Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dynamics of naphthenic acids and microbial community structures in a membrane bioreactor treating oil sands process-affected water: impacts of supplemented inorganic nitrogen and hydraulic retention time†
Jinkai Xuea,
Yanyan Zhangab,
Yang Liu*a and
Mohamed Gamal El-Din *a
*a
aDepartment of Civil and Environmental Engineering, University of Alberta, Edmonton, Alberta T6G 1H9, Canada. E-mail: yang.liu@ualberta.ca; mgamalel-din@ualberta.ca; Fax: +1-780-492-0249; Fax: +1-780-492-0249; Tel: +1-780-492-5115 Tel: +1-780-492-5124
bDepartment of Civil Engineering, New Mexico State University, Las Cruces, New Mexico 88003, USA
First published on 22nd March 2017
Abstract
This study was focused on how different operating conditions affected the biodegradation of naphthenic acids (NAs) and the microbial community architectures in an anoxic–aerobic membrane bioreactor (MBR) for oil sands process-affected water (OSPW) treatment. After 442 days of continuous optimization, a supplemented NH4–N concentration of 25 mg L−1 and a hydraulic retention time (HRT) of 12 h demonstrated the best removal rates of total classical NAs (37.6%) and total oxidized NAs (23.9%). Neither higher HRTs nor higher supplemented NH4–N concentrations resulted in a better overall removal of NAs. In addition, NAs with larger carbon numbers were generally better degraded, whereas higher cyclicity tended to lessen the biodegradability of NAs. MiSeq sequencing analysis disclosed that orders under Proteobacteria (i.e., Rhodocyclales, Burkholderiales and Nitrosomonadales), Bacteroidetes (i.e., Cytophagales, [Saprospirales] and Flavobacteriales), and Nitrospirae (i.e., Nitrospirales) were the major microbes over the whole study though their relative abundances varied. The results of this study provide insightful information for future studies and application of biological processes for OSPW treatment on a large scale.
1. Introduction
Due to the huge water demand (3.1 barrels of water per barrel of bitumen produced) in the modified Clarke caustic hot water extraction process in the oil sands industry, the total area of existing tailings ponds (storing oil sands process-affected water, OSPW) in Canada reached 182 km2 (including associated structures) by the year 2013.1 Among all the organic acids in OSPW, naphthenic acids (NAs) have been reported as the major contributors to OSPW toxicity and persistency. They are known to be acutely toxic to a range of organisms, including microorganisms, algae, vertebrates and invertebrates.2–5 Therefore, NAs are regarded as the target pollutants that need to be treated urgently.6–9NAs are a wide group of aliphatic and alicyclic, alkyl-substituted carboxylic acids with a generalized formula CnH2n+zOx, in which n refers to molecular carbon number, z is zero or a negative even integer indicating hydrogen deficiency caused by the formation of rings or double bond equivalents,4,10 and x refers to the number of oxygen atoms. Species with x = 2 are referred to as classical NAs, while those with x ≥ 3 are termed oxidized NAs (oxy-NAs).11 It has been estimated that more than 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individual NAs are potentially associated with OSPW.12 Hanging on the ore composition, extraction processes, the tailings age and quantification approaches, the NAs concentrations reportedly varied greatly within the range of 20–120 mg L−1.3,13–15 It has been suggested that NAs with a carbon number n ≤ 21 and/or linearly grouped carbon rings may be more toxic in OSPW;4,16–18 whereas NAs with ring structures tend to be more persistent in environment.2 It is, therefore, desirable to remove NAs from OSPW to mitigate its environment impact.
000 individual NAs are potentially associated with OSPW.12 Hanging on the ore composition, extraction processes, the tailings age and quantification approaches, the NAs concentrations reportedly varied greatly within the range of 20–120 mg L−1.3,13–15 It has been suggested that NAs with a carbon number n ≤ 21 and/or linearly grouped carbon rings may be more toxic in OSPW;4,16–18 whereas NAs with ring structures tend to be more persistent in environment.2 It is, therefore, desirable to remove NAs from OSPW to mitigate its environment impact.
It is suggested that biodegradation is responsible for the natural, but slow dissipation of NAs in tailings ponds.19 However, the in situ biodegradation half-lives of NAs are within a range of 12.8–13.6 years.20 To accelerate the biodegradation of OSPW NAs, it is crucial to develop engineered biological processes that are efficient in mitigating OSPW organic compounds. Choi and Liu performed a study on the treatment of OSPW by using two sequencing batch reactors (SBRs) and achieved an acid extractable fraction (AEF) removal rate of 8.7% and 16.6% at a hydraulic retention time (HRT) of 24 h with the inoculation of activated sludge and mature fine tailings (MFT), respectively.6 Hwang et al. reported that 13.8% of OSPW AEF and 18.5% of the OSPW parent NAs were eliminated by OSPW indigenous microorganisms in a biofilm reactor with an HRT of 19 h.7 Hence, it is suggestive that disparate reactor configurations have different capabilities in degrading OSPW organic substances.6,21,22
Membrane bioreactor (MBR) is finding wider application for municipal and industrial wastewater treatment because of its compactness, high treatment capacity, low sludge production and excellent effluent quality.23,24 To investigate the feasibility of MBR for OSPW treatment, an anoxic–aerobic MBR system was performed at an HRT of 48 h, and attained an OSPW NA removal of 25% after the startup period (day 1–425).25 To further improve its performance for OSPW treatment, more effort is needed to optimize the operating conditions.
