Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Maghemite-containing PLGA–PEG-based polymeric nanoparticles for siRNA delivery: toxicity and silencing evaluation
Emmanuel Lellouche†
ad,
Erica Locatelli† b,
Liron Limor Israelcd,
Maria Naddakae,
Ella Kurlanderad,
Shulamit Michaeli*ad,
Jean-Paul Lellouche*cd and
Mauro Comes Franchini
b,
Liron Limor Israelcd,
Maria Naddakae,
Ella Kurlanderad,
Shulamit Michaeli*ad,
Jean-Paul Lellouche*cd and
Mauro Comes Franchini *b
*b
aThe Mina and Everard Goodman Faculty of Life Sciences, Bar-Ilan University, Ramat-Gan, Israel
bDepartment of Industrial Chemistry “Toso Montanari”, University of Bologna, Viale Risorgimento 4, 40136 Bologna, Italy. E-mail: mauro.comesfranchini@unibo.it
cDepartment of Chemistry Faculty of Exact Sciences, Bar-Ilan University, Ramat-Gan, Israel
dInstitute of Nanotechnology & Advanced Materials, Bar-Ilan University, Ramat-Gan, Israel
eStephenson Institute for Renewable Energy, University of Liverpool, Peach Street, L69 7ZF, Liverpool, UK
First published on 19th May 2017
Abstract
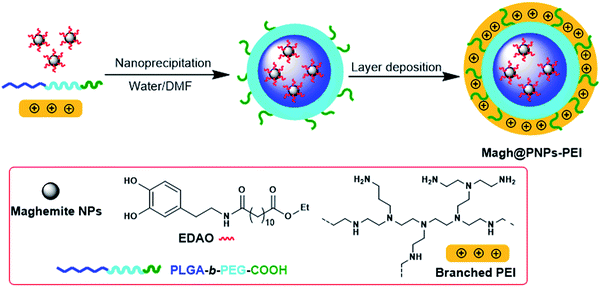
Gene therapy based on small interfering RNA (siRNA) has emerged as an exciting new therapeutic approach. In this work, incorporation of PEI into poly(D,L-lactide-co-glycolide)-poly(ethylene glycol) (PLGA-b-PEG) particles has been shown to be quite effective in the development of corresponding gene delivery systems, and encapsulation of magnetic nanoparticles as an MRI contrast agent, resulted in unique theranostic nanoparticles.
Introduction
It is known that synthetic small interfering RNA (siRNA) can inhibit specific protein expression by suppressing a target gene selectively by a mechanism called RNA interference (RNAi). However, the development of appropriate nanocarriers to deliver siRNA is crucial for practical applications as therapeutics. For instance several carrier systems for siRNA, including cationic polymers, lipids and peptides, have been utilized to form nanosized polyelectrolyte complexes via electrostatic interactions, but have shown deficiencies associated with specific gene inhibition and biocompatibility.1–3 Among the most suitable delivery systems, there are those derived from biodegradable and biocompatible synthetic polymers4 and in this view the polymeric nanomicelles derived from the poly(D,L-lactide-co-glycolide)-poly(ethylene glycol) (PLGA-b-PEG) play a now well-established key role.5,6Moreover, the tracking of siRNA delivery would be very important and Magnetic Resonance Imaging (MRI) is certainly the technique of choice due to the ease of the synthesis of magnetic nanoparticles and their possible surface modifications that allows incorporation into biodegradable polymeric nanoparticles.7,8 This feature rendered these nanoparticles over the years a potent nano-tool, which, after incorporation of other therapeutics as siRNA, can be used in nanomedicine. One of the major obstacles in delivery of siRNA using nanoparticles is in the trapping of the carrier in the endosome. This critical step can be overcome by using polymers such as PEI that induce endosomal escape, or using a polymer carrier that mimic viral escape mechanism from the endosome.9–12
Polo-like kinase 1 (PLK1) is a well-established mitotic regulator involved in multiple biological functions throughout cell cycle progression. PLK-1 was found to be over-expressed in several cancer cells and inhibition of PLK-1 activity has emerged as a promising therapeutic target.13
In this study, we developed “all-in-one” polymeric nanoparticle probes containing maghemite for the delivery of siRNA and we evaluated our nanosystem both with in vitro test and toxicity in vivo experiments. We report the successful delivery of siRNA by PEI-capped PLGA-b-PEG polymeric nanoparticles to reduce the PLK1 expression in pancreatic cancer cells, and cause subsequent cell cycle arrest and induction of cell apoptosis, thus demonstrating its potential in cancer therapy. Also, we report preliminary in vivo data demonstrating that following the intravenously injection of 1 mg kg−1 dose of siRNA, no acute toxicity was observed as no animal death occurred as well as no changes were observed in various biochemical and hematological parameters.
