were identified by searching for the names of the annotated genes. A total of 5 unigenes involved in fructose and mannose metabolism were found, including one encoding phosphomannomutase (PMM) and 4 annotated to mannose-1-phosphate guanylyltransferase (GMPP) (ESI Table 2†). Regarding the pathways associated with respiration, the genes involved in the citrate cycle and glycolysis, including the pyruvate dehydrogenase E1 component beta subunit (PDHB), the pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase; DLAT), citrate synthase (CS), the succinate dehydrogenase (ubiquinone) flavoprotein subunit (SDHA), and 6-phosphofructokinase 1 (PFK), were mined (Fig. 8 and ESI Table 2†).| Direction of carbon flow | Pathway | Gene name | Unigene ID | log2fold2 | Padj3 |
|---|---|---|---|---|---|
| Biosynthesis | Photosynthesis | Ferredoxin (petF) | c72738_g2 | 1.4667 | 0.016632 |
| Cytochrome c6 (petJ) | c75563_g2 | 1.3971 | 0.019929 | ||
| Starch and sucrose metabolism | Sucrose synthase (SUS) | c89528_g2 | 6.2099 | 8.74 × 10−9 | |
| Cellulose synthase (UDP-forming) (bcsA) | c92372_g2 | 1.067 | 0.042397 | ||
| Starch synthase (glgA) | c85783_g1 | 1.7936 | 0.000142 | ||
| Consumption | Glycolysis | Phosphoglycerate kinase (PGK) | c89109_g2 | 5.0264 | 3.63 × 10−7 |
Validation and expression analysis of genes involved in polysaccharide accumulation
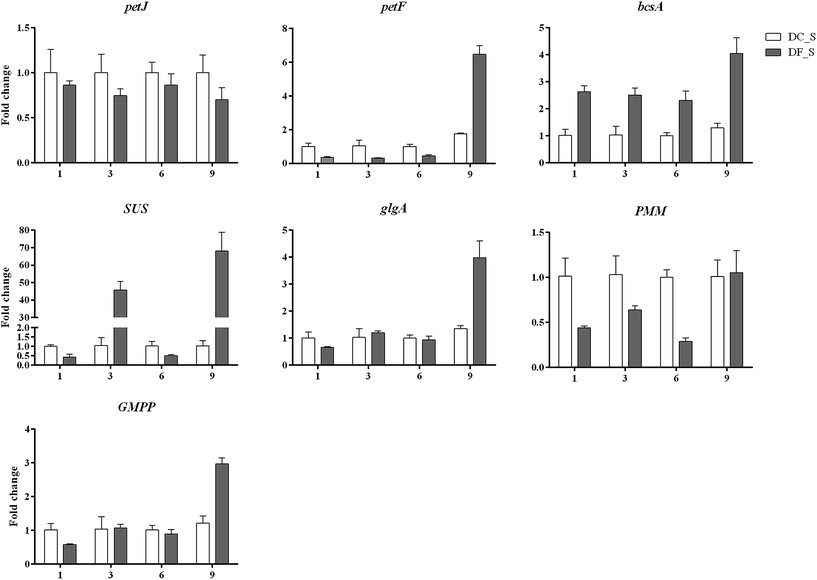
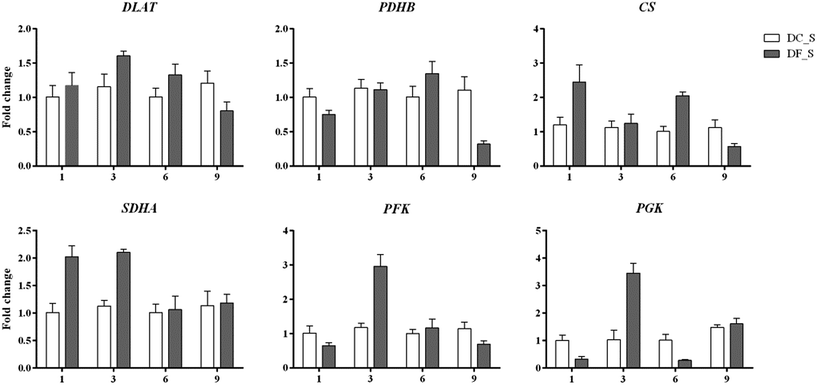
In this study, 13 key enzyme-encoding genes associated with polysaccharide accumulation, including both the biosynthetic and consumption pathways, were chosen for qRT-PCR analysis. Except for petJ and PMM, all chosen key enzyme-coding genes involved in polysaccharide biosynthesis exhibited >2-fold higher expression in D. nobile in response to MF23 than in the control at the 9th week, when the content of polysaccharides was high (Fig. 9). In contrast, the expression levels of most genes associated with polysaccharide consumption were not significantly changed at 9 weeks (Fig. 10), similar to the result obtained using Illumina sequencing technology, which indicated that our experimental results were valid.In addition, the expression level of petF, which encodes a key enzyme of photosynthesis, was markedly repressed before 9 weeks, while genes involved in respiration, PDHB, CS, SDHA and PFK, were activated to different degrees at different stages. This finding implied that D. nobile consumed more polysaccharides in the early period after induction by MF23.