HRT is one of the essential parameters in MBR operation. It not only affects the extent to which the target contaminants are degraded,26 but also profoundly impacts membrane fouling development.27,28 Longer HRTs are usually required for industrial wastewater, especially those containing complex organic contaminants.29 However, the effect of HRT on the degradation of organic pollutants was still unclear as indicated by some studies.28,30–32 On the other hand, a shorter HRT is favorable as it reduces the required reactor volume (less investment) for the same treatment capacity. In addition, HRT has an indirect yet profound impact on membrane fouling through altering of sludge properties (i.e., the concentration of mixed liquor suspended solids (MLSS), the production of extracellular polymeric substances (EPS) and the viscosity of sludge).33–35 Due to increased membrane flux, shortened HRTs usually result in severer membrane fouling,27,28,33 which could increase the operating cost substantially. Therefore, a balance between contaminant removal and operation cost must be considered for HRT selection.29
Further, ammonia monooxygenase (amo) produced by ammonia oxidizing bacteria (AOB) is able to degrade a wide spectrum of refractory organic substrates (including hydrocarbons) through co-metabolism.36–40 Nitrifiers are not necessarily stimulated by higher ammonium nitrogen (NH4–N) concentrations as free ammonia (FA) is inhibitory to many microbial activities including nitrification.41 Given the alkaline nature of OSPW, the effect of FA should not be ignored. Therefore, it is needed to find out an optimal supplemented NH4–N concentration to maximize the MBR's performance of NA degradation.
The objectives of this study were: (1) to explore the optimal operation conditions to improve the MBR's NA degradation efficiency; and (2) to characterize the microbial community structure in the MBR with the aim of uncovering the most effective bacterial groups associated with NA removal. According to our literature search, this is the very first study evaluating the performance (particularly the biodegradation of classical and oxy-NAs) and characterizing the microbial communities of an MBR for OSPW treatment under various operating conditions.
2. Methodology
2.1 Source waters, external carbon source and supplemented nutrients
The OSPW was collected from the same tailings pond in Northern Alberta region. The OSPW used for the optimization operation had a pH of 8.8 ± 0.3, chemical oxygen demand (COD) of 224.1 ± 46.3 mg L−1, 5 day biochemical oxygen demand (BOD5) of 5.4 ± 0.8 mg L−1, classical NAs of 49.9 ± 4.0 mg L−1, and oxy-NAs of 39.8 ± 4.2 mg L−1. Raw OSPW was received in 200 liter barrels and preserved in dark at 4 °C prior to treatment. To facilitate the growth of microorganisms in the system, sodium acetate was supplied as an external carbon (C) source at a constant organic loading rate (OLR) of 125 g COD per (m3 d) throughout the whole operation. To ensure the sufficiency of nitrogen (N), phosphorus (P), and other trace nutrients, supplemented KNO3, NH4Cl, NaH2PO4, and modified Bushnell-Haas medium (BHM)17 without KH2PO4, Na2HPO4, NH4NO3, and (NH4)2SO4 were added in the feed water. To avoid undesirable microbial growth in the feed tank, the stock solution of external C source, and the stock solution of external N (to achieve equivalently 75 mg N per L in the feed) and P (to achieve equivalently 10 mg P per L in the feed) source were continuously injected in the MBR at a pre-set rate by using a syringe pump (KDS Infusion Pump, Model 220, KDS Scientific Inc., USA).2.2 MBR configuration and operating conditions
The configuration of the MBR is illustrated in Fig. S1.† Detailed description on the system configuration, biomass inoculation and acclimatization is available elsewhere.25,42 Dissolved oxygen (DO) concentrations in the anoxic tank and the aerobic tank were maintained at a level of <0.3 mg L−1 and >4.0 mg L−1, respectively. The retentate in the aerobic tank was recirculated to the anoxic tank by overflow at a recycling ratio of 2.The different operating conditions examined in this study are tabulated in Table 1. Despite the changes made to NO3–N and NH4–N concentrations, the total inorganic nitrogen (TIN, i.e., NH4–N, NO2–N and NO3–N) concentration supplemented to the system was maintained at 75 mg N per L throughout the study. The system automatically performed a 30 second backwashing after every 9.5 minutes of membrane filtration, comprising a 10 minute working phase. The relaxation duration between two working phases was adjusted to achieve a desired equivalent HRT. According to the manufacturer's suggestion, chemical cleaning (soaking the membrane module in 0.1% HClO solution for 1 hour followed by rinsing with running tap water) was applied once the transmembrane pressure (TMP) exceeded −35 kPa (severe fouling event). Sludge in the MBR was wasted through weekly sampling (30 mL from each tank for solids content measurements) to achieve a calculated solid retention time (SRT) of ∼187 days.
