Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Decolorization of Color Index Acid Orange 20 buffer solution using horseradish peroxidase immobilized on modified PAN-beads
Zhu Yincan,
Liu Yan,
Guo Xueyong,
Wu Qiao and
Xu Xiaoping *
*
College of Chemistry, Fuzhou University, Fuzhou, 350108, P. R. China. E-mail: xu@fzu.edu.cn
First published on 29th March 2017
Abstract
In the present work, horseradish peroxidase (HRP) is utilized to be immobilized onto polyacrylonitrile based beads (PAN-beads) for decolorization of Color Index (C. I.) Acid Orange 20 (AO20) in aqueous solution. Sodium hydroxide and hydrochloric acid were used to treat PAN-beads to obtain carboxyl groups, followed by being modified with ethanediamine to get more amino groups, and then were activated with glutaraldehyde. The enzyme was immobilized onto the activated beads by covalent crosslinking. Optimum factors for decolorization of AO20 were investigated both on free horseradish peroxidase (F-HRP) and immobilized horseradish peroxidase (I-HRP), including pH values, H2O2 amount, enzyme amount, temperature and so on. Under optimum conditions, percent decolorization was about 90% or 98% respectively for F-HRP or I-HRP. A kinetic study showed that F-HRP possessed more affinity compared to I-HRP, but reusability experiments clarified that I-HRP was cost-efficient as about 90% decolorization was obtained after three cycles. The human normal hepatocytes cell line (L02 cell line) was utilized to assess cytotoxicity of metabolites, and the result was acceptable.
1. Introduction
In 1857, W. H. Perkin invented the first synthetic dye called mauveine and put it into industrial production. Since then more and more synthetic dyes have appeared and have been widely used in industrial production sectors, such as textiles, paper, plastics, printing, leather, cosmetics, pharmaceuticals, and petrochemicals. But the wide usage of the various kinds of dyes in industries has led to discharging effluents containing dyes, rich in complex aromatic structures into the environment. There are more than 105 kinds of commercially available dyes with over 8 × 105 metric tons of dye stuff leaked and discharged to industrial effluents,1 and nearly half of these are azoic dyes.2 The dyes are hardly degraded in the environment because oxidizing reagents, light and water can't damage their chemical structure. The chemical structure is characterized by the azo group N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N which is a chromophore, associated with an auxochrome such as amine or hydroxyl groups. In natural degradation processes, microorganisms decolor azo dyes by reductive cleavage of azo bond under anaerobic conditions, leading to the formation of mutagenic aromatic amines, and these compounds become contaminants.3–6 When dye-containing effluents are released into the water bodies, dyes are visible pollutants at concentration of 0.005 mg L−1,7 and the passage of light decreases along with increasing the chemical oxygen demand (COD). The azo dyes in the environment are toxic and potential carcinogens.8 Considering the harm to humans' health and environment, how to remove the synthetic dyes is always an important challenge and many researchers have reported their attempts, for example, Li and his co-workers used activated carbon as absorbent to adsorb Acid Orange 7
N which is a chromophore, associated with an auxochrome such as amine or hydroxyl groups. In natural degradation processes, microorganisms decolor azo dyes by reductive cleavage of azo bond under anaerobic conditions, leading to the formation of mutagenic aromatic amines, and these compounds become contaminants.3–6 When dye-containing effluents are released into the water bodies, dyes are visible pollutants at concentration of 0.005 mg L−1,7 and the passage of light decreases along with increasing the chemical oxygen demand (COD). The azo dyes in the environment are toxic and potential carcinogens.8 Considering the harm to humans' health and environment, how to remove the synthetic dyes is always an important challenge and many researchers have reported their attempts, for example, Li and his co-workers used activated carbon as absorbent to adsorb Acid Orange 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and Gokce et al. found the O3–H2O2–UV process could dispose the wastewater of reactive dyeing of cotton fabric efficiently.10 Though physical and chemical methods have been employed in many studies,11–13 they have some noticeable weaknesses, for instance, physical methods for color removal can't completely dispel recalcitrant azo dyes so it needs subsequent disposal, and chemical methods can degrade dyes but it needs to add other reagents which may be produce secondary pollution. As for activated sludge methods, it's not effective for dye removal.14–16
9 and Gokce et al. found the O3–H2O2–UV process could dispose the wastewater of reactive dyeing of cotton fabric efficiently.10 Though physical and chemical methods have been employed in many studies,11–13 they have some noticeable weaknesses, for instance, physical methods for color removal can't completely dispel recalcitrant azo dyes so it needs subsequent disposal, and chemical methods can degrade dyes but it needs to add other reagents which may be produce secondary pollution. As for activated sludge methods, it's not effective for dye removal.14–16
Based on the situation above, enzyme-based methods are more advantageous, which produce lower toxicity and make minimal impact on ecosystem. In addition, during enzyme-based treatment process there is not need high energy requirements, difficult process control and rigorous pH, temperature and ionic strength conditions. It is sufficient to use enzyme-based treatments alone as the toxic compounds can be transformed to less harmful compounds that means complete degradation is not necessary.17–20
It is crucial to find a proper enzyme to degrade the dyes. It is reported that peroxidases and laccases from plants or fungi have the ability to oxidize dyes.21–23 Heme-containing peroxidases possess similar amino acid sequences and similar catalytic activities, but are slightly different in subsequent reaction products and substrate specificities.24 In the terms of decoloring or degrading the dyes, laccases and horseradish peroxidase have been extensively studied.25–28
Horseradish peroxidase (HRP, EC 1.11.1.7) is an efficient, environmentally friendly and widely-researched biocatalyst-enzyme, which mainly exists in the roots of horseradish plants. HRP is a monomeric heme-obtaining glycoprotein (44 kDa) having approximately 18% N-linked oligosaccharides with function of two Ca2+ ions to maintain enzyme conformation.29–31 In the presence of HRP, hydrogen peroxide (H2O2) can oxidize aromatic compounds or azo dyes without high temperature or harsh chemical conditions. It has been proved by many researchers that phenolic compounds and textile dyes can be degraded or decolored by peroxidase, which has shown that HRP plays an important role in the removal of aqueous phenols and dyes.32–35 Compared to another effective biocatalyst-laccase (benzenediol: oxygen oxidoreductase, EC 1.10.3.2), HRP is more economical to produce, inactivates slowly, or acts on a wider variety of substrates.