Materials and methods
All chemicals were purchased from Sigma-Aldrich (St Louis, MO, USA) and used as received. Poly(D,L-lactide-co-glycolide) (50/50) with carboxylic acid end group (PLGA–COOH, pharmaceutical grade, inherent viscosity 0.12 dL g−1, molecular weight (MW) ∼ 7 kDa or ∼43.3 kDa) was purchased from Lakeshore Biomaterials (Lakeshore Biomaterials, Inc, Birmingham, AL, USA). PEG with carboxylic acid end groups (NH2–PEG–COOH, MW ∼ 3 kDa) was purchased from Rapp Polymere GmbH (Tübingen, Germany). All aqueous solutions were prepared with deionized water obtained using an ultrafiltration system (Milli-Q, Millipore Corporation, Billerica, MA, USA) with a measured resistivity above 18 MΩ cm−1. Dynamic light scattering (DLS) measurements were performed on a Malvern Zetasizer nano-S working with a 532 nm laser beam (Malvern Instruments Ltd, Malvern, UK). Moreover, ζ-potential measurements were conducted in DTS1060C-clear disposable zeta cells at 25 °C. SpectraAA 100 Varian was used for atomic absorption spectroscopy (AAS) analyses and iron determination. Transmission Electron Microscopy (TEM) was conducted on a Jeol JEM 2010 at 200 keV. Samples for TEM analyses were prepared by spreading a small drop of the nanoparticle dispersion on amorphous carbon-coated copper grids (Formvar carbon 400 mesh grids) followed by air-drying.Synthesis of PLGA-b-PEG–COOH (7–3 or 43.3–3 kDa)
The synthesis of the copolymer was performed according to our already reported procedure.14 Briefly, PLGA–NHS was obtained after reaction of PLGA–COOH (7 or 43.3 kDa, 1 eq.), N-hydroxysuccinimide (NHS, 4 eq.), and N,N′-dicyclohexylcarbodiimide (DCC, 4.5 eq.) in dichloromethane for 24 h, and precipitation in diethyl ether. A subsequent addition of NH2–PEG–COOH (3 kDa, 1 eq.) and N,N′-diisopropylethylamine (DIPEA, 3 eq.) in chloroform brought, after 24 h precipitation and washing in diethyl ether, to the final copolymer.Fabrication of PNPs
For the preparation PLGA-b-PEG polymeric nanoparticles (PNPs), a nanoprecipitation technique was exploited. A total of 100 mg of PLGA-b-PEG–COOH (10 kDa) and were solubilized in 10 mL of dimethylformamide (DMF) and added into 100 mL of water under vigorous stirring, maintaining the water/organic ratio of 10/1 with constant removal of the solution. The mixture was kept for 2 h under vigorous stirring. The so-obtained PNPs were purified with a centrifugal filter device (Amicon Ultra, Ultracel membrane with 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 NMWL, Millipore, USA) following by filtration using a syringe filter Sterivex™-GP of polyethersulfone (0.22 μm, Millipore Corporation). The final volume was adjusted to 5 mL.
000 NMWL, Millipore, USA) following by filtration using a syringe filter Sterivex™-GP of polyethersulfone (0.22 μm, Millipore Corporation). The final volume was adjusted to 5 mL.
Fabrication of Magh@PNPs
CAN–maghemite NPs were synthesized and coated with the specifically designed organic ligand ethyl 12-([3,4-dihydroxyphenethyl]amino)-12-oxododecanoate (EDAO), as already reported by us.15 For the entrapment of lipophilic maghemite particles into PNPs, a total of 100 mg of PLGA-b-PEG–COOH (10 kDa) were solubilized in 10 mL of dimethylformamide (DMF), which already contain the re-dispersed lipophilic maghemite particles. The solution was added into 100 mL of water under vigorous stirring, maintaining the water/organic ratio of 10/1 with constant removal of the solution. The mixture was kept for 2 h under vigorous stirring. The particles were purified with a centrifugal filter device (Amicon Ultra, Ultracel membrane with 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 NMWL, Millipore, USA) following by filtration using a syringe filter Sterivex™-GP of polyethersulfone (0.22 μm, Millipore Corporation). The final volume was adjusted to 5 mL.
000 NMWL, Millipore, USA) following by filtration using a syringe filter Sterivex™-GP of polyethersulfone (0.22 μm, Millipore Corporation). The final volume was adjusted to 5 mL.
Fabrication of PNPs–PEI
For the preparation of PEI-coated PLGA-b-PEG polymeric nanoparticles (PNPs–PEI), a simultaneous nanoprecipitation technique and layer deposition was exploited. A total of 68 mg of PLGA-b-PEG–COOH (46 or 10 kDa) and 34 mg of PEI (branched, 25 kDa) were solubilized in 3.8 mL and 3.0 mL of dimethylformamide (DMF) respectively, mixed together and added into 68 mL of water under vigorous stirring, maintaining the water/organic ratio of 10/1 with constant removal of the solution. The mixture was kept for 2 h under vigorous stirring. The so-obtained PNPs–PEI were purified with a centrifugal filter device (Amicon Ultra, Ultracel membrane with 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 NMWL, Millipore, USA) following by filtration using a syringe filter Sterivex™-GP of polyethersulfone (0.22 μm, Millipore Corporation). The final volume was adjusted to 5 mL.
000 NMWL, Millipore, USA) following by filtration using a syringe filter Sterivex™-GP of polyethersulfone (0.22 μm, Millipore Corporation). The final volume was adjusted to 5 mL.
Fabrication of Magh@PNPs–PEI
CAN–maghemite NPs were synthesized and coated with the specifically designed organic ligand ethyl 12-([3,4-dihydroxyphenethyl]amino)-12-oxododecanoate (EDAO), as already reported by us.16 For the entrapment of corresponding lipophilic maghemite particles into PNPs–PEI, a slight modification of the above reported procedure was exploited. A total of 100 mg of PLGA-b-PEG–COOH (46 or 10 kDa) were solubilized in 5.5 mL of dimethylformamide (DMF), which already contain the re-dispersed lipophilic maghemite particles, while 50 mg of PEI (branched, 25 kDa) were solubilized in 4.5 mL of DMF. The two solutions were mixed together and added into 100 mL of water under vigorous stirring, maintaining the water/organic ratio of 10/1 with constant removal of the solution. The mixture was kept for 2 h under vigorous stirring. The particles were purified with a centrifugal filter device (Amicon Ultra, Ultracel membrane with 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 NMWL, Millipore, USA) following by filtration using a syringe filter Sterivex™-GP of polyethersulfone (0.22 μm, Millipore Corporation). The final volume was adjusted to 5 mL.
000 NMWL, Millipore, USA) following by filtration using a syringe filter Sterivex™-GP of polyethersulfone (0.22 μm, Millipore Corporation). The final volume was adjusted to 5 mL.