| Stage | Period (days) | Supplemented nitrogen | HRT (h) | |
|---|---|---|---|---|
| NH4–N (mg L−1) | NO3–N (mg L−1) | |||
| A25H48 | 300–449 | 25 | 50 | 48 |
| A75H48 | 449–546 | 75 | 0 | 48 |
| A50H48 | 546–630 | 50 | 25 | 48 |
| A25H72 | 630–680 | 25 | 50 | 72 |
| A25H12 | 680–712 | 25 | 50 | 12 |
| A25H24 | 712–742 | 25 | 50 | 24 |
2.3 Analyses
Methods of conventional water chemistry analyses and acute aquatic toxicity bioassay are described in the (ESI†).3. Results and discussion
3.1 Classical and oxidized NA removal performance
Table 2 presents total classical NAs concentrations of the feeds and permeates under different conditions and the estimated half-life of classical NAs under each condition. By degrading 37.6% of classical NAs within 12 h, A25H12 stage demonstrated the shortest half-life (i.e., 17.6 h) of the contaminants among the six stages. In contrast, A25H72 achieved the longest half-life (i.e., 159.7 h) of classical NAs by removing 26.8% within 72 h. The half-lives of classical NAs in all the six operating conditions were substantially shorter than the estimated half-life of 44–240 days in Mahdavi et al.43 and Han et al.'s14 suspended biological systems, and even negligible compared with the estimated ∼13 year in situ half-life in tailings ponds.20 The shortest half-life exhibited by our previous batch suspended reactors was 20 days.44 In Hwang et al.'s continuous biofilm reactor, 18.5% of classical NAs was removed within 19 h (half-life = 64.4 h).7 More recently Huang et al.'s, integrated fixed-film activated sludge system (IFAS) demonstrated a half-life of 59.0 h of classical NAs.45 Therefore, the performance of MBR for OSPW NA degradation could be advantageous over conventional activated sludge systems and biofilm reactors.| Operating condition | Classical NAs (mg L−1) | Removal (%) | Estimated half-life (h) | |
|---|---|---|---|---|
| Feed | Permeate | |||
| A75H48 | 45.3 | 34.9 | 23.0 | 127.6 |
| A50H48 | 51.8 | 35.9 | 30.7 | 90.7 |
| A25H48 | 51.4 | 34.1 | 33.7 | 81.1 |
| A25H72 | 44.7 | 32.7 | 26.8 | 159.7 |
| A25H24 | 55.3 | 35.4 | 36.0 | 37.3 |
| A25H12 | 44.7 | 27.9 | 37.6 | 17.6 |
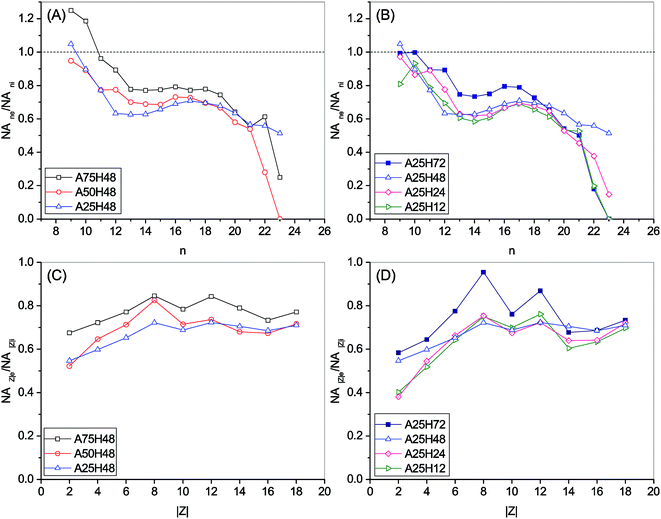
Fig. 1 is prepared to examine the effect of n and Z on biodegradation of NAs. A relative abundance of residual classical NAs (NA∑ne/NA∑ni) is used to indicate the extent of biodegradation of NAs with each n. NA∑ne denotes the total concentration of NAs with the same carbon number n in the effluent, while NA∑ni is the total concentration of NAs with the same n in the influent. Therefore, a lower NA∑ne/NA∑ni means a better removal of the group of NAs with n. Similarly, a relative abundance of residual classical NAs (NA∑|Z|e/NA∑|Z|i) is used to indicate the extent of biodegradation of NAs with each |Z|. To better disclose how molecular n and |Z| affected the degradation of NAs, Fig. 2 and 3, and Fig. S3 and S4† are plotted based on the NA concentration profiles.
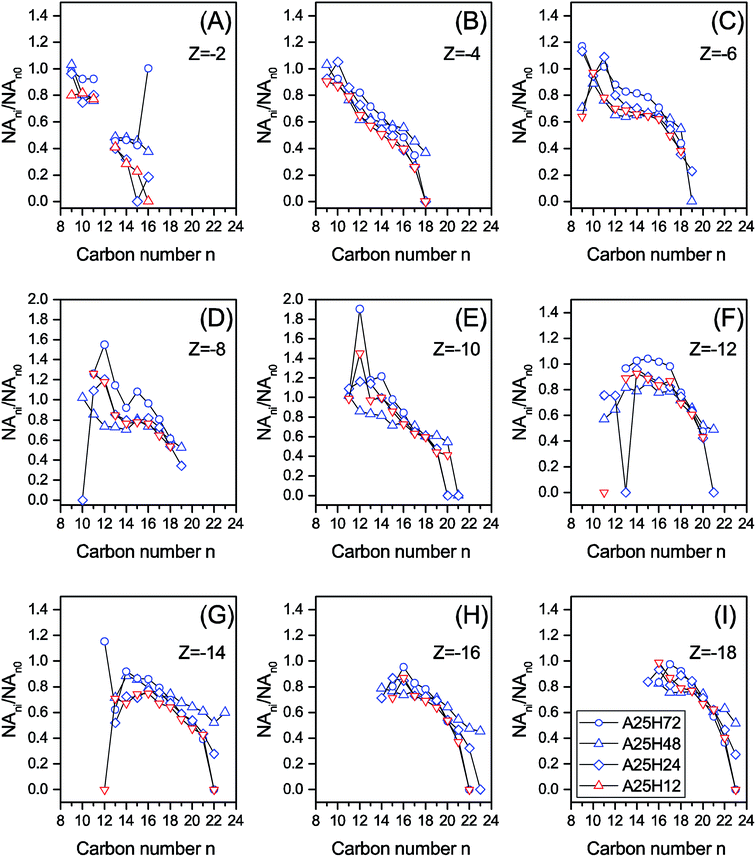 | ||
| Fig. 2 The relative remaining abundance of NAs with different carbon numbers (NAni/NAn0) within each Z series of the permeate samples at different HRTs. | ||
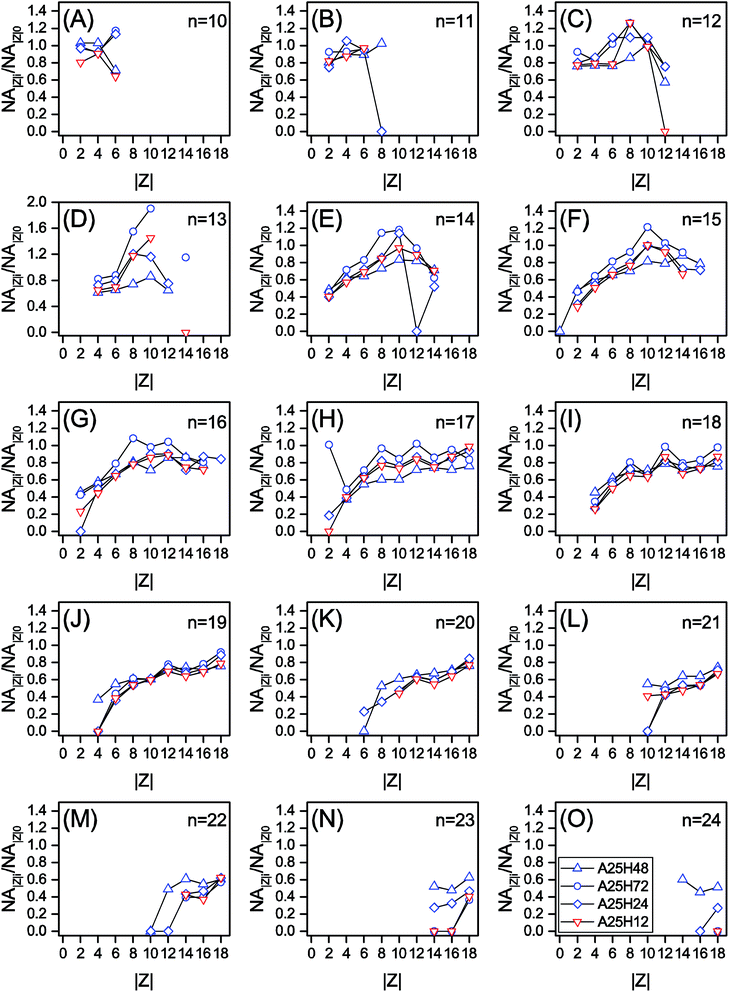 | ||
| Fig. 3 The relative remaining abundance of NAs with different cyclicity (NAZi/NAZ0) within each n series of the permeate samples at different HRTs. | ||