Though enzyme-catalyzed process has more advantages in terms of handling effluents containing dyes, free enzymes have drawbacks of low stability, low production yield and unreusability.36 Immobilized enzymes on solid supports could overcome the limits above, which is capable of reusing immobilized enzymes and, as a result, reduces the overall cost of enzymatic waste.37,38 Immobilizing enzymes has many techniques, such as entrapment, encapsulation, self-immobilization and covalent binding of enzymes to solid support and so on, where covalent cross-linking gives enzyme more rigid structure and reduces protein unfolding, consequently makes enzymes significantly more stable.37 Polyacrylonitrile (PAN) are widely used as supporting materials of enzymes because of its good mechanical stability and its active nitrile groups. Different PAN modifications can be used to improve its properties as catalyst supports.
Our work was designed to immobilize horseradish peroxidase by covalent cross-linking with glutaraldehyde on polyacrylonitrile beads followed by the application of immobilized HRP for the elimination of AO20 from waste water. To establish an optimal operational process, some variables (pH, H2O2, contact time, temperature, dye concentration) involved in the oxidative process to remove AO20 from wastewater as well as the reusability experiment was evaluated particularly. The enzyme kinetic parameters (Km and Vmax) on the applied AO20 were also calculated. In addition, the toxicity of HRP-treated samples was evaluated using MTT cytotoxicity assay. MTT is the abbreviation of methylthiazolyldiphenyl-tetrazolium bromide, and the succino dehydrogenase in mitochondria of living cells can make exogenous MTT reduced to insoluable violet crystal – formazan.
2. Experimental
2.1 Materials and reagents
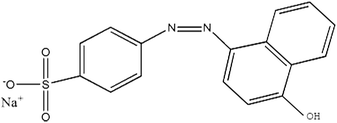
Polyacrylonitrile (PAN) was purchased from Heowns Biochem Technologies LLC, China. C. I. Acid Orange (shown in Table 1), hydrogen peroxide (H2O2), sodium hydroxide (NaOH), hydrochloric acid (HCl), phosphoric acid (H3PO4), ethylenediamine, N,N-dimethylformamide (DMF), glutaraldehyde, guaiacol, coomassie brilliant blue G-250, ethanol, sodium dihydrogen phosphate and disodium hydrogen phosphate were all obtained from Sinopharm Chemical Reagents CO., Ltd. Horseradish peroxidase (HRP) ((EC 1.11.1.7), >300 U mg−1) and bovine serum albumin (BSA) were purchased from Shanghai SANGON Biological Engineering Co., Ltd, China. The aforementioned chemicals were of analytical grade and were used without further purification during follow-up study. Solutions were confected with distilled water (prepared by Fuzhou University). In this experiment, AO20 buffer solution was used to simulate industrial wastewater containing an azo dye.2.2 Preparation of PAN-based beads
The preparation of PAN-based beads was performed as follows. 2 g polyacrylonitrile was dissolved in 23 g DMF and was kept in constant mechanical stirring until the solution became a homogenous gel. The PAN-based beads were prepared by one-step extrusion similar to that described by Nigma.39 With the help of a peristaltic pump, the homogenous gel was dropped through a silicon tube into a beaker containing 200 mL of distilled water. 10 cm dropping height was chose to avoid the droplets sticking together and trailing. The obtained PAN-based beads were spherical beads with the same size, and the diameter of beads was about 3 mm. Then these beads were washed three times using distilled water. Afterwards the PAN-beads were filtrated and stored in the phosphate buffer (pH 7.4) under 4 °C for further utilization.2.3 Modifications of PAN-beads