Cell lines and culture
SK-OV-3 human ovarian adenocarcinoma cells were obtained from the American Type Culture Collection (ATCC). The dual luciferase-expressing U2OS human osteosarcoma cell line was generated as previously described17 and enzyme activities of the Renilla and Firefly luciferases were determined using Dual-Luciferase® Assay System (Promega). Cells were cultured in Dulbecco's Modified Eagle Medium (U2OS) or RPMI-1640 medium (SK-OV-3) and grown at 37 °C in a 5% CO2 atmosphere.Gel retardation assay
Magh@PNPs–PEI suspensions were diluted in water at different concentrations to reach different NP/siRNA w/w ratios (0.5, 1, 2, 4, 8 and 10). To each NP suspension and control tube (absence of particles), 2 μg of Firefly luciferase siRNA (IDT Technologies) were added, incubated for 15 min at room temperature for complex formation, and then, the resulting NP/siRNA complexes (10 μL) were mixed with 2 × RNA loading buffer (10 μL). Complexes were loaded into 1.5% agarose gel that was pre-stained with ethidium bromide. The samples were electrophoresed at 100 mA for 30 min in Tris-acetate (TAE) running buffer and bands were visualized using a UV imaging system (MiniLumi; DNR Bio-Imaging Systems Ltd.). The amount of free siRNA in each ratio was normalized to control tube and densitometry was performed using ImageJ software.Luciferase assay
U2OS-Luc cells were seeded (1 × 104 cells per well) in a 96 well optical bottom plate (Thermo) and incubated overnight at 37 °C with 5% CO2. Cells were transfected with Firefly luciferase siRNA (0.166 μg, 100 nM) mixed with Magh@PNPs–PEI NPs at different NP/siRNA w/w ratios (1, 2, 4, 6, 8 and 10) or without NPs (control). After 48 hours, cells were assayed for both Firefly and Renilla luciferase activities using the Dual® Luciferase Assay System (Promega). Briefly, cells were lysed and the Firefly luciferase substrate added (50 μL per well Dual® substrate/buffer). Firefly luciferase activity was measured after 10 min using a luminometer (Synergy 4, Biotek). Next, the Renilla luciferase substrate was added (50 μL per well Stop & GLO® substrate/buffer) and the luminescence was measured after further 10 min incubation. Silencing efficacy is reflected by luciferase activities normalized to control luciferase activities. The following oligonucleotide sequences (sense/antisense) were used: 5′-GGACAUCACCUAUGCCGAGUACUTC-3′/5′-GAAGUACUCGGCAUAGGUGAUGUCCAC-3′ (IDT Technologies).MTT assay
U2OS-Luc cells were seeded (1 × 104 cells per well) in a 96 well plate (Greiner) and incubated overnight at 37 °C with 5% CO2. Cells were treated with Firefly luciferase siRNA (0.166 μg) (100 nM) mixed with Magh@PNPs–PEI NPs at different NP/siRNA w/w ratios (1, 2, 4, 6, 8 and 10) or without NPs (control), and then incubated for 48 h at 37 °C with 5% CO2. After 48 h incubation, the medium was removed, and 100 μL of fresh medium containing the MTT reagent (Sigma-Aldrich) (0.5 mg mL−1) was added to each well followed by 20 min incubation. After incubation, MTT was removed, and 50 μL of DMSO (Sigma-Aldrich) was added to each well. All the absorption values were measured at 570 nm (Synergy 4, Biotek) and normalized to control.Silencing experiments
SK-OV-3 cells were seeded in a 6 well plate (6 × 105 cells per well) and incubated overnight at 37 °C with 5% CO2. Cells were transfected with PLK-1 or non-specific (control) siRNAs (QBI Enterprises, Ltd., Ness Ziona, Israel) mixed with Magh@PNPs–PEI at a 6/1 NP/siRNA w/w ratio. After 48 h incubation, cells and total RNA were isolated for real-time RT-PCR and cell cycle analysis.Total RNA isolation and quantitative estimation of mRNA by real-time RT-PCR
SK-OV-3 cells were collected after trypsinization into a single cell suspension and total RNA was isolated with TRI Reagent (Sigma) according to the manufacturer's instructions. RNA quality and quantity were determined with a spectrophotometer (Nanodrop 1000, Fisher Scientific). Then, mRNAs (0.5 μg) were reverse-transcribed with reverse transcriptase using oligo(dT)18 primers (First Strand cDNA Synthesis Kit, Thermo). Levels of target genes and of the housekeeping gene GAPDH were determined by quantitative PCR on the StepOnePlus Real-Time PCR System (Applied Biosystems) using the Fast SYBR Green Master Mix (Applied Biosystems) under the following conditions: activation step at 95 °C for 20 s, denaturation step at 95 °C for 3 s and annealing/extension step at 60 °C for 30 s (40 cycles). The target gene levels were normalized to those of GAPDH in the same samples. The primers used were (forward/reverse): PLK-1-5′-ACCAGCACGTCGTAGGATTC-3′/5′-CAAGCACAATTTGCCGTAGG-3′ (QBI Enterprises, Ltd., Ness Ziona, Israel), and GAPDH-5′-CATGTTCCAATATGAT TCCACC-3′/5′-GATGGGATTTCCATTGATGAC-3′ (IDT Technologies).Cell cycle analysis
SK-OV-3 cells were collected after trypsinization into a single cell suspension, and fixed with cold ethanol at 4 °C overnight. Before analysis, fixed cells were washed and re-suspended in 1 mL of PBS containing 200 μg mL−1 RNase A (10-109-169, Roche Applied Science) and 5 μg mL−1 propidium iodide (P4170, Sigma). After incubation of 20 min at 37 °C, cells were analyzed for DNA content by flow cytometry (Gallios, Beckman Coulter). For each sample 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were acquired and cell cycle distribution was determined using cell cycle analysis software (ModFit LT).
000 events were acquired and cell cycle distribution was determined using cell cycle analysis software (ModFit LT).