Although the stages differed in operating conditions, similar patterns could be seen for NA∑ne/NA∑ni over the n range in Fig. 1A and B. For C9–12 and C19–23, a higher NA∑ne/NA∑ni ratio is associated with a lower n, whereas a plateau or a slightly climbing trend of the NA∑ne/NA∑ni ratio is observed when n increases from 13 to 18 in Fig. 1A and B. As demonstrated in Fig. 2 and S3,† species with more carbon numbers were generally better degraded within each Z series, which is in agreement with our previous batch study.44 In contrast, previous studies indicated that n has either negatively proportional correlation to or minor influence on biodegradation of NAs.14,17,46 This discrepancy evidences that the capabilities of biological systems are considerably influenced by their configurations. In addition, it is noticed that, within each Z series, some NAs with smaller carbon numbers (e.g., n = 9 and Z = −6 in Fig. 2) were even increased after biodegradation. It is thus suggested that some parent NAs with larger carbon numbers were biologically converted into NA species with smaller carbon numbers, which is consistent with our previous observation.25,42
In Fig. 1C and D, for |Z| = 2 to 8, the NA∑|Z|e/NA∑|Z|i ratio is positively proportional to |Z|, whereas the |Z| range of 9 to 18 witnesses a relatively stable region. It is noticed that within each n series, an increased degree of cyclization (larger |Z|) generally resulted in poorer removal efficiency (Fig. 3 and S4†). This trend is not surprising as the biodegradation rate of NAs decreased with increasing cyclicity in both of commercial NAs and OSPW NAs according to previous research.14 A reduction was observed for all the NAs with specific cyclicity (Fig. 1), which suggested the occurrence of ring cleavage. Some NAs (e.g., n = 14 and |Z| = 10 in Fig. 3) were even enriched after biodegradation. Hence, it is speculated that microorganisms might convert some parent NAs with higher cyclicity into species with less rings.7,25,42
According to previous studies, the potential biodegradation pathways of aliphatic and alicyclic carboxylic acids include β-oxidation, combined α- and β-oxidation, and aromatization pathways.14,47 These researchers suggested that β-oxidation is the most probable degradation pathways of OSPW NAs in engineered biological systems. Quaternary and tertiary carbons, at β or α position to the carboxylic group, would prevent or impede the proceeding of β-oxidation due to steric hindrance. This explains why the presence of alkyl branches and ring structures result in increased bio-resistance of NAs. In addition, it is claimed that a tertiary carbon in a ring is less favorable than tertiary carbons in an acyclic NAs as the ring creates further steric hindrance. Moreover, the previous researchers suggested that the intermediate product species generated during biodegradation of NAs could be even more recalcitrant to microbes depending on the occurrence and position of tertiary carbons.14 Furthermore, NAs with larger n are generally more hydrophobic.48 Within the same n series, a higher cyclicity tends to decrease the NA hydrophobicity.48 Organic compounds of higher hydrophobicity (or higher lipophilicity) could easily attach onto or penetrate the bacterial cell envelope, possibly leading to better biodegradation under certain circumstances.49–51 For those particular species, stronger hydrophobic interactions of NAs–NAs, NAs-flocs, and NAs-fouling layer were expected, which would result in elevated local concentrations of hydrophobic NAs. And the locally elevated concentrations of NAs would benefit overall biodegradation of them as a threshold level of NAs exists for microorganisms to initiate the biodegradation process.52
Oxy-NAs concentrations of the MBR feeds and permeates were also monitored (Fig. 4). O3 and O4 oxy-NAs are conceivably worth more attention than O5 and O6 oxy-NAs owing to their dominating abundance. O4, O5, and O6 oxy-NAs were all degraded to a certain extent at each stage. In contrast, more O3 oxy-NAs were generated after the MBR biological process at all stages except A25H48 and A25H12. O3 (probably hydroxylated NAs) and O4 oxy-NAs (probably hydroxylated NAs) are intermediates during aerobic biodegradation of classical NAs.9,14,20,53,54 According to Martin et al., O3 species may be byproducts of more cyclic classical NAs.9 The poorer removal of oxy-NAs might be attributed to the bioconversion of classical NAs. Some researchers reported unappreciable changes of O3 and O4 oxy-NAs after 98 days![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14 and 84 days
14 and 84 days![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55 of incubation. Therefore, the MBR in this study is competent for classical NAs and oxy-NA degradation.
55 of incubation. Therefore, the MBR in this study is competent for classical NAs and oxy-NA degradation.