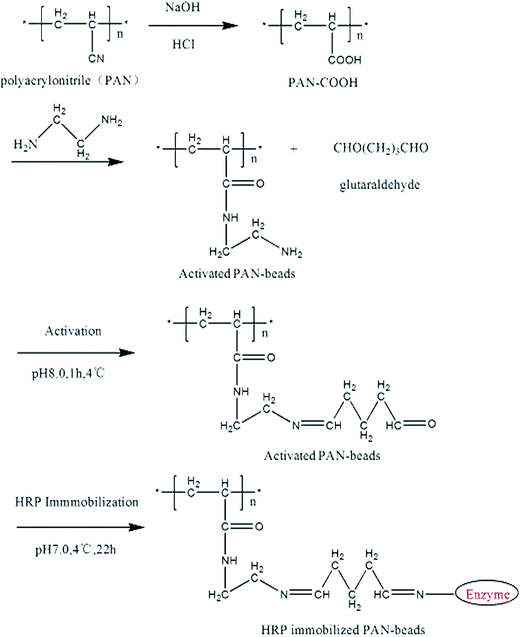
This step of the modifications of PAN-based beads was aimed to transform the nitrile group in PAN into amino or introduce amino groups so that these PAN-based beads could crosslinked with glutaraldehyde. Firstly, these beads were immersed in 100 mL of 12% (w) NaOH aqueous solution for 1 h at 50 °C, and the nitrile groups in PAN were transformed into sodium carboxylate groups. Secondly, they were washed with deionized water and placed in 1 mol L−1 HCl aqueous solution at room temperature for 2 h,40 and the sodium carboxylate groups were transformed into carboxylic groups. Thirdly, the beads were immersed in 10% (v/v) solution of ethylenediamine for 1 h at room temperature, a large number of amino groups were introduced and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N was formed at the moment. Lastly, the obtained beads were washed thoroughly with deionized water to remove the unreacted ethylenediamine and then rinsed with phosphate buffer.
N was formed at the moment. Lastly, the obtained beads were washed thoroughly with deionized water to remove the unreacted ethylenediamine and then rinsed with phosphate buffer.
2.4 Glutaraldehyde activation and horseradish peroxidase immobilization
The modified beads were immersed in 10% (v/v) solution of glutaraldehyde for 60 min at 4 °C. Then, the beads were thoroughly washed with distilled water and phosphate buffer. After that the beads were kept in the solution of horseradish peroxidase (1 mg mL−1) for 24 h at 4 °C. Finally, the resulting beads were washed with distilled water thoroughly once again. After immobilization, the color of the initial white beads were turned into brownish red, but almost no changes in the sizes of the beads. The immobilized beads were stored in phosphate buffer (0.1 M, pH 7.0) at 4 °C.2.5 Characterization of the beads
2.6 Measuring methods of protein loading and horseradish peroxidase activity
The total amount of protein bound with the modified beads was determined by the method of Coomassie light blue staining assay (also called Bradford assay).42 The amount of immobilized enzyme were obtained by subtracting the amount of enzyme determined in the residual solutions and washings from the total amount of protein. The tested solution was determined at 595 nm. A calibration curve was obtained using BSA as a standard.43The activity of HRP denoted as the decomposition rate of H2O2 by HRP, with guaiacol as hydrogen donor, was determined colorimetrically by measuring the rate of color development at 470 nm and at 25 °C.44 In this assay, HRP needed to be dissolved in 0.1 M sodium phosphate buffer at pH 6.0. The reaction mixture was prepared as follows: 0.1 M sodium phosphate buffer (pH 6.0) was placed in a 50 mL beaker, adding 28 μL guaiacol, and stirred on the heated magnetic stirrer until guaiacol was completely dissolved. When the before-mentioned solution cooled, 19 μL 30% (v/v) H2O2 was added and mixed uniformly and then stored in the refrigerator. When measuring the absorbance, 3 mL reaction mixture and 1 mL HRP solution (1 mL 0.1 M sodium phosphate buffer (pH 6.0) for the control group) would be mixed together and then measured at 470 nm every 30 s for 5 min. The activity of immobilized HRP was calculated by subtracting the amount of activity determined in the residual solutions and washings from the total amount of activity.