In vivo experiments
All animal experiments were performed in compliance with the Guidelines for the Care and Use of Research Animals established by the Bar-Ilan University Animal Studies Committee. Head of the Bar-Ilan University Animal Studies Committee, Dr Motro Benny and Study Approval no. 06-01-2015.BALB/c mice (Harlan Laboratories Israel Ltd., Jerusalem, Israel) aged 8–9 weeks were intravenously injected with a 1 mg kg−1 non-specific siRNA dose (QBI Enterprises, Ltd., Ness Ziona, Israel) mixed with Magh@PNPs–PEI NPs at a 6 NP/siRNA w/w ratio or with water alone. Each group was composed of 8 mices. Acute in vivo toxicity was assessed by mortality within 24 h following NP injection and by the evaluation of diverse hematological and biochemical parameters after 7 days. For the evaluation of hematological parameters, ∼150 μL of blood were collected in EDTA-coated tubes. For the evaluation of biochemical parameters, ∼350 μL of blood were collected in non-coated gel tubes, centrifuged (4000g, 4 min, RT) and serum separated. Then, EDTA-coated tubes and serum samples were transferred to American Medical Laboratories (AML, Herzliya Medical Center, Israel) for further analyses.
Statistical analyses
Statistical analysis of the data was performed with the GraphPad Prism software (GraphPad Software).Results and discussion
The hybrid PNPs–PEI particles were prepared in a single step using a modified nanoprecipitation technique. During this process the core nanosystem, derived from the self-assembling of PLGA-b-PEG, is formed as already described by us.16,18 PNPs composed of pure PLGA–PEG–COOH and obtained with this technique generally present a mean diameter of 70 ± 5 nm, PDI between 0.1–0.2 and a negative ζ-potential value of −40 mV.In this case, the PEI amine groups interact with the PEG free carboxylic acid ones, which remained exposed toward the external environmental, thus creating a positively charged layer of PEI adsorbed onto the outer shell of PNPs. Particle formation was confirmed by both Dynamic Light Scattering (DLS) and ζ-potential measurements (Table 1). Overall, the size of PNPs–PEI dropped to around 50 nm in comparison to the typical 70 nm of pure PLGA-b-PEG–COOH NPs prepared under similar conditions,16,19 most likely because of the strong electrostatic interactions between PEI and the carboxylic acid groups on the surface of the PLGA-b-PEG–COOH core. The ζ-potential drastically increased from around −50 mV to above +40 mV at pH around 6.5–7, confirming the presence of the positively charged PEI on the particle surface. Particularly, the effect of PLGA molecular weight (MW) and the w/w ratio between PLGA–PEG–COOH and PEI on size and ζ-potential of the resulting PNPs were investigated. It was noted that a reduction of the MW of PLGA (entry 1) did not affected the size of the resulting PNPs (always 40–50 nm), however a slightly higher ζ-potential value was obtained when a 43.3 kDa PLGA was used (+63 mV, entry 2) instead of PLGA of 7 kDa (+38 mV). A big w/w ratio (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) between PLGA-b-PEG–COOH and PEI leads to the formation of highly poly-dispersive PNPs (entry 3), probably due to inhomogeneous coating of the PLGA-b-PEG–COOH core by PEI, which minimized the size reducing effect of the electrostatic interactions. Due to these reasons, a 43.3 kDa MW for PLGA and 2/1 w/w PLGA-b-PEG/PEI ratio were selected for all the following preparations and studies (entry 4). The concentration in the final solution of the nanosystem obtained with the selected procedure was estimated by weighting the residual organic matter after solvent evaporation (gravimetric analysis) and was found to be 8.44 mg mL−1.
1) between PLGA-b-PEG–COOH and PEI leads to the formation of highly poly-dispersive PNPs (entry 3), probably due to inhomogeneous coating of the PLGA-b-PEG–COOH core by PEI, which minimized the size reducing effect of the electrostatic interactions. Due to these reasons, a 43.3 kDa MW for PLGA and 2/1 w/w PLGA-b-PEG/PEI ratio were selected for all the following preparations and studies (entry 4). The concentration in the final solution of the nanosystem obtained with the selected procedure was estimated by weighting the residual organic matter after solvent evaporation (gravimetric analysis) and was found to be 8.44 mg mL−1.
| Entry | Name | PLGA-b-PEG/PEI (w/w) | Diameter [nm] | PDI | ζ-Pot [mV] |
|---|---|---|---|---|---|
| 1 | PNPs–PEI (7 kDa) | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
49.6 ± 0.2 | 0.207 ± 0.007 | 38.6 ± 5.3 |
| 2 | PNPs–PEI (43 kDa) | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
48.5 ± 0.3 | 0.194 ± 0.004 | 63.5 ± 5.4 |
| 3 | PNPs–PEI | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
50.4 ± 0.3 | 0.365 ± 0.013 | 42.6 ± 7.5 |
| 4 | PNPs–PEI | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
44.8 ± 0.1 | 0.125 ± 0.007 | 47.6 ± 10.4 |
| 5 | Magh@PNPs–PEI | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
147.8 ± 1.9 | 0.227 ± 0.013 | 51.4 ± 7.3 |
Once selected the optimal conditions for the fabrication of PNPs–PEI, the entrapment of magnetic NPs in the inner core of the nanosystem was also attempted. Specific metal cation (Ce3/4+)-doped CAN–maghemite NPs were selected as magnetic particles due to their well-established properties as MRI contrast agent enhancer.20,21
Since the inner core of PLGA-b-PEG PNPs can only host lipophilic moieties,5 it was compulsory to modify the CAN–maghemite NPs surface from a hydrophilic to a lipophilic one. Surface chemistry of magnetic iron oxide-based NPs as well as possibility to coat them with suitable organic molecules and their resulting properties' modifications have been widely investigated by us.22,23 In particular for CAN–maghemite NPs coating, we have already synthesized a specific ligand, ethyl 12-([3,4-dihydroxyphenethyl]amino)-12-oxododecanoate (EDAO), able to bind onto the iron oxide surface thanks to its catechol moiety and to confer lipophilicity to the entire system. With a ligand exchange procedure, organo-soluble maghemite NPs have been obtained and characterized.7 The obtained particles are re-dispersible in DMF, thus they can be mixed with the PLGA-b-PEG solution and entrapped into the inner core of PNPs–PEI during the core nanoparticles formation. The entire procedure is summarized in Scheme 1.