Discussion
Polysaccharides are among the main active constituents of D. nobile. Because of the positive effects of these constituents on human health, research on D. nobile polysaccharides has been of great interest, especially studies on their content,47 composition48,49 and pharmacological effects.50 However, with the exception of two reports by Guo's research group,23,24 there are few reports on technology that can elevate the polysaccharide content of D. nobile or on the molecular mechanisms by which its polysaccharide content increases. In this study, we assessed the possible mechanism of the MF23-mediated induction of polysaccharide content, which helps us to understand the relationship between a mycorrhizal fungus and its host and provides a new strategy to promote the active components of D. nobile.Pelotons indicate the formation of orchid mycorrhizae. A previous isotope feeding experiment showed that in mycorrhizae, C flows into senescent pelotons and is then transferred to the host.51 In some respects, pelotons are considered pools of nutrition and centres of nutrition transformation that deliver nutrients from the host and environment to the energy-dissipating area of plant.52 Thus, MF23 might promote the biosynthesis of polysaccharides by forming pelotons to supply nutrition to its host, D. nobile.
A mycorrhizal fungus may influence its host even before colonization. Many studies have shown that the secretions of mycorrhizal fungi can rapidly and specifically induce the expression of specific genes in medicinal plants, activating a series of specific secondary metabolic pathways.53,54 For instance, when cultured on MS medium supplied with fermentation broths of mycorrhizal fungus Th-9.7-1, the expression of expansin gene in Tetrastigma hemsleyanum was significantly up – regulated (>3-fold), though there was no fungal colonization.55 The similar pattern was also found in the present study during the early period of MF23 infection. Although there was no significant change in the light microscopy and the polysaccharide content in the 1st week after inoculation, a total of 1388 DEGs were observed between DF1_S and DC1_S. In addition, 25% of the top 20 KEGG pathways in the 1st week were involved in polysaccharide metabolism. These results indicate that the effect of MF23 on the polysaccharide metabolism of D. nobile occurred much earlier than colonization. MF23 may release a ‘fungal elicitor’56,57 into the culture medium to produce physiological changes in D. nobile and then promote polysaccharide metabolism.
Photosynthesis is the main pathway by which plants obtain carbohydrates. Among the key enzymes involved in photosynthesis, petF, the terminal electron acceptor of photosystem I (PSI) seems more important.58 It delivers electrons to FDX-NADPH-oxidoreductase (FNR) to generate NADPH, which is required for carbon assimilation in the Calvin cycle.59 Furthermore, evidence has shown that Thalassiosira oceanica expressing exogenous petF has increased tolerance to severe environments by maintaining the electron transport interface of photosynthesis intact.60 Since polysaccharides are the main carbohydrates in plant, petF plays a vital role in the biosynthesis of polysaccharides. In this study, the expression of petF markedly increased by more than 6-fold in symbiotic D. nobile 9 weeks after inoculation, when the content of polysaccharides was elevated; prior to this time point, petF expression was at a very low level. These findings strongly support the importance of petF and imply that MF23 may stimulate polysaccharide biosynthesis by activating the expression of petF to assimilate more carbon.