2.7 Optimization of AO20 oxidation by immobilized HRP (I-HRP) and free HRP (F-HRP)
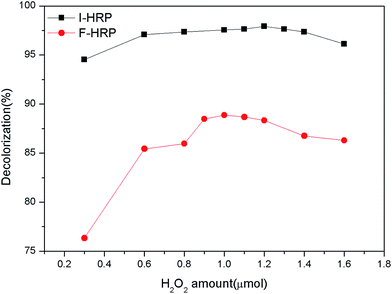
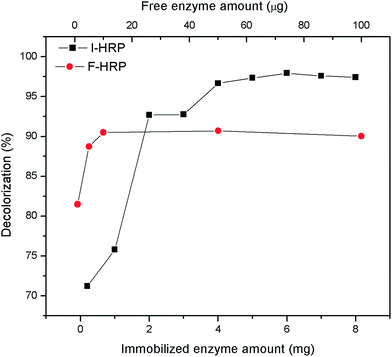
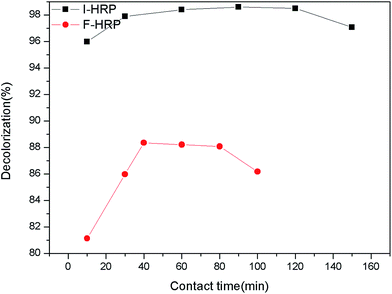
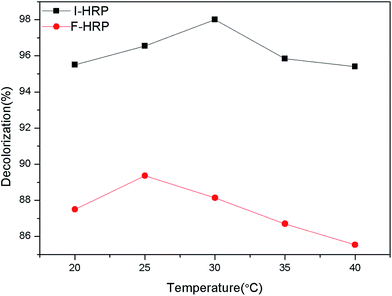
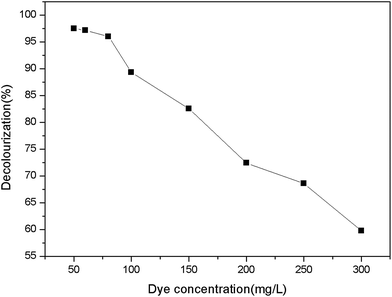
The assay was conducted to obtain an optimum condition for the removal of AO20 in aqueous phase using PAN-based immobilized HRP. Some crucial factors vastly impacted the removal of AO20: pH value, contact time, the amount of immobilized HRP, H2O2 amount, dye concentration, temperature. To obtain the optimization of AO20 oxidation by immobilized HRP must obtain the optimum values of above-mentioned factors. The experiments were carried out equipped with a constant temperature oscillation incubator. The effects of pH on the dye removal ability of the immobilized and free enzymes were determined using the pH range 3.0–10.0 at 25 °C, in more detail, the percent dye removal for F-HRP was determined in buffer solutions of pH 3, 5, 6.5, 7, 7.5, 8, 9 and 10, and the percent dye removal for I-HRP was determined in buffer solutions of pH 3, 4, 5, 6, 6.5, 7, 8, 9 and 10. The effects of contact time were determined using the time range 10–150 min for I-HRP and the samples were withdrawn every twenty or thirty minutes, while the time range for F-HRP was 10–100 min and the samples were tested every ten or twenty minutes. The effects of H2O2 amount for I-HRP and F-HRP were determined using the range 0.3–1.6 μmol with an interval of 0.1 μmol or 0.2 μmol. The effect of the amount of immobilized enzyme was determined using the range 0.2–8 mg (10 mg immobilized enzyme equaled to 1 g PAN-based beads) with an interval of 1 mg, while the effect of free enzyme dosage was designed to use the range 10–100 μg and the dye removal was determined respectively when there was 1, 5, 10, 50 and 100 μg free enzyme in the dye solution. The effects of the dye concentration on both free and immobilized enzyme were determined at the range 50–300 mg L−1 with an interval of 50 mg L−1. The effects of the temperature were determined at the range 20–40 °C with an interval of 5 °C. Every obtained optimum value was applied for other tests of influencing factors and these experiments were conducted in triplicate.2.8 The reusability of PAN-based immobilized HRP
The reusability of PAN-based immobilized beads was measured following the steps below. After each cycle of reaction, the enzyme immobilized beads were removed and washed with phosphate buffer (0.1 M, pH 7.0) to remove any residual substrate adhering to immobilized beads. After that, the immobilized beads were transferred into fresh dye solution and percent removal was determined. The percent removal in every cycle was used to indicate immobilized enzyme activity. All the experiments were conducted in triplicate.2.9 Cytotoxicity test
3. Results and discussion
3.1 Preparation of HRP modified PAN-beads