Generally, PNPs of pure PLGA–PEG with maghemite nanoparticles in the inner core, obtained with the described technique, present a mean diameter of 150 ± 10 nm, PDI between 0.15 and 0.25 and ζ-potential values of −25 ± 5 mV.
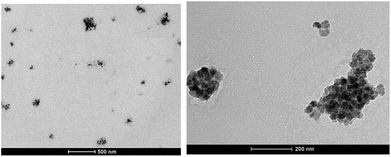
Also in this case, as expected the so obtained Magh@PNPs–PEI showed an increase in size (entry 5) due to presence of maghemite NPs in the core, while the ζ-potential remained highly positive and practically unchanged confirming the non-interaction of EDAO-coated maghemite NPs with PEI during the nanoprecipitation nor their adsorption onto the surface. Iron concentration was determined by means of Atomic Absorption Spectroscopy (AAS) and found to be 0.81 mg mL−1, while the concentration of the entire nanosystem determined with gravimetric analysis was equal to 23.4 mg mL−1. TEM images confirmed the entrapment of the maghemite NPs into well-defined confined of the polymeric matrix and the obtainment of a homogeneous sample (Fig. 1).
Prior to the encapsulation phase, the basic magnetism-based properties of (Ce3/4+) cation-doped CAN–maghemite nanoparticles, MRI relaxivity values, have been measured and recently published:20 basically, it has been clearly shown that both 1/T1 and 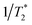 relaxation rates varied linearly with Fe concentrations affording both corresponding longitudinal and transverse r1 and
relaxation rates varied linearly with Fe concentrations affording both corresponding longitudinal and transverse r1 and 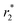 relaxivity (curve slopes in Fig. 2) values of 0.0015 and 189 mmol−1 s−1 respectively, which characterize such strong
relaxivity (curve slopes in Fig. 2) values of 0.0015 and 189 mmol−1 s−1 respectively, which characterize such strong 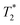 contrast relaxation maghemite-based NPs that will be quite useful for effective in vivo MRI. After the incorporation step, the same properties for maghemites containing PLGA–PEG nanoparticles have been also measured:15 also in this case, it has been clearly shown that both r1 and
contrast relaxation maghemite-based NPs that will be quite useful for effective in vivo MRI. After the incorporation step, the same properties for maghemites containing PLGA–PEG nanoparticles have been also measured:15 also in this case, it has been clearly shown that both r1 and 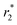 relaxivities, with values of 0.9 and 134 mmol−1 s−1 respectively, remain effective for future MRI purpose.
relaxivities, with values of 0.9 and 134 mmol−1 s−1 respectively, remain effective for future MRI purpose.
 | ||
Fig. 2 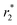 relaxivity curve of (Ce3/4+) cation-doped CAN–maghemite nanoparticles at variable Fe concentrations. relaxivity curve of (Ce3/4+) cation-doped CAN–maghemite nanoparticles at variable Fe concentrations. | ||
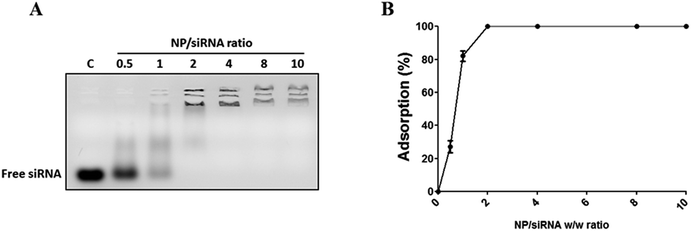
First, to evaluate if Magh@PNPs–PEI NPs can be used as a siRNA nanocarrier for gene silencing, siRNA binding was evaluated by a gel retardation assay. NP suspensions at different concentrations were incubated with a constant amount of siRNA to reach specific NP/siRNA w/w ratios, and then, loaded in an agarose gel to separate free siRNA from NP/siRNA complexes (Fig. 3). As can be seen in Fig. 3A, results suggest that the optimal binding NP/siRNA w/w ratio for these particles is 2 demonstrating an efficient binding of 100% since no residual free siRNA, which migrates faster through the gel matrix compared to NP/siRNA complexes, can be observed. Similar results were obtained in three different experiments (Fig. 3B).
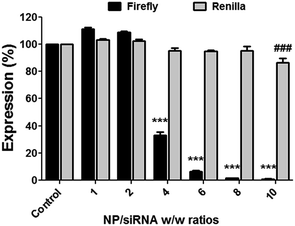
Next, to examine if Magh@PNPs–PEI NPs can efficiently induce gene silencing, we used a dual-luciferase reporter assay based on the luciferase proteins, Firefly and Renilla, stably transfected in human U2OS cancer cells. This assay allowed us to measure the specific silencing of the Firefly luciferase, while the Renilla luciferase was used as an internal control for cell viability. Based on the previously described binding results, silencing was conducted with NP/siRNA w/w ratios from 1 to 10 using a constant amount of siRNA (100 nM) (Fig. 4). A knockdown of 94 ± 1% was observed with a ratio of 6 while no significant decrease in the Renilla levels was observed indicating that no toxicity occurs under these conditions. Better silencing was obtained with higher NP/siRNA w/w ratios; however, silencing using a ratio of 10 resulted in minimal toxicity as can be seen in the Renilla level (86 ± 3%).
In addition, lower NP/siRNA w/w ratio of 4 resulted in a significant silencing of 68 ± 2%, while no silencing occurred with ratios of 1 and 2. Indeed, following cell internalization, siRNA molecules need to escape the endocytic pathway in order to reach the RNAi machinery found in cell cytosol. Therefore, a polycationic 25 kDa PEI polymer was used to enable lysosomal escape based on its well known “proton sponge” effect. However, sufficient amount of PEI is always needed to enable siRNAs escape without causing cytotoxic effects due to the massive destruction of cell endosome/lysosome compartments. Therefore, silencing was observed in a dose dependent manner and efficient silencing was obtained only with higher NP/siRNA w/w ratios, meaning higher amounts of PEI.