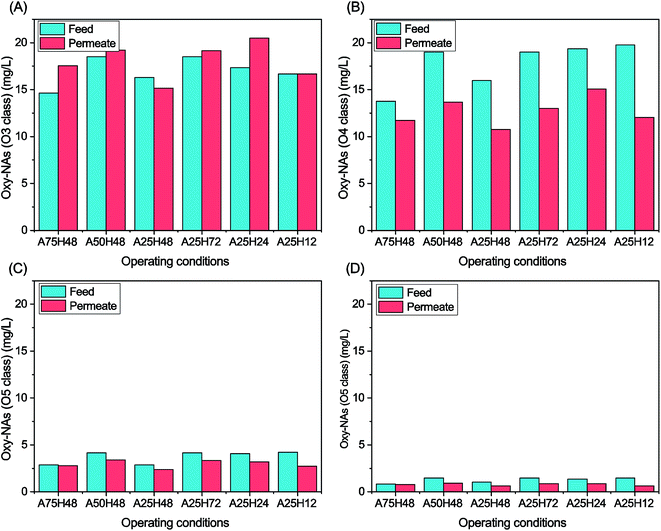 | ||
| Fig. 4 Oxy-NA concentrations of the MBR feed and permeate under different operating conditions: (A) oxy-NAs with O3; (B) oxy-NAs with O4; (C) oxy-NAs with O5; and (D) oxy-NAs with O6. | ||
Discussion within the following paragraphs will therefore be focused on how different influent NH4–N concentrations affected the MBR's efficiency in degrading OSPW NAs when the HRT was 48 h.
Based on the removal efficiency of total classical NAs in Table 2, NA∑ne/NA∑ni and NA∑|Z|e/NA∑|Z|i in Fig. 1, A75H48 showed the lowest effectiveness for OSPW treatment, which may be attributed to the toxicity of free ammonia (FA) caused by high NH4–N concentration. It is known that a higher total ammonia (NH3–N and NH4–N) concentration under an alkaline pH environment results in a higher concentration of FA, which has been reported to be inhibitory to nitrifiers and other microbes.56,57 Based on Ford et al.'s study,58 an FA concentration of ∼13.5 mg L−1 is estimated in the MBR at A75H48 stage.
In Fig. 1A, the two lines of A50H48 and A25H48 are weaved over the n range. The difference between the lines for A50H48 and A25H48 is insignificant (P = 0.3360 > 0.05). For NAs of C12–17, A25H48 removed more NAs than A50H48 did, whereas the latter achieved higher removal of C9 and C20–23 NA species. Moreover, it is interesting to notice the residual abundance of C23 NAs at A75H48 was lower than that at A25H48. Therefore, it is suggested that higher concentrations of influent NH4–N might be helpful to degradation of NAs with higher carbon numbers (i.e., n > 21 in this study). It is noticed that for |Z| of 4, 6, 8 and 10, A50H48 resulted in more (P = 0.034 < 0.05) residual NAs than A25H48 (Fig. 1C).
Among the three stages with an HRT of 48 h, A25H48 was the only stage that reduced the O3 oxy-NAs, whereas A75H48 and A50H48 increased the abundance of that group of oxy-NAs by 20% and 4%, respectively (Fig. 4). As for O4 oxy-NAs, A25H48 showed the best degradation (33%). Regarding the removal of O5 and O6 oxy-NAs, A25H48 demonstrated either similar or higher performance than the other two stages.
Therefore, based on the higher removal rates of total classical NAs (33.7%) and total oxy-NAs (20%), the nitrogen composition used at A25H48 (25 mg NH4–N per L and 50 mg NO3–N per L) was selected for further trials on different HRTs.
No clear correlation between HRT and oxy-NA degradation rate could be elucidated based on the data in this study. Among the four stages, A25H12 exhibited the highest overall removal of oxy-NAs (O3 + O4 + O5 + O6) (23.9%). Therefore, an HRT of 12 h is considered as the optimal on the basis of overall classical NA and oxy-NA removal.
Furthermore, it is not unconceivable that NA species with diverse molecular structures may have different bio-persistence, thus requiring different optimal operating conditions. Thousands of different NAs were present in the system, making it intricate to decipher the kinetics underlying. Previous researchers have reported enhancement of contaminant rejection performance by membrane biofouling. It has been suggested that the formation of biofouling may promote the microfiltration (MF) membrane rejection performance up to the nanofiltration (NF) efficiency.59 Solutes with higher molecular weights and more complex molecular structures tend to be better rejected by NF membrane.60 In addition, it is apparent that the negatively charged deprotonated NA molecules confronted electrostatic repulsion from the membrane fouling layer.60 Thus, NAs could be either repelled or adsorbed by the fouling layer, depending on the permeate dragging force, hydrophobic interaction and electrostatic repulsion. In our study, HRT of 12 hours led to the removal of NA with higher hydrophobicity (n > 21 and |Z| < 6) to a bigger extent (Fig. 1), suggesting the contribution of fouling layer. Microbial community structure analyses indicate that the potential major degraders of NAs were also abundantly present in the membrane's fouling layer (biofilm) (Section 3.2). Thus, the fouling layer formed on membrane surface may enhance the biodegradation of NAs through rejecting NAs thereby elevating the NAs concentration in the MBR sludge and on membrane surface. Organic and biological fouling in the MBR was not necessarily unfavorable in light of the improved NA degradation. It is therefore imperative to contain membrane fouling within a certain level to help mitigation of recalcitrant organic pollutants without significantly increasing the demand of chemical cleaning and membrane replacement. Further study could be performed to look into the influences of membrane/fouling-layer filtration on the removal of NAs in MBR.
Regarding the aquatic toxicity reduction, all six operating conditions effectively reduced the OSPW's inhibition effect to Vibrio fischeri (Fig. S5†). Although the best detoxification percentage (37%) was achieved at the A25H12 stage based on the Microtox bioassay, the correlation between detoxification efficiency and NA (or oxy-NA) removal was vague. That is not unexpected as the acute toxicity is not only determined by NAs but also associated with other toxic organic compounds, such as aromatic compounds.
3.2 Microbial community characterization
Out of the great similarity caused by the internal recirculation between the anoxic and aerobic tanks, the OTU numbers of the two compartments were averaged for following analyses. As is shown in Fig. 5A–F, Rhodocyclales, Burkholderiales and Nitrosomonadales of the phyla Proteobacteria, Cytophagales, [Saprospirales] and Flavobacteriales of the phyla Bacteroidetes, Nitrospirales of the phyla Nitrospirae, and RB41 of the phyla Acidobacteria were arguably the most abundant microorganisms in the MBR throughout the whole operation.It is found that the variations in relative abundances of some microbial species corresponded to the changes in MBR performance over the different operating conditions. The relative abundance of Rhodocyclales of sludge samples over the stages was positively correlated to the MBR's denitrification performance with an rp of 0.790. More interestingly, Rhodocyclales demonstrated strong positive correlations (rp > 0.700) to removal rates of C12–17 NAs and 3–4 ringed NAs (Tables S4 and S5†). In comparison, Cytophagales showed positive correlations to removal rates of C19–23 NAs (rp ≥ 0.755) and 7–8 ringed (rp ≥ 0.680) NAs. In addition, relative abundances of Burkholderiales, RB41, and Deinococcales showed moderate positive correlation with removal rate of C23 classical NAs. It is, therefore, suggested that degradation of NAs with different molecular structures were completed by different microbial species either synergistically or independently. To confirm this, further effort should be made by using isolated bacterial cultures and model compounds.