Cellulose contributes to the structural support of the cell wall and is the most common polysaccharide in plants.61 The expression of bcsA, which encodes cellulose synthase, was clearly and significantly induced (over 4-fold) in the model group in the 9th week. This induction might be one of the most important reasons for the accumulation of polysaccharides. Interestingly, although the expression levels of bcsA in symbiotic D. nobile in the 1st, 3rd, and 6th weeks were lower than those in the 9th week, they were higher than those of the control group all time points. The plant cell wall thickens in response to stress.62 During resistance responses, the secondary walls of the xylem in rice seedlings thicken within 48 h, effectively reducing the area of the pit membrane that may be accessed by bacteria.63 Similarly, the induction of bcsA in symbiotic D. nobile might also be associated with plant defence mechanisms.
SUS is one of the key enzymes of sucrose metabolism, primarily affecting the degradation of sucrose to provide precursors for polysaccharides.64 For Anabaena sp., the accumulation of polysaccharide in a mutant strain constitutively overexpressing SUS increase to 2-fold more than those of the mutant strain with an insertion inactivation of SUS and the wild strain, which demonstrated the importance of SUS in the biosynthesis of polysaccharides.65 In recent years, many studies have indicated that for D. huoshanense and D. officinale, an increase in the polysaccharide content was always accompanied by the activation of SUS.66–68 Therefore, in D. nobile, SUS may play the same role in polysaccharide biosynthesis is in these related plant species. This conjecture was supported by the 68.06-fold higher expression of SUS over that in the controls in the 9th week. This result means that MF23 might increase the polysaccharide content by activating the expression of SUS.
Starch is the final product of photosynthesis, and it stores energy from photosynthesis for many types of metabolism. Starch is complex. On the one hand, starch itself is a polysaccharide; on the other hand, starch can be degraded to glucose, the main constituent of different types of polysaccharides. Regardless of the role of starch, its accumulation has a positive impact on the accumulation of polysaccharides. Evidence has suggested a positive correlation between the contents of starch and polysaccharides in Dendrobium.69 Thus, the effect of the starch synthase gene glgA on polysaccharide biosynthesis is non-negligible. In this study, the expression of glgA rose sharply in the 9th week when the polysaccharide content was high, implying that glgA plays a vital role in the influence of MF23 on polysaccharide biosynthesis.
By using precolumn derivation HPLC, more than 60% monosaccharice compositions in polysaccharide of D. nobile were identified as mannose, which implied that mannose-containing polysaccharides are main types of polysaccharides in D. nobile.46 GDP-mannose is a donor for the biosynthesis of mannose-containing polysaccharides.70 PMM catalyses the conversion of mannose-6-phosphate to mannose-1-phosphate, which is the precursor for the biosynthesis of GDP-mannose.71 GMPP catalyses the formation of GDP-mannose directly.72 Thus, changes in PMM and GMPP might make a substantial difference in the proportion of mannose-containing polysaccharides in D. nobile and lead to an increase in polysaccharide accumulation. In this study, the expression level of PMM was significantly repressed at the first three stages, but returned to the control level in the 9th week after inoculation. This result suggested that PMM might play an indirect but essential role in the regulation of polysaccharide biosynthesis in symbiotic D. nobile. Unlike PMM, GMPP was expressed at a high level in response to MF23 when the polysaccharide content was high. This corresponding relationship demonstrated that MF23 might increase the polysaccharide content by stimulating the expression of GMPP.