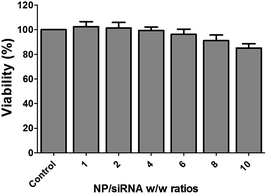
To further investigate the toxicity of Magh@PNPs–PEI NPs, we treated U2OS-Luc cells with increasing NP/siRNA w/w ratios in a similar way as it was previously done for the dual-luciferase reporter assay (Fig. 5). Mitochondrial activity was determined using the MTT assay. After 48 h, no significant changes in cell proliferation and viability were observed at a ratio of 6, however, minor toxicity can be observed using higher ratios. These results are in accordance to the cell toxicity measured by the Renilla luciferase levels in the dual-luciferase reporter assay. These results indicate that no adverse toxicity occurs using a NP/siRNA w/w ratio of 6 which was found to be optimal for silencing.
The dual-luciferase reporter assay is quick and efficient, but is extremely sensitive due to low mRNA levels and short half-life of the Firefly luciferase protein. We therefore explored if these NPs can silence the overexpressed and abundant PLK-1 kinase and whether the silencing results in downstream effects.
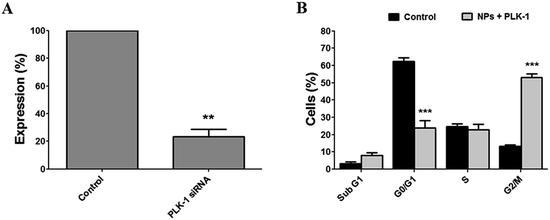
Based on the luciferase silencing results, we used a NP/siRNA w/w ratio of 6 since it is the lowest amount of NPs that still gave a very efficient silencing with no toxicity. SK-OV-3 human ovarian cancer cells were transfected with PLK-1 siRNA complexed to Magh@PNPs–PEI following by mRNA level and cell cycle distribution analyses (Fig. 6). A significant knockdown of 77 ± 5% was observed after PLK-1 silencing (Fig. 6A) resulting in a major cell cycle arrest at the G2/M phase (from 13 ± 1% to 53 ± 2%), a decrease in cell percentage in the G0/G1 phase (from 62 ± 2% to 23 ± 4%) and an important Sub-G1 increase from 3 ± 1 to 8 ± 1% (Fig. 6B).
Despite the well-known in vivo toxicity of the PEI polymer, it is still considered as the best endosomal escape-enabling polycationic polymer for nucleic acid delivery. PEI toxicity is well documented in the literature and is mainly due to its ability to interact with negatively charged membranes of red blood cells (RBCs) causing RBC aggregation and lysis, leading to thrombosis, and finally, to animal death.24,25 Different covalent and non-covalent chemical modification approaches have been employed to modify original PEI polymers in order to mitigate their toxicity, for example when incorporating/linking poly(ethylene glycol) polymers (PEGs), polysaccharides, and/or various hydrophobic moieties.26
In previous illustrative works, CAN–maghemite–PEI NPs have been intravenously injected into mice resulting in animal death within two hours due to PEI toxicity, while no animal death was observed when injecting the iron core particles alone.27,28 This PEI phase generating toxicity was successfully mitigated using diverse chemical modifications by reducing the overall number of positively charged PEI amine species (polycationic feature controlled reduction). Here, to determine if Magh@PNPs–PEI NPs can be potentially used as a therapeutic tool in vivo, we evaluate the acute NP toxicity by injecting mice intravenously with a 1 mg kg−1 non-specific siRNA dose mixed with NPs. Surprisingly, following injection, no signs of animal distress or death have been observed within 24 h, despite the fact that no chemical modifications of the PEI component have been performed. In addition, various biochemical and hematological parameters were collected after 7 days following injection to deeper investigate the acute NP toxicity (Tables 2 and 3, respectively) with an emphasis on liver and kidney toxicity.
| Water | Magh@PNPs–PEI NPs | |||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| Total protein (g dL−1) | 5.48 | 0.21 | 5.50 | 0.22 |
| Albumin (g dL−1) | 4.05 | 0.18 | 4.08 | 0.18 |
| Total bilirubin (mg dL−1) | 0.14 | 0.19 | 0.06 | 0.03 |
| ALP (IU L−1) | 227.00 | 18.14 | 235.75 | 8.24 |
| AST = SGOT (IU L−1) | 111.00 | 30.22 | 124.25 | 38.66 |
| ALT = SGPT (IU L−1) | 58.25 | 33.59 | 67.50 | 20.14 |
| Creatinine (mg dL−1) | <0.2 | NA | <0.2 | NA |
| Urea (mg dL−1) | 49.83 | 4.57 | 57.80 | 5.83 |
| Water | Magh@PNPs–PEI NPs | |||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| WBC (103 μL) | 6.69 | 1.46 | 6.21 | 0.75 |
| RBC (106 μL) | 10.50 | 0.28 | 10.75 | 0.16 |
| HGB (g dL−1) | 16.71 | 0.49 | 17.21 | 0.42 |
| HCT (%) | 49.41 | 1.95 | 50.69 | 2.08 |
| MCV (fL) | 47.09 | 1.91 | 47.19 | 1.95 |
| MCH (pg) | 15.93 | 0.45 | 16.01 | 0.44 |
| MCHC (%) | 33.84 | 0.61 | 33.96 | 0.61 |
| Neutrophils (%) | 15.41 | 5.53 | 18.74 | 6.41 |
| Stab (%) | 0.13 | 0.35 | 0.00 | 0.00 |
| Lymphocytes (%) | 80.66 | 6.40 | 77.15 | 6.70 |
| Monocytes (%) | 1.15 | 0.64 | 1.11 | 0.96 |
| Eosinophils (%) | 2.09 | 1.56 | 2.54 | 2.45 |
| Basophils (%) | 0.14 | 0.12 | 0.10 | 0.09 |
| Platelets (103 μL) | 1009.38 | 111.04 | 1036.88 | 152.70 |
Liver toxicity can be evaluated by various blood tests while some tests are associated with cellular integrity and other with liver functionality. Typical biochemical parameters of cellular toxicity include the liver enzymes alanine aminotransferase (ALT), aspartate aminotransferase (AST) and alkaline phosphatase (ALP) which elevated serum levels can be detected due to their release from damaged liver cells, while elevated bilirubin serum levels as well as decreased total protein and albumin serum levels can indicate liver dysfunction. In addition, kidney function can be evaluated by creatinine and urea serum levels.29 Following Magh@PNPs–PEI NPs, no significant changes can be observed in all liver and kidney parameters (Table 2).