Fig. 5G and H delineate the 10 most abundant bacterial orders of the two samples collected from membrane cake layers. All the major orders were commonly found in both of sludge samples and their corresponding cake layer samples. Some bacteria, such as Rhodocyclales and Cytophagales, were found more in the sludge samples, whereas some others, such as Nitrospirales, were more abundant in the cake layer samples. It is also found that the microbial community of membrane cake layer was affected by the suspended growth community. For instance, stage A25H24 witnessed more Rhodocyclales in both of sludge and cake layer microbial communities than stage A50H48 did. Furthermore, the similar dominant microorganisms in the sludge samples and their corresponding cake layer samples imply that the microorganisms inhabiting membrane cake layer might contribute to degradation of NAs given that biofilm systems have proven their effectiveness on OSPW NA degradation.21,45
Researchers have reported dominating β-Proteobacteria in oil sands tailings ponds as degrader of NAs and aromatic hydrocarbons,61 which explains the high abundance of β-Proteobacteria detected in this study. Rhodocyclales are responsible for denitrification and degradation of aromatic hydrocarbons.62 Burkholderiales, Cytophagales, Nitrosomonadales and Flavobacteriales have been linked to degradation of bio-recalcitrant hydrocarbons including NAs.63–69 The high population of Nitrosomonadales (ammonium oxidizers) and Nitrospirales (nitrite oxidizers) justifies the excellent nitrification performance of the MBR over the whole study.
Alpha diversity indices, Chao1, observed species, and Shannon were computed through rarefaction (Table S6†). A25H12 had the lowest bacterial richness among the six stages. It is interesting to notice that 5% of the count of genera constituted over 70% of the total bacterial abundance (Table S7†) for every operating condition, suggesting that the majority of the species richness was originated from numerically minor microbial species. Given that the A25H12 stage demonstrated the best classical and oxidized NA removal rates, it is suggested that numerically minor microbial species might not contribute to the MBR's NA degradation performance though their existence largely affected the species richness of the community. This is in accordance to previous research.70
The MBR also demonstrated excellent anti-fouling performance. The first severe fouling was not observed until after 433 days of continuous operation. Within the whole 742 day operation, only six severe fouling events occurred. Detailed results on membrane fouling behavior, major foulants, and fouling layer microbial community structure are available elsewhere.71
4. Conclusion
After more than one year of continuous operation, the MBR's performance on OSPW NA degradation was successfully improved through changing the supplemented inorganic nitrogen composition and HRT. The study showed that neither higher supplemented NH4–N concentrations nor longer HRTs guaranteed a better NA removal. Instead, an NH4–N concentration of 25 mg L−1 and an HRT of 12 h demonstrated the best removal rates of total classical NAs (37.6%) and total oxy-NAs (23.9%). Classical NAs with larger carbon numbers were generally better mitigated in the MBR; and species with higher cyclicity were more persistent to microorganisms. Moreover, MiSeq sequencing analyses revealed that orders of Proteobacteria (e.g., Rhodocyclales and Burkholderiales), Bacteroidetes (e.g., Cytophagales and Flavobacteriales), and Nitrospirae (i.e., Nitrospirales) were the dominating microbial groups over the whole study duration though their relative abundances varied as the operating conditions changed. In the next stage, biofilm process would be added into the reactor to further enhance the removal efficiency. For instance, biodegradation of commercial NA model compounds has been studied based on biochar and granular activated carbon (GAC), which demonstrated quite encouraging performance. It is interesting to investigate whether the integration of bio-adsorption into the MBR would enhance its OSPW NA degradation.72,73 Further efforts may also be made using model compounds and isolated bacteria cultures to verify the contribution and specificity of those microbes to degradation of NAs with certain molecular structural features. The findings from this study could be illuminating to future studies and projects on MBRs and other biological processes for not only OSPW treatment but also other impaired waters containing recalcitrant organic contaminants, such as shale gas produced water.Acknowledgements
The authors acknowledge financial support for this project provided by research grants from the Helmholtz-Alberta Initiative (Theme 5) through the Alberta Environment and Parks' ecoTrust Program and Meidensha Corporation (Y. L. and M. G. E. D.) and funding through an NSERC Industrial Research Chair Program in Oil Sands Tailings Water Treatment (M. G. E. D.) through the support by Syncrude Canada Ltd., Suncor Energy Inc., Shell Canada, Canadian Natural Resources Ltd., Total E&P Canada Ltd., EPCOR Water Services, IOWC Technologies Inc., Alberta Innovates – Energy and Environment Solution, and Alberta Environment and Parks. The authors would also thank the help from Ms Shimiao Dong for her temporary participation in water quality measurement experiments.References
- CAPP, The facts on: oil sands, Canadian Association of Petroleum Producers, http://oilsandstoday.ca, 2014 Search PubMed.
- J. C. Anderson, S. B. Wiseman, N. Wang, A. Moustafa, L. Perez-Estrada, M. G. El-Din, J. W. Martin, K. Liber and J. P. Giesy, Environ. Sci. Technol., 2012, 46, 486–493 CrossRef CAS PubMed.
- J. S. Clemente and P. M. Fedorak, Chemosphere, 2005, 60, 585–600 CrossRef CAS PubMed.
- J. Headley and D. McMartin, J. Environ. Sci. Health, Part A: Environ. Sci. Eng., 2004, 39, 1989–2010 CrossRef.