The preparation of HRP modified PAN-beads were began with the conversion of the nitrile groups on the surface of the PAN-beads to carboxyl groups treated in NaOH solution and HCl solution and the color of these beads became yellow. Then the carboxylic PAN-beads were rinsed in ethanediamine solution and the color became flavescens, which indicated a certain chemical change on these beads in a way. The aforementioned PAN-beads were crosslinked in glutaraldehyde solution and then covalent bound with HRP and the color of these beads ultimately became brown. The whole process was shown as Fig. 1.3.2 Characterization of modified PAN-beads
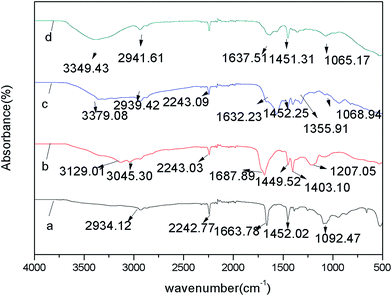
FT-IR spectra of beads of every stage during immobilization process were depicted in Fig. 2 to verify the whole changes on the surface of PAN-beads. Line (a) represents initial beads, the absorption peak at 2242 cm−1 is attributed to the stretching vibrations band of nitrile groups (–CN). The bands at 2934 cm−1 and 1452 cm−1 are attributed by stretching and bending vibration bands of methylene (–CH2–) on the bead surface. Line (b) represents carboxylated PAN formed by using NaOH and HCl to dispose initial PAN. The peaks at 3129 cm−1 and 1687 cm−1 can be ascribed to the presence of O–H bond and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond of carboxylic acids, and both the two peaks verify the presence of –COOH. Line (c) is representative of cross-linked PAN-beads formed by using ethylenediamine and glutaraldehyde to treat carboxylated PAN. The peak of 3379 cm−1 is attributed to stretching vibrations band of –NH because of reaction with ethylenediamine on the bead surface. The absorption peak of 1632 cm−1 is ascribed to C
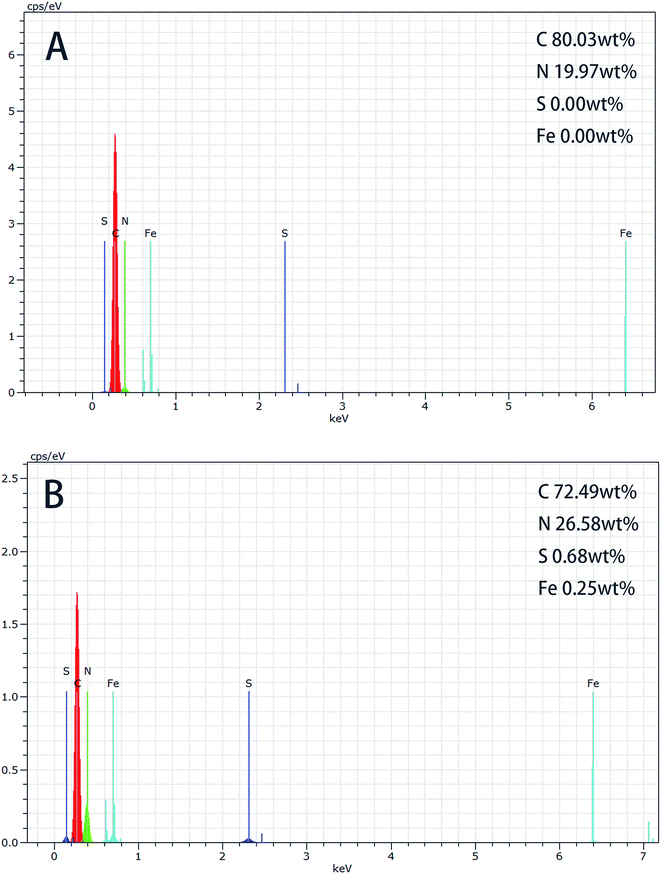
O bond of carboxylic acids, and both the two peaks verify the presence of –COOH. Line (c) is representative of cross-linked PAN-beads formed by using ethylenediamine and glutaraldehyde to treat carboxylated PAN. The peak of 3379 cm−1 is attributed to stretching vibrations band of –NH because of reaction with ethylenediamine on the bead surface. The absorption peak of 1632 cm−1 is ascribed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N formed by the reaction between –CHO and –NH2. Line (d) shows the spectra of immobilization HRP (I-HRP). Line (d) is similar to line (a), because there is a same reaction happening between –CHO and –NH2 during this course and it can't testify the presence of HRP obviously, so undermentioned EDS analysis is to add to prove it.
N formed by the reaction between –CHO and –NH2. Line (d) shows the spectra of immobilization HRP (I-HRP). Line (d) is similar to line (a), because there is a same reaction happening between –CHO and –NH2 during this course and it can't testify the presence of HRP obviously, so undermentioned EDS analysis is to add to prove it.
3.3 The amount of HRP onto the activated PAN-beads and the activity of immobilized beads
The biocatalytic performance was evaluated regarding the immobilization amount (IA) and relative activity (RA). Eqn (2) was used to calculate the amount of enzyme immobilized. The protein standard curve was obtained by Bradford assay and the fitting equation was described as eqn (1), which was used to count the amount of enzyme in solution. Eqn (3) was used to count relative activity (RA).| A = 0.0062C + 0.2364 (R2 = 0.9996) | (1) |
| IA = enzyme added − enzyme in residual solution − enzyme in washings | (2) |
 | (3) |
The results were that IA was 10.0 mg g−1 and RA was 67%.