In addition to biochemical parameters, hematological parameters including red and white blood cell counts as well as hemoglobin concentrations, are widely used as clinical indicators of general health and disease. Also in this case, no significant changes can be observed in all tested hematological parameters following injection of Magh@PNPs–PEI NPs (Table 3).
Altogether, these results indicate that a 1 mg kg−1 siRNA dose mixed with Magh@PNPs–PEI NPs can be safely injected since no acute toxicity was observed following injection.
Although further investigations should be performed, one supposes that the PEI toxicity mitigation in these Magh@PNPs–PEI NPs is likely due to their unique chemical properties and nanofabrication mode. Compared to former CAN–maghemite–PEI NPs, where the PEI layer was incorporated/attached onto the NP surface via cerium coordinative chemistry, this same 25 kDa PEI polymer component in these Magh@PNPs–PEI NPs has been incorporated via electrostatic interactions of the PEI amino groups with the free carboxylic acid groups of the PEG polymer component, most likely resulting in effective neutralization of most reactive PEI primary amines. In addition, PEI toxicity mitigation might also be a direct consequence of the well-known hydrophilic nature of PEG polymers enabling charge interactions/complexation of positively charged PEI ammonium salt species.
Conclusions
In this work, incorporation of PEI onto the surface of PLGA-b-PEG nanoparticles has been shown as a promising strategy for the building up of a novel gene delivery system. Easy introduction of magnetic nanoparticles into the core of the same system has been demonstrated and could lead to the obtainment of an efficient theranostic agent. Strong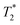 contrast (Ce3/4+) cation-doped CAN–maghemite NPs (longitudinal & transverse r1 and
contrast (Ce3/4+) cation-doped CAN–maghemite NPs (longitudinal & transverse r1 and 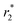 relaxivity values of 0.0015 and 189 mmol−1 s−1) also when encapsulated (longitudinal & transverse r1 and
relaxivity values of 0.0015 and 189 mmol−1 s−1) also when encapsulated (longitudinal & transverse r1 and 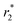 relaxivity values of 0.9 and 134 mmol−1 s−1) will clearly justify and promote strong potentially effective magnetism-based tumor targeting. Moreover, these manuscript novel functional composite nanoparticles are potentially highly versatile for 2nd step surface engineering with any well-know tumor-targeting species together with siRNA delivery capabilities (folic acid binding for example, PEI covalent modifications before encapsulation, etc.). Finally, silencing of ∼95% was observed using a dual-luciferase reporter assay without any toxicity. In addition, preliminary in vivo results suggest that NPs can be injected with a dose of up to 1 mg kg−1 with no acute toxicity. These preliminary in vivo results together with the very efficient in vitro silencing suggest that Magh@PNPs–PEI NPs can be potentially used as a powerful platform for the in vivo delivery of siRNAs in cancer treatment.
relaxivity values of 0.9 and 134 mmol−1 s−1) will clearly justify and promote strong potentially effective magnetism-based tumor targeting. Moreover, these manuscript novel functional composite nanoparticles are potentially highly versatile for 2nd step surface engineering with any well-know tumor-targeting species together with siRNA delivery capabilities (folic acid binding for example, PEI covalent modifications before encapsulation, etc.). Finally, silencing of ∼95% was observed using a dual-luciferase reporter assay without any toxicity. In addition, preliminary in vivo results suggest that NPs can be injected with a dose of up to 1 mg kg−1 with no acute toxicity. These preliminary in vivo results together with the very efficient in vitro silencing suggest that Magh@PNPs–PEI NPs can be potentially used as a powerful platform for the in vivo delivery of siRNAs in cancer treatment.
Acknowledgements
This work was supported by the European Union's Seventh Programme for research, technological development and demonstration [VIIth Framework RTD European Project Save Me, grant number 263307]. We would also like to thank the company QBI Enterprises for kindly providing us with PLK-1 siRNA species for silencing, cell cycle investigation, and in vivo experiments.Notes and references
- A. Aigner, Curr. Opin. Mol. Ther., 2007, 9, 345–352 CAS.
- A. Akinc, A. Zumbuehl, M. Goldberg, E. S. Leshchiner, V. Busini, N. Hossain, S. A. Bacallado, D. N. Nguyen, J. Fuller, R. Alvarez, A. Borodovsky, T. Borland, R. Constien, A. de Fougerolles, J. R. Dorkin, K. N. Jayaprakash, M. Jayaraman, M. John, V. Koteliansky, M. Manoharan, L. Nechev, J. Qin, T. Racie, D. Raitcheva, K. G. Rajeev, D. W. Y. Sah, J. Soutschek, I. Toudjarska, H.-P. Vornlocher, T. S. Zimmermann, R. Langer and D. G. Anderson, Nat. Biotechnol., 2008, 26, 561–569 CrossRef CAS PubMed.
- P. Kumar, H. Wu, J. L. McBride, K.-E. Jung, M. H. Kim, B. L. Davidson, S. K. Lee, P. Shankar and N. Manjunath, Nature, 2007, 448, 39–43 CrossRef CAS PubMed.
- A. J. Gavasane and H. A. Pawar, Clin. Pharmacol. Biopharm., 2014, 3(2), 121–128 Search PubMed.