As shown above, nearly all the genes that we chose that were involved in the polysaccharide biosynthesis pathway were up-regulated in the 9th week. However, polysaccharide accumulation also depends on the carbohydrate consumption rates, which are affected by glycolysis and the citrate cycle. Early studies showed that the exogenous expression of PFK, encoding a key enzyme of the glycolysis pathway, reduced the sugar content of potato tubers.73 For higher organisms, there is a positive correlation between the respiration rate and the enzyme activity in the citrate cycle.74–76 In this study, we assessed the relationship between 6 key genes in these two pathways, including DLAT, PDHB, CS and SDHA, PFK and PGK, as well as the polysaccharide content (Fig. 8). No genes were markedly induced in the 9th week. Instead, PDHB was significantly down-regulated to levels 3.46-fold lower than those of controls. These results suggested that the consumption of polysaccharides was at a normal or low level in the 9th week. Thus, these results that the activation of genes involved in polysaccharide biosynthesis in the 9th week might be the main reason for polysaccharide levels to increase in D. nobile infected with MF23.
Before the 9th week, CS, SDHA, PFK and PGK in symbiotic D. nobile were affected to different degrees at different stages. For instance, the expression levels of SDHA, PFK and PGK were up-regulated more than 2-fold in the 3rd week, implying that MF23 might influence the rate of respiration at the early stage of inoculation; this result was in accordance with the results from Cymbidium sinense and C. ensifolium infected with a mycorrhizal fungus.77 CS, PFK and PGK catalyse the pivotal irreversible reaction that leads to the consumption of carbohydrates.78 SDHA participates in both the citrate cycle and the electron transport chain.79 Their stimulation would undoubtedly result in a reduction in polysaccharides. However, there was no significant change in the polysaccharide content before the 9th week. A further analysis of recent experimental data showed that the high expression levels of two key genes, SUS and bcsA, which were associated with starch and sucrose metabolism before the 9th week, might account for this lack of change. These findings strongly suggested that MF23 might influence the polysaccharide content by regulating both the biosynthesis and consumption pathways of its host and that the combination of the effects of these two pathways determines the accumulation of polysaccharides. However, the polysaccharide metabolic network is complex, and further research regarding the mechanisms that regulate it should be performed.
Conclusions
To determine the mechanism by which polysaccharide accumulation is regulated in D. nobile infected with the mycorrhizal fungus MF23, the first large-scale transcriptome dataset from D. nobile (control and model groups) was generated using RNA-Seq. Using the results of RNA-Seq and qRT-PCR, we showed that the regulatory network by which MF23 acts on D. nobile is complex. MF23 might regulate both the biosynthesis and consumption of polysaccharides to change polysaccharide accumulation. The combined effects of the activated biosynthetic pathway and the depressed consumption pathway might result in the accumulation of polysaccharides. Among the analysed genes, petF, SUS, bcsA, PMM, GMPP, PDHB, CS, SDHA, PGK and PFK might play vital roles in this regulatory network. This study provides a good example of an endophyte promoting the formation of bioactive compounds in its host and paves the way for further investigations of the associated regulatory mechanisms using molecular methods.Acknowledgements
The research was financially supported by the National Natural Science Foundation of China (No. 31170314, No. 81473331).Notes and references
- X. S. Bao, Q. S. Shun and C. L. Zhuan, The medicinal plants of dendrobium(shi-hu)in china a coloured atlas, Fudan University Press, Shanghai, 2001 Search PubMed.
- S. Chen, Y. Li, Y. Wu, Z. Zhou and L. Sun, China J. Chin. Mater. Med., 1995, 20, 181–182 CAS.
- M. Y. Yoon, J. H. Hwang, J. H. Park, M. R. Lee, H. J. Kim, E. Park and H. R. Park, J. Med. Food, 2011, 14, 120 CrossRef PubMed.
- Q. Huang, X. Liao, W. U. Qin, L. I. Fei, L. Y. Wang and J. S. Shi, Chin. J. New Drugs Clin. Rem., 2013, 32, 490–493 CAS.
- M. M. Li, B. X. Zhang, S. B. He, R. Zheng, Y. L. Zhang and Y. Wang, China J. Chin. Mater. Med., 2015, 40, 3709–3712 Search PubMed.
- Q. Ye, G. Qin and W. Zhao, Phytochemistry, 2002, 61, 885–890 CrossRef CAS PubMed.
- Y. P. Li, J. H. Jiang, Y. Liu and Y. G. Chen, Lishizhen Med. Mater. Med. Res., 2010, 21, 39–40 CAS.