- D. C. Herman, P. M. Fedorak, M. D. MacKinnon and J. W. Costerton, Can. J. Microbiol., 1994, 40, 467–477 CrossRef CAS PubMed.
- J. Choi and Y. Liu, Int. Biodeterior. Biodegrad., 2014, 92, 79–85 CrossRef CAS.
- G. Hwang, T. Dong, M. S. Islam, Z. Sheng, L. A. Pérez-Estrada, Y. Liu and M. G. El-Din, Bioresour. Technol., 2013, 130C, 269–277 CrossRef PubMed.
- M. MacKinnon, T. V. Meer and A. Verbeek, Assessment of biological impact from a wet landscape option for the reclamation of fine tails from oil sands, Edmonton, 1993 Search PubMed.
- J. W. Martin, T. Barri, X. Han, P. M. Fedorak, M. G. El-Din, L. Perez, A. C. Scott and J. T. Jiang, Environ. Sci. Technol., 2010, 44, 8350–8356 CrossRef CAS PubMed.
- F. M. Holowenko, M. D. Mackinnon and P. M. Fedorak, Water Res., 2001, 35, 2595–2606 CrossRef CAS PubMed.
- M. S. Islam, J. Moreira, P. Chelme-Ayala and M. G. El-Din, Sci. Total Environ., 2014, 493, 282–290 CrossRef CAS PubMed.
- S. J. Rowland, A. G. Scarlett, D. Jones, C. E. West and R. A. Frank, Environ. Sci. Technol., 2011, 45, 3154–3159 CrossRef CAS PubMed.
- E. W. Allen, J. Environ. Eng. Sci., 2008, 7, 123–138 CrossRef CAS.
- X. Han, A. C. Scott, P. M. Fedorak, M. Bataineh and J. W. Martin, Environ. Sci. Technol., 2008, 42, 1290–1295 CrossRef CAS PubMed.
- A. C. Scott, M. D. Mackinnon and P. M. Fedorak, Environ. Sci. Technol., 2005, 39, 8388–8394 CrossRef CAS PubMed.
- F. M. Holowenko, M. D. MacKinnon and P. M. Fedorak, Water Res., 2002, 36, 2843–2855 CrossRef CAS PubMed.
- J. S. Clemente, M. D. MacKinnon and P. M. Fedorak, Environ. Sci. Technol., 2004, 38, 1009–1016 CrossRef CAS PubMed.
- R. A. Frank, H. Sanderson, R. Kavanagh, B. K. Burnison, J. V. Headley and K. R. Solomon, J. Toxicol. Environ. Health, Part A, 2009, 73, 319–329 CrossRef PubMed.
- E. Quagraine, H. Peterson and J. Headley, J. Environ. Sci. Health, Part A: Environ. Sci. Eng., 2005, 40, 685–722 CrossRef CAS.
- X. Han, M. D. MacKinnon and J. W. Martin, Chemosphere, 2009, 76, 63–70 CrossRef CAS PubMed.
- J. Choi, G. Hwang, M. G. El-Din and Y. Liu, Int. Biodeterior. Biodegrad., 2014, 89, 74–81 CrossRef CAS.
- G. Hwang, S. Kang, M. G. El-Din and Y. Liu, Colloids Surf., B, 2012, 91, 181–188 CrossRef CAS PubMed.
- N. S. A. Mutamim, Z. Z. Noor, M. A. A. Hassan, A. Yuniarto and G. Olsson, Chem. Eng. J., 2013, 225, 109–119 CrossRef CAS.
- Y. Tian, L. Chen, S. Zhang and S. Zhang, Chem. Eng. J., 2011, 168, 1093–1102 CrossRef CAS.
- J. Xue, Y. Zhang, Y. Liu and M. G. El-Din, Water Res., 2016, 88, 1–11 CrossRef CAS PubMed.
- G. Wen, J. Ma, L. Zhang and G. Yu, in Integrated Membrane Operations in Various Industrial Sectors, Case Studies, Elsevier B.V., 2010, pp. 195–209 Search PubMed.
- N. Fallah, B. Bonakdarpour, B. Nasernejad and M. A. Moghadam, J. Hazard. Mater., 2010, 178, 718–724 CrossRef CAS PubMed.
- S. R. P. Shariati, B. Bonakdarpour, N. Zare and F. Z. Ashtiani, Bioresour. Technol., 2011, 102, 7692–7699 CrossRef CAS PubMed.
- A. F. Viero and G. L. Sant'Anna, J. Hazard. Mater., 2008, 150, 185–186 CrossRef CAS PubMed.
- J.-S. Chang, C.-Y. Chang, A.-C. Chen, L. Erdei and S. Vigneswaran, Desalination, 2006, 191, 45–51 CrossRef CAS.
- J. Qin, M. Oo, G. Tao and K. Kekre, J. Membr. Sci., 2007, 293, 161–166 CrossRef CAS.
- J. Sipma, B. Osuna, N. Collado, H. Monclús, G. Ferrero, J. Comas and I. Rodriguez-Roda, Desalination, 2010, 250, 653–659 CrossRef CAS.
- F. Meng, B. Shi, F. Yang and H. Zhang, Bioprocess Biosyst. Eng., 2007, 30, 359–367 CrossRef CAS PubMed.
- X. Qu, W. Gao, M. Han, A. Chen and B. Liao, Sep. Sci. Technol., 2013, 48, 1529–1536 CrossRef CAS.
- Y. J. Chan, M. F. Chong, C. L. Law and D. G. Hassell, Chem. Eng. J., 2009, 155, 1–18 CrossRef CAS.
- B. De Gusseme, B. Pycke, T. Hennebel, A. Marcoen, S. E. Vlaeminck, H. Noppe, N. Boon and W. Verstraete, Water Res., 2009, 43, 2493–2503 CrossRef CAS PubMed.
- H. Roh, N. Subramanya, F. Zhao, C.-P. Yu, J. Sandt and K.-H. Chu, Chemosphere, 2009, 77, 1084–1089 CrossRef CAS PubMed.
- N. H. Tran, T. Urase and O. Kusakabe, J. Hazard. Mater., 2009, 171, 1051–1057 CrossRef CAS PubMed.
- J. Deni and M. J. Penninckx, Appl. Environ. Microbiol., 1999, 65, 4008 CAS.