- E. Locatelli and M. Comes Franchini, J. Nanopart. Res., 2012, 14, 1–17 CrossRef.
- J. Cheng, B. A. Teply, I. Sherifi, J. Sung, G. Luther, F. X. Gu, E. Levy-Nissenbaum, A. F. Radovic-Moreno, R. Langer and O. C. Farokhzad, Biomaterials, 2007, 28, 869–876 CrossRef CAS PubMed.
- M. Barrow, A. Taylor, P. Murray, M. J. Rosseinsky and D. J. Adams, Chem. Soc. Rev., 2015, 44(19), 6733–6748 RSC.
- T. K. Jain, M. K. Reddy, M. A. Morales, D. L. Leslie-Pelecky and V. Labhasetwar, Mol. Pharmaceutics, 2008, 5(2), 316–327 CrossRef CAS PubMed.
- M. Gillard, Z. Jia, J. J. C. Hou, M. Song, P. P. Gray, T. P. Munro and M. J. Monteiro, Biomacromolecules, 2014, 15, 3569–3576 CrossRef CAS PubMed.
- N. P. Truong, W. Gu, I. Prasadam, Z. Jia, R. Crawford, Y. Xiao and M. J. Monteiro, Nat. Commun., 2013, 4, 1902–1908 CrossRef PubMed.
- N. P. Truong, Z. Jia, M. Burgess, L. Payne, N. A. J. McMillan and M. J. Monteiro, Biomacromolecules, 2011, 12, 3540–3548 CrossRef CAS PubMed.
- N. T. D. Tran, N. P. Truong, W. Gu, Z. Jia, M. A. Cooper and M. J. Monteiro, Biomacromolecules, 2013, 14, 495–502 CrossRef CAS PubMed.
- S. Kumar, A. R. Sharma, G. Sharma, C. Chakraborty and J. Kim, Biochim. Biophys. Acta, 2016, 1865, 190–203 CAS.
- E. Strocchi, F. Fornari, M. Minguzzi, L. Gramantieri, M. Milazzo, V. Rebuttini, S. Breviglieri, C. M. Camaggi, E. Locatelli, L. Bolondi and M. Comes-Franchini, Eur. J. Med. Chem., 2012, 48, 391–401 CrossRef CAS PubMed.
- E. Locatelli, L. Gil, L. L. Israel, L. Passoni, M. Naddaka, A. Pucci, T. Reese, V. Gomez-Vallejo, P. Milani, M. Matteoli, J. Llop, J. P. Lellouche and M. Comes Franchini, Int. J. Nanomed., 2012, 7, 6021–6033 CAS.
- G. Baldi, C. Ravagli, F. Mazzantini, G. Loudos, J. Adan, M. Masa, D. Psimadas, E. A. Fragogeorgi, E. Locatelli, C. Innocenti, C. Sangregorio and M. Comes Franchini, Int. J. Nanomed., 2014, 9, 3037–3056 CAS.
- Y. K. Buchman, E. Lellouche, S. Zigdon, M. Bechor, S. Michaeli and J. P. Lellouche, Bioconjugate Chem., 2013, 24(12), 2076–2087 CrossRef CAS PubMed.
- M. Comes Franchini, B. F. Bonini, C. M. Camaggi, D. Gentili, A. Pession, M. Rani and E. Strocchi, Eur. J. Med. Chem., 2010, 45, 2024–2033 CrossRef CAS PubMed.
- M. Comes Franchini, J. Ponti, R. Lemor, M. Fournelle, F. Broggi and E. Locatelli, J. Mater. Chem., 2010, 20, 10908–10914 RSC.
- L. L. Israel, E. Lellouche, R. Kenett, O. Green, S. Michaeli and J. P. Lellouche, J. Mater. Chem. B, 2014, 2(37), 6215–6225 RSC.
- R. Ben Ishay, L. L. Israel, E. Levy, D. M. Partouche and J. P. Lellouche, J. Mater. Chem. B, 2016, 4, 3801–3814 RSC.
- D. Psimadas, G. Baldi, C. Ravagli, M. Comes Franchini, E. Locatelli, C. Innocenti, C. Sangregorio and G. Loudos, Nanotechnology, 2014, 25, 025101–025110 CrossRef CAS PubMed.
- G. Baldi, C. Ravagli, F. Mazzantini, G. Loudos, J. Adan, M. Masa, D. Psimadas, E. A. Fragogeorgi, E. Locatelli, C. Innocenti, C. Sangregorio and M. Comes Franchini, Int. J. Nanomed., 2014, 9, 3037–3056 CAS.
- C. Li, D. Zhong, Y. Zhang, W. Tuo, N. Li, Q. Wang, Z. Liu and W. Xue, J. Mater. Chem. B, 2013, 1, 1885–1893 RSC.
- R. L. Kanasty, K. A. Whitehead, A. J. Vegas and D. G. Anderson, Mol. Ther., 2012, 20(3), 513–524 CrossRef CAS PubMed.
- S. Höbel and A. Aigner, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5(5), 484–501 Search PubMed.
- E. Lellouche, L. L. Israel, M. Bechor, S. Attal, E. Kurlander, V. A. Asher, A. Dolitzky, L. Shaham, S. Izraeli, J. P. Lellouche and S. Michaeli, Bioconjugate Chem., 2015, 26(8), 1692–1701 CrossRef CAS PubMed.
- L. L. Israel, E. Lellouche, S. Ostrovsky, V. Yarmiayev, M. Bechor, S. Michaeli and J. P. Lellouche, ACS Appl. Mater. Interfaces, 2015, 7(28), 15240–15255 CAS.
- M. L. Bishop, E. P. Fody and L. E. Schoeff, Clinical chemistry : techniques, principles, correlations, Lippincott Williams & Wilkins, Philadelphia, Pa., London, 6th edn, 2010 Search PubMed.
Footnote |
| † Contributed equally. |
| This journal is © The Royal Society of Chemistry 2017 |