- X. M. Zhang, Z. R. Sun, L. Chen, X. X. Wei and W. L. Liu, Chin. J. Mod. Appl. Pharm., 2014, 31, 895–899 CAS.
- F. J. An and Y. X. He, J. Anhui Agric. Sci., 2014, 42, 3857–3862 CAS.
- L. N. Wang, Q. H. Gong, F. Li, Q. Wu and J. S. Shi, Acta Neuropharmacologica, 2016, 6, 89 Search PubMed.
- C. J. Li, Yunnan Journal of Traditional Chinese Medicine and Meteria Medica, 2012, 33, 61–62 Search PubMed.
- Z. H. Wang, J. Li, J. H. Zang, Y. C. Xu and Q. S. Ye, Chinese Journal of Tropical Crops, 2016, 37, 2333–2337 Search PubMed.
- X. J. Ge, L. M. Zheng, Y. L. Wang and Y. P. Tang, Chongqing Med., 2015, 44, 1305–1310 CAS.
- S. Z. Zheng, Effects of the proliferation and apoptosis of K562 cell affects by Dendrobium nobile polysaccharides, Zunyi Medical University, 2009 Search PubMed.
- A. Luo and Y. Fan, Afr. J. Pharm. Pharmacol., 2011, 5, 625–631 CrossRef CAS.
- J. H. Wang, Carbohydr. Polym., 2010, 79, 114–118 CrossRef CAS.
- Y. Y. Xu, L. Y. Wang, B. Huang, X. L. Xie, Q. Wu and J. S. Shi, West China J. Pharm. Sci., 2014, 29, 288–291 CAS.
- M. A. Selosse and M. Roy, Trends Plant Sci., 2009, 14, 64–70 CrossRef CAS PubMed.
- Q. X. Wang, N. Yan, D. G. Ji, S. Y. Li, J. M. Hu and H. Hu, Plant Diversity Resour., 2014, 36, 321–330 CAS.
- L. Q. Chen, X. M. Wang and Z. D. Pei, Forest Res., 2005, 18, 163–168 Search PubMed.
- L. C. Zhang and S. X. Guo, J. Chin. Pharm. Sci., 2009, 44, 1540–1543 Search PubMed.
- J. Chen, K. X. Hu, X. Q. Hou and S. X. Guo, World J. Microbiol. Biotechnol., 2011, 27, 1009–1016 CrossRef.
- H. L. Yan, X. M. Chen, F. H. Liao, X. F. Deng, C. L. Wang, L. X. Tian, S. X. Guo and J. Y. Yang, Chin. Pharm. J., 2016, 51, 1450–1454 Search PubMed.
- X. M. Chen and S. X. Guo, China J. Chin. Mater. Med., 2005, 30, 253–257 CAS.
- Y. L. Wang, W. H. Chen, Y. X. Xie, J. Xu and J. Wang, Soil Fert. Sci. China, 2000, 32–34 Search PubMed.
- Z. Y. Chen, Correlation between polysaccharides accumulation and sugar metabolism in Dendrobium officinale, Zhejiang A & F University, 2015 Search PubMed.
- F. C. Zhao, L. Q. Jing, D. L. Lu, G. Y. Wang and W. P. Lu, J. Nucl. Agric. Sci., 2014, 28, 2230–2237 CAS.
- X. C. Liu and A. Vonshak, J. Guangxi Norm. Univ., 2003, 21, 91–95 CAS.
- L. Zhang, J. Chen, Y. Lv, C. Gao and S. Guo, Mycol. Progr., 2012, 11, 395–401 CrossRef.
- X. M. Chen, H. L. Yan, C. L. Wang, L. X. Tian, A. R. Wang and S. X. Guo, Sci. China: Life Sci., 2016, 59, 974–976 CrossRef CAS PubMed.
- N. P. C., Pharmacopoeia of the People's Republic of China, China Medical Science Press, 2015, vol. 1, pp. 282–283 Search PubMed.