3.4 Kinetics parameters of free and immobilized horseradish peroxidase
To obtain the kinetics parameters of both free and immobilized enzyme, five kinds of concentrations of dye solution were employed for our kinetics experiment. The linear parts of their curves were depicted in Fig. 5 to calculate the kinetics parameters. Kinetics parameters of free and immobilized horseradish peroxidase towards AO20 were elucidated in Table 2. Km value of free horseradish peroxidase was 0.288 mM and that of immobilized horseradish peroxidase was 1.458 mM, which was 5 folds to free horseradish peroxidase. Vmax of immobilized horseradish peroxidase was 14.2 μM min−1 and that of free horseradish peroxidase was 89.7 μM min−1, which was about 6 folds to immobilized horseradish peroxidase. The higher Km value of immobilized horseradish peroxidase towards the applied dye means lower affinity of immobilized horseradish peroxidase for the applied dye compared to free ones. At the same time, the lower Vmax value also means lower affinity, corresponding with Km value. Some reasons explaining the difference in behavior of the free and immobilized horseradish peroxidase are listed here: immobilized enzyme is located in a quite different microenvironment from that of free enzyme in the bulk solution. Steric hindrance and diffusional limitation result in less chance to react with active sites of immobilized horseradish peroxidase due to the change of enzyme conformation.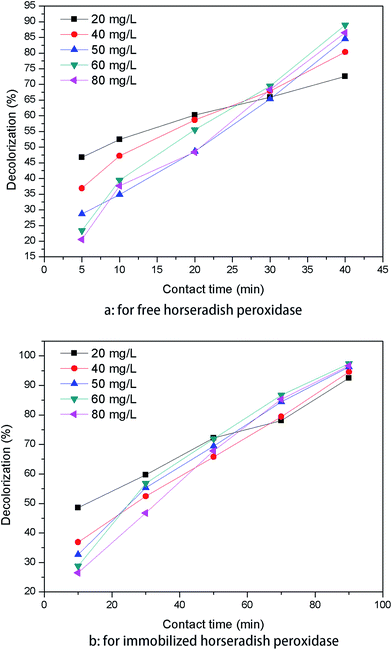 | ||
| Fig. 5 The relation between the percent dye removal and contact time for five kinds of concentrations of dye solution. | ||
| Horseradish peroxidase | Km (mM) | Vmax (μM min−1) |
|---|---|---|
| Free enzyme | 0.288 | 89.7 |
| Immobilized enzyme | 1.458 | 14.2 |
3.5 The optimum conditions to treat C. I. Acid Orange 20 (AO20) solution
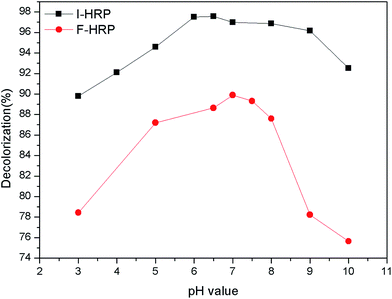
The optimum 89.89% of AO20 decolorization was obtained with neutral medium (buffer pH 7.0) for F-HRP. Dye removal efficiency increased from pH 3.0 to 7.0, followed by a decrease down to about 75% from pH 7.0 to 10.0. The reason could be that pH 7.0 may provide F-HRP with appropriate environment, where F-HRP possessed a higher activity so that dye removal efficiency was high. On the other hand, for I-HRP, about 97% of AO20 was removed with light acidulous medium (buffer pH 6.5). And the percent decolorization of I-HRP was higher over a wide range of varying pKa value compared to F-HRP, which could be attributed to the bared activated centers of enzyme because of the changes in the structure of I-HRP. In addition, pH could influence the form of dye in water which could impact degradation, and we could clearly know I-HRP behaved better at wide range of pH from Fig. 6.
3.6 Reusability of immobilized horseradish peroxidase
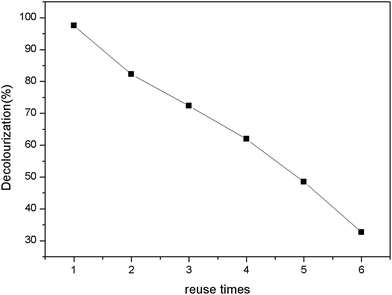
The reusability can offer advantages and economy during practical process. The reusability was observed by assessing the percent decolorization of immobilized enzyme under same conditions after successive several times.47 After each recycle, the same enzyme immobilized PAN particles were washed with phosphate buffer (0.1 M, pH 7.0) to remove any residual reactant within the enzyme-support system. The results were summarized as Fig. 12. The percent decolorization decreased with increase of recycle times. The immobilized beads still decreased 50% chromaticity up to 4 times recycle. In this work, there was ∼30% decolorization after 6 times reuse.3.7 Cytotoxicity analysis
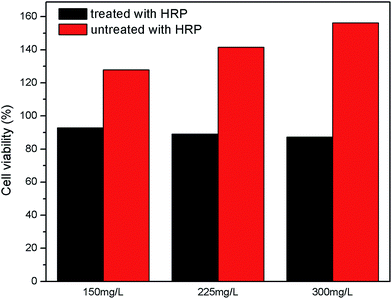
L02 cell line is a kind of immortalized human normal hepatocytes which possess similar biological properties to normal human hepatocytes in vivo and is an ideal cell model for studying cell toxicity.48 These cells have been widely used in studying liver cancer, genotoxicity, apoptosis and cytotoxicity in vitro study.49,50 MTT is a kind of assay which has been utilized in many study of cellular response to a toxicant.51 Cell toxicity is expressed as percentage cell viability which is indicated by value of optical density (OD) at 570 nm, because formazan belonging to living cells is absorbed at 570 nm on a multiscan spectrum.52 The results were presented in Fig. 13. According to the Fig. 9, the amounts of those cells cultivated in normal culture containing different concentrations of dye solution (150 mg L−1, 225 mg L−1, 300 mg L−1) untreated with enzyme increased by 27.96%, 41.52%, 56.36% respectively after four-day culture, which was different from previous reports, but this couldn't indicate that this kind of dye was harmless to human cells because cell proliferation in the experiment was faster than control set. So there were reasons to doubt that the dye tended to induce malignant cellular proliferation, of course this conjecture needed further study.53 As for dye solutions treated with enzyme, the amounts of those cells cultivated with different concentrations of treated solutions decreased by small range for no more than 14%, which was comparable to the result reported by K. Klemola and his co-workers.544. Conclusion
In the present work, the preparation and application of immobilized horseradish peroxidase on the PAN-based beads for AO20 removal from aqueous solution was investigated. Horseradish peroxidase was designed to be immobilized onto PAN-beads by crosslinking with glutaraldehyde accompanying with immobilized amounts of 10 mg genzyme−1 and retention activity of 67%. The performance of AO20 decolourization was found to be dependent on some factors that were pH values, H2O2 amounts, enzyme amounts and temperature, and the optimum conditions were obtained in this study. In addition, reusability experiment showed that the immobilized beads can be used up to three cycles remaining higher percentage decolourization of about 90%, which provided a cost-efficient advantage for large-scale applications. Cell toxicity was also studied in this work, and the results indicated that the toxicity of metabolites of AO20 was acceptable compared to control group. In conclusion, this study provided a feasible potential method for economic, continuous and relatively safe disposal of azo dyes.References
- G. Palmieri, G. Cennamo and G. Sannia, Enzyme Microb. Technol., 2005, 36, 17–27 CrossRef CAS.