- W. Verstraete and S. Philips, Environ. Pollut., 1998, 102, 717–726 CrossRef CAS.
- A. Anthonisen, R. Loehr, T. Prakasam and E. Srinath, Water Res., 1976, 835–852 CAS.
- Y. Zhang, J. Xue, Y. Liu and M. G. El-Din, Chem. Eng. J., 2016, 302, 485–497 CrossRef CAS.
- H. Mahdavi, V. Prasad, Y. Liu and A. C. Ulrich, Bioresour. Technol., 2015, 187, 97–105 CrossRef CAS PubMed.
- J. Xue, Y. Zhang, Y. Liu and M. G. El-Din, Biodegradation, 2016, 27, 247–264 CrossRef CAS PubMed.
- C. Huang, Y. Shi, M. G. El-Din and Y. Liu, Water Res., 2015, 85, 167–176 CrossRef CAS PubMed.
- N. Wang, P. Chelme-Ayala, L. Perez-Estrada, E. Garcia-Garcia, J. Pun, J. W. Martin, M. Belosevic and M. G. El-Din, Environ. Sci. Technol., 2013, 47, 6518–6526 CAS.
- B. E. Smith, C. A. Lewis, S. T. Belt, C. Whitby and S. J. Rowland, Environ. Sci. Technol., 2008, 42, 9323–9328 CrossRef CAS PubMed.
- P. Pourrezaei, Doctor of Philosophy Dissertation, University of Alberta, 2013 Search PubMed.
- R. Peltola, Bioavailability aspects of hydrophobic contaminant degradation in soils, 2008 Search PubMed.
- S. N. Singh, Microbial degradation of xenobiotics, Springer, 2012 Search PubMed.
- J. Parsons and H. Govers, Ecotoxicol. Environ. Saf., 1990, 19, 212–227 CrossRef CAS PubMed.
- T. M. Misiti, U. Tezel, M. Tandukar and S. G. Pavlostathis, Water Res., 2013, 47, 5520–5534 CrossRef CAS PubMed.
- D. M. Grewer, R. F. Young, R. M. Whittal and P. M. Fedorak, Sci. Total Environ., 2010, 408, 5997–6010 CrossRef CAS PubMed.
- J. V. Headley, K. M. Peru, S. A. Armstrong, X. Han, J. W. Martin, M. M. Mapolelo, D. F. Smith, R. P. Rogers and A. G. Marshall, Rapid Commun. Mass Spectrom., 2009, 23, 515–522 CrossRef CAS PubMed.
- L. D. Brown, L. Pérez-Estrada, N. Wang, M. G. El-Din, J. W. Martin, P. M. Fedorak and A. C. Ulrich, Chemosphere, 2013, 93, 2748–2755 CrossRef CAS PubMed.
- B. Calli, B. Mertoglu, B. Inanc and O. Yenigun, Process Biochem., 2005, 40, 1285–1292 CrossRef CAS.
- S.-F. Yang, J.-H. Tay and Y. Liu, Biochem. Eng. J., 2004, 17, 41–48 CrossRef CAS.
- D. L. Ford, R. L. Churchwell and J. W. Kachtick, Water Res., 1980, 2726–2746 CAS.
- S. L. Khor, D. D. Sun, Y. Liu and J. O. Leckie, Process Biochem., 2007, 42, 1641–1648 CrossRef CAS.
- C. Bellona, J. E. Drewes, P. Xu and G. Amy, Water Res., 2004, 38, 2795–2809 CrossRef CAS PubMed.
- E. Yergeau, J. R. Lawrence, S. Sanschagrin, M. J. Waiser, D. R. Korber and C. W. Greer, Appl. Environ. Microbiol., 2012, 78, 7626–7637 CrossRef CAS PubMed.
- A. Loy, C. Schulz, S. Lücker, K. Stoecker, C. Baranyi, A. Lehner, M. Wagner, S. Lu and A. Scho, Appl. Environ. Microbiol., 2005, 71, 1373 CrossRef CAS PubMed.
- T. H. Bell, E. Yergeau, C. Maynard, D. Juck, L. G. Whyte and C. W. Greer, ISME J., 2013, 7, 1200–1210 CrossRef CAS PubMed.
- D. Pérez-Pantoja, R. Donoso, L. Agulló, M. Córdova, M. Seeger, D. H. Pieper and B. González, Environ. Microbiol., 2012, 14, 1091–1117 CrossRef PubMed.
- W. F. Röling, M. G. Milner, D. M. Jones, K. Lee, F. Daniel, R. J. Swannell and I. M. Head, Appl. Environ. Microbiol., 2002, 68, 5537–5548 CrossRef.
- X. Wang, M. Chen, J. Xiao, L. Hao, D. E. Crowley, Z. Zhang, J. Yu, N. Huang, M. Huo and J. Wu, PLoS One, 2015, 10, e0132881 Search PubMed.
- W. V. den Tweel, J. Smits and J. De Bont, Arch. Microbiol., 1988, 149, 207–213 CrossRef.
- B. Cao, K. Nagarajan and K.-C. Loh, Appl. Microbiol. Biotechnol., 2009, 85, 207–228 CrossRef CAS PubMed.
- C. Whitby, Adv. Appl. Microbiol., 2010, 70, 93–125 CAS.
- J. Ma, Z. Wang, Y. Yang, X. Mei and Z. Wu, Water Res., 2013, 47, 859–869 CrossRef CAS PubMed.
- J. Xue, Y. Zhang, Y. Liu and M. G. El-Din, Water Res., 2016, 105, 444–455 CrossRef CAS PubMed.
- M. L. Frankel, T. I. Bhuiyan, A. Veksha, M. A. Demeter, D. B. Layzell, R. J. Helleur, J. M. Hill and R. J. Turner, Bioresour. Technol., 2016, 216, 352–361 CrossRef CAS PubMed.
- M. S. Islam, T. Dong, Z. Sheng, Y. Zhang, Y. Liu, M. G. El-Din and M. G. El-Din, Int. Biodeterior. Biodegrad., 2014, 91, 111–118 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra01836c |
| This journal is © The Royal Society of Chemistry 2017 |