- M. Dubois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Anal. Chem., 1956, 28, 350–356 CrossRef CAS.
- N. Feder and T. P. O'Brien, Am. J. Bot., 1968, 55, 123–142 CrossRef.
- M. M. Liu, Y. M. Xing, D. W. Zhang and S. X. Guo, Sci. Rep., 2015, 5, 16075 CrossRef CAS PubMed.
- M. Pei, J. Niu, C. Li, F. Cao and S. Quan, BMC Genomics, 2016, 17, 132 CrossRef PubMed.
- M. G. Grabherr, B. J. Haas, M. Yassour, J. Z. Levin, D. A. Thompson, I. Amit, X. Adiconis, L. Fan, R. Raychowdhury and Q. Zeng, Nat. Biotechnol., 2011, 29, 644–652 CrossRef CAS PubMed.
- L. Wang, Z. Feng, X. Wang, X. Wang and X. Zhang, Bioinformatics, 2010, 26, 136 CrossRef CAS PubMed.
- J. D. Storey and R. Tibshirani, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9440–9445 CrossRef CAS PubMed.
- M. D. Young, M. J. Wakefield, G. K. Smyth and A. Oshlack, Genome Biol., 2010, 11, 1–12 CrossRef PubMed.
- X. Mao, T. Cai, J. G. Olyarchuk and L. Wei, Bioinformatics, 2005, 21, 3787 CrossRef CAS PubMed.
- W. Chen, X. Cheng, Z. Zhou, J. Liu and H. Wang, Mol. Biol. Rep., 2013, 40, 1145–1154 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- G. Hadley, Mycorrhizas and plant growth and development, Springer, Netherlands, 1988 Search PubMed.
- M. Kanehisa, M. Araki, S. Goto, M. Hattori, M. Hirakawa, M. Itoh, T. Katayama, S. Kawashima, S. Okuda and T. Tokimatsu, Nucleic Acids Res., 2008, 36, D480–D484 CrossRef CAS PubMed.
- M. Kanehisa, Y. Sato, M. Kawashima, M. Furumichi and T. Mao, Nucleic Acids Res., 2016, 44, D457 CrossRef PubMed.
- X. U. Li, L. Guo, F. L. Luo, T. M. Zhang, X. W. Liu and Y. Han, Lishizhen Med. Mater. Med. Res., 2014, 25, 1725–1727 Search PubMed.
- Y. Y. Wang, H. Xu, S. S. Shi, L. S. Wang and Z. T. Wang, J. Chin. Med. Mater., 2009, 32, 493–495 CAS.
- A. X. Luo, Carbohydr. Polym., 2010, 79, 1014–1019 CrossRef CAS.
- J. H. Wang, Carbohydr. Polym., 2010, 81, 1–7 CrossRef CAS.
- Y. L. Zhang, X. R. He, Y. Y. Liu, L. X. Qiong and Y. P. Tang, Lishizhen Med. Mater. Med. Res., 2013, 24, 7–9 CAS.
- Y. Kuga, N. Sakamoto and H. Yurimoto, New Phytol., 2014, 202, 594–605 CrossRef CAS PubMed.
- R. L. Wang, H. Hu and S. Y. Li, Acta Bot. Yunnanica, 2004, 26, 445–450 Search PubMed.
- X. Zhai, M. Jia, L. Chen, C. J. Zheng, K. Rahman, T. Han and L. P. Qin, Crit. Rev. Microbiol., 2016, 43, 238–261 CrossRef PubMed.
- Y. Tan, R. Jia, J. H. Tao, N. Y. Yang, J. A. Duan, D. W. Qian and S. Jiang, Chin. Tradit. Herb. Drugs, 2013, 44, 2004–2008 CAS.
- Y. L. song, Molecular cloning of expansion gene, isolation and identification of emdophytic fungi from root tuber of Tetrastigma hemsleyanum, Hangzhou Normal University, 2016 Search PubMed.
- X. M. Chen, S. X. Guo and Z. X. Meng, Chin. Pharm. J., 2006, 41, 423–426 Search PubMed.