- K. Selvam, K. Swaminathan and K. S. Chae, Bioresour. Technol., 2003, 88, 115–119 CrossRef CAS PubMed.
- S. Sandhya, in Biodegradation of Azo Dyes, ed. H. Atacag Erkurt, Springer, Berlin, 2010, pp. 39–57 Search PubMed.
- K. Golka, S. Kopps and Z. W. Myslak, Toxicol. Lett., 2004, 151, 203 CrossRef CAS PubMed.
- A. Gnanamani, M. Bhaskar, R. Ganga, G. Sekaran and S. Sadulla, Chemosphere, 2004, 56, 833 CrossRef CAS PubMed.
- J. Binkley and A. Kandelbauer, in Textile Processing with Enzymes, ed. A. Cavaco-Paulo and G. M. Guebitz, CRC Press, Boca Raton, 2003, pp. 199–221 Search PubMed.
- C. O'Neill, F. Hawkes, D. Hawkes, N. Lourenco, H. Pinheiro and W. Delee, J. Chem. Technol. Biotechnol., 1999, 74, 1009–1018 CrossRef.
- H. Moawad, W. M. Abd El-Rahim and M. Khalafallah, J. Basic Microbiol., 2003, 43, 218–229 CrossRef CAS PubMed.
- G. T. Güyera, K. Nadeemb and N. Dizgec, J. Cleaner Prod., 2016, 139, 488–494 CrossRef.
- J. Li, Y. Du, B. Deng, K. M. Zhu and H. Zhang, Environ. Sci. Pollut. Res., 2017, 24, 4932–4941 CrossRef CAS PubMed.
- T. Robinson, G. McMullan, R. Marchant and P. Nigam, Bioresour. Technol., 2001, 77, 247–255 CrossRef CAS PubMed.
- S. H. Lin and M. L. Chen, Water Res., 1997, 31, 868–876 CrossRef CAS.
- H. Zhang, J. H. Zhang, C. Y. Zhang, F. Liu and D. B. Zhang, Ultrason. Sonochem., 2009, 16, 325–330 CrossRef CAS PubMed.
- E. V. C. Rosa, E. L. Simionatto, M. M. Sierra, S. L. Bertoli and C. M. Radetski, Environ. Toxicol. Chem., 2001, 20, 839–845 CrossRef CAS.
- M. Hassan, W. M. Abd and M. Khalafallah, J. Basic Microbiol., 2003, 43, 218–229 CrossRef PubMed.
- C. Wang, A. Yediler, D. Lienert, Z. Wang and A. Kettrup, Chemosphere, 2002, 46, 339–344 CrossRef CAS PubMed.
- S. M. A. G. U. De Souza, E. Forgiarini and A. A. U. De Souza, J. Hazard. Mater., 2007, 147, 1073–1078 CrossRef PubMed.
- I. Khouni, B. Marrot, P. Moulin and A. R. Ben, Desalination, 2011, 268, 27–37 CrossRef CAS.
- Q. Husain, Crit. Rev. Biotechnol., 2006, 26, 201–221 CrossRef CAS PubMed.
- T. S. Shaffiqu, J. J. Roy, R. A. Nair and T. E. Abraham, Appl. Biochem. Biotechnol., 2002, 102, 315–326 CrossRef PubMed.
- H. Qayyum, H. Maroof and K. Yasha, Crit. Rev. Biotechnol., 2009, 29, 94 CrossRef CAS PubMed.
- E. Torres, I. Bustos-Jaimes and S. Le Borgne, Appl. Catal., B, 2003, 46, 1 CrossRef CAS.
- C. Regalado, B. E. García-Almendárez and M. A. Duarte-Vázquez, Phytochem. Rev., 2004, 3, 243 CrossRef CAS.
- K. G. Welinder, Curr. Opin. Struct. Biol., 1992, 2, 388–393 CrossRef CAS.
- M. Bilal, H. M. N. Iqbal, S. Z. H. Shah, H. Hu, W. Wang and X. H. Zhang, J. Environ. Manage., 2016, 183, 836–842 CrossRef CAS PubMed.