- J. H. Chen, C. Wang, L. Shi and J. M. Yin, Hunan Agricultural Sciences, 2014, 28–31 Search PubMed.
- B. Munge, S. K. Das, R. Ilagan, Z. Pendon, J. Yang, H. A. Frank and J. F. Rusling, J. Am. Chem. Soc., 2003, 125, 12457 CrossRef CAS PubMed.
- K. Fukuyama, Photosynth. Res., 2004, 81, 289–301 CrossRef CAS PubMed.
- M. Lommer, A. S. Roy, M. Schilhabel, S. Schreiber, P. Rosenstiel and J. Laroche, BMC Genomics, 2010, 11, 718 CrossRef CAS PubMed.
- D. L. Song, J. H. Shen and L. G. Li, Plant Physiol. Commun., 2008, 44, 791–796 CAS.
- G. H. Le, F. Philippe, J. M. Domon, F. Gillet, J. Pelloux and C. Rayon, Plants, 2015, 4, 112–166 CrossRef PubMed.
- E. Hilaire, S. A. Young, L. H. Willard, J. D. Mcgee, T. Sweat, J. M. Chittoor, J. A. Guikema and J. E. Leach, Mol. Plant-Microbe Interact., 2001, 14, 1411–1419 CrossRef CAS PubMed.
- S. Chalivendra, S. C. Huber, M. M. Sachs and D. Rhoads, Plant Signaling Behav., 2007, 2, 28–29 CrossRef.
- L. Curatti, L. E. Giarrocco, A. C. Cumino and G. L. Salerno, Planta, 2008, 228, 617–625 CrossRef CAS PubMed.
- M. Wei, S. T. Jiang and J. P. Luo, Biotechnol. Lett., 2007, 29, 495–499 CrossRef CAS PubMed.
- J. Yang, H. L. Meng, S. C. Yang, W. Zhang, Y. H. Zha and G. S. Wen, J. West China For. Sci., 2012, 41, 62–67 CAS.
- M. Wei, S. T. Jiang and J. P. Luo, Chin. J. Biotechnol., 2007, 23, 327–331 CAS.
- H. Zhu, J. Teng, Y. Cai, J. Liang, Y. Zhu and T. Wei, China J. Chin. Mater. Med., 2011, 36, 3262–3264 CAS.
- C. M. He, J. X. Zhang, C. Wang, K. L. Wu, S. J. Zeng and J. Duan, The Congress of Botanical Society of China and the 80th Anniversary, 2013 Search PubMed.
- C. He, S. Zeng, J. A. T. D. Silva, Z. Yu, J. Tan and J. Duan, Protoplasma, 2016, 1–12 Search PubMed.
- T. K. Hyun, J. D. Lim, J. K. Kim, E. S. Seong, J. G. Lee, B. S. Yoon, M. J. Kim, D. H. Cho and C. Y. Yu, Br. J. Orthod., 2005, 13, 155–158 Search PubMed.
- O. Navrátil, P. Bucher and J. Vacek, Czech J. Genet. Plant Breed., 2012, 48, 42–45 Search PubMed.
- L. J. Liu, J. Cang, H. W. Li, j. Yu, X. Wang, J. F. Wang, R. Huang and C. Xu, J. Triticeae Crops, 2013, 33, 65–72 CAS.
- A. Nunes-Nesi, L. J. Sweetlove and A. R. Fernie, Physiol. Plant., 2007, 129, 45–56 CrossRef CAS.
- X. G. Zhu, Photosynthetic carbon assimilation and respiration, Higher Education Press, Beijing, 2012 Search PubMed.
- C. Pan, R. Chen and L. Li, J. Trop. Subtrop. Bot., 2002, 10, 46–50 CAS.
- R. C. Pan, X. J. Wang and N. H. Li, Plant Physiology, Higher Education Press, Beijing, 2004 Search PubMed.
- S. B. Huang, N. L. Taylor, E. Ströher, R. Fenske and A. H. Millar, Plant J., 2013, 73, 429–441 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra02010d |
| This journal is © The Royal Society of Chemistry 2017 |