- M. R. Silva, L. R. Vasconcelos, C. Russo, E. Scio and V. S. Ferreira-Leitao, Enzyme Res., 2010, 2010, 703824–703831 Search PubMed.
- H. Zhang, S. Zhang, F. He, X. Qin, X. Y. Zhang and Y. Yang, J. Hazard. Mater., 2016, 320, 265–277 CrossRef CAS PubMed.
- F. Zheng, B. K. Cui, X. J. Wu, G. Meng, H. X. Liu and J. Si, Int. Biodeterior. Biodegrad., 2016, 110, 69–78 CrossRef CAS.
- L. Hassani, B. Ranjbar, K. Khajeh, H. N. Manesh, M. N. Manesh and M. Sadeghi, Enzyme Microb. Technol., 2006, 38, 118–125 CrossRef CAS.
- J. Z. Liu, T. L. Wang and L. N. Ji, J. Mol. Catal. B: Enzym., 2006, 41, 81–86 CrossRef CAS.
- N. C. Veitch, Phytochemistry, 2004, 65, 249–259 CrossRef CAS PubMed.
- E. Kalaiarasan and T. Palvannan, Clean: Soil, Air, Water, 2015, 43, 846–856 CrossRef CAS.
- G. Bayramoglu and M. Y. Arica, J. Hazard. Mater., 2008, 156, 148–155 CrossRef CAS PubMed.
- M. Celebi, M. Altikatoglu, Z. M. Akdeste and H. Yildrim, J. Biochem., 2012, 37, 200–206 Search PubMed.
- S. Khanahmadi, F. Yusof, A. Amid, S. S. Mahmod and M. K. Mahat, J. Biotechnol., 2015, 202, 153–161 CrossRef CAS PubMed.
- P. Sathishkumar, J. C. Chae, A. R. Unnithan, T. Palvannan, H. Y. Kim, K. Lee and J. M. Kamala-Kannana, Enzyme Microb. Technol., 2012, 51, 113–118 CrossRef CAS PubMed.
- M. Fernandez-Fernandez, M. A. Sanroman and D. Moldes, Biotechnol. Adv., 2013, 31, 1808–1825 CrossRef CAS PubMed.
- A. Sadighi and M. A. Faramarzi, J. Taiwan Inst. Chem. Eng., 2013, 44, 156–162 CrossRef CAS.
- S. C. Nigma, I. F. Tsao, A. Sakoda and H. Y. Wang, Biotechnol. Tech., 1988, 2, 271–276 CrossRef.
- Z. Wang, Z. L. Chen, J. Chang, J. M. Shen, J. Kang and Q. Chen, Chem. Eng. J., 2015, 262, 904–912 CrossRef CAS.
- S. F. Li, J. P. Chen and W. T. Wu, J. Mol. Catal. B: Enzym., 2007, 47, 117–124 CrossRef CAS.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- F. Anjum, V. Rishi and F. Ahmad, Biochim. Biophys. Acta, 2000, 1476, 75–84 CrossRef CAS.
- H. U. Bergmeyer, Methods of Enzymatic Analysis, Academic Press, New York, 2nd edn, 1974, vol. 1, p. 495 Search PubMed.
- Z. Wang, Z. L. Chen, J. Chang, J. M. Shen, J. Kang and Q. Chen, Chem. Eng. J., 2015, 262, 904–912 CrossRef CAS.
- M. L. Shuler and F. Kargi, Bioprocess Engineering—Basic Concepts, Prentice Hall PTR, Englewood Cliffs, NJ, 1992, pp. 58–102 Search PubMed.
- M. Bilal, M. Asgher and B. Asgher, Chem. Cent. J., 2015, 9, 47 CrossRef PubMed.
- X. M. Huang, C. Chen, Y. Shang, Y. F. Zhong, G. F. Ren, Z. Q. Yu and J. An, Chemosphere, 2016, 161, 251–258 CrossRef CAS PubMed.
- C. S. Dai, D. W. Li, L. J. Gong, X. L. Xiao and S. S. Tang, Molecules, 2016, 21, 106 CrossRef PubMed.
- G. P. Lv, L. Z. Menga, D. Q. Han, H. Y. Li, J. Zhao and S. P. Li, J. Pharm. Biomed. Anal., 2015, 109, 105–111 CrossRef CAS PubMed.
- M. V. Berridge, P. M. Herst and A. S. Tan, Biotechnol. Annu. Rev., 2005, 11, 127–152 CAS.
- R. F. Hussain, A. M. E. Nouri and R. T. D. Oliver, J. Immunol. Methods, 1993, 160, 89–96 CrossRef CAS PubMed.
- C. Chen, Z. H. Lu, J. Yang, W. C. Hao, Y. J. Qin and H. Y. Wang, Cancer Med., 2016, 5, 3489–3499 CrossRef CAS PubMed.
- K. Klemola, J. Pearson and P. Lindstrom-Seppa, Autex Res. J., 2007, 7, 217–223 Search PubMed.
| This journal is © The Royal Society of Chemistry 2017 |