Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
L-aspartate oxidase magnetic nanoparticles: synthesis, characterization and L-aspartate bioconversion
Ilaria Armenia a,
Riccardo Balzaretti
a,
Riccardo Balzaretti a,
Cristina Pirronea,
Chiara Allegrettib,
Paola D'Arrigobcd,
Mattia Valentinocd,
Rosalba Gornatiac,
Giovanni Bernardini
a,
Cristina Pirronea,
Chiara Allegrettib,
Paola D'Arrigobcd,
Mattia Valentinocd,
Rosalba Gornatiac,
Giovanni Bernardini *ac and
Loredano Pollegioniac
*ac and
Loredano Pollegioniac
aDipartimento di Biotecnologie e Scienze della Vita, Università degli Studi dell'Insubria, Via J.H. Dunant 3, 21100 Varese, Italy. E-mail: giovanni.bernardini@uninsubria.it
bDipartimento di Chimica, Materiali e Ingegneria Chimica “Giulio Natta”, Politecnico di Milano, Piazza Leonardo da Vinci 32, 20133 Milano, Italy
cThe Protein Factory, Politecnico di Milano, ICRM CNR Milano, Università degli Studi dell'Insubria, Via Mancinelli 7, 20131 Milano, Italy
dCNR – Istituto di Chimica del Riconoscimento Molecolare (ICRM), Via Mancinelli 7, 20131 Milano, Italy
First published on 12th April 2017
Abstract
The FAD-containing enzyme L-aspartate oxidase (LASPO) catalyzes the stereospecific oxidative deamination of L-aspartate and L-asparagine functioning under both aerobic and anaerobic conditions. LASPO possesses distinctive features that make it attractive for biotechnological applications. In particular, it can be used for the production of D-aspartate from a racemic mixture of D,L-aspartate, a molecule employed in the pharmaceutical industry, for parenteral nutrition, as a food additive and in sweetener manufacture. Since the industrial application of LASPO is hampered by the high cost per enzymatic unit, several attempts have been performed to improve its reusability, such as LASPO immobilization on various matrices. In this context, magnetic nanoparticles (NPs) have recently become available for the immobilization of enzymes. In this work, we have covalently immobilized LASPO from the thermophilic archaea Sulfolobus tokodaii on iron oxide NPs using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and hydroxysuccinimide as cross-linking agents. The NP-LASPO system showed a better stability than the free enzyme, was reused five times reaching full L-aspartate conversion and yielded a productivity (>3 μmol per h per unit) similar to that obtained with the free enzyme or with the enzyme immobilized on classical chromatographic supports. The NP-LASPO system can be easily recovered after each cycle. These results indicate that the prepared NP-LASPO system has promising industrial applications.
Introduction
The FAD-containing enzyme L-aspartate oxidase (LASPO, EC 1.4.3.16) catalyzes the stereospecific oxidative deamination of L-aspartate and L-asparagine to the corresponding imino acid and, following spontaneous hydrolysis, an α-keto acid (i.e. oxaloacetate from L-aspartate) along with the production of ammonia and hydrogen peroxide. The ability of LASPO to use both O2 and fumarate in reoxidation of the reduced flavin cofactor enables it to function under both aerobic and anaerobic conditions.1 This agrees with the physiological role of the enzyme in prokaryotes, which is to catalyze the first step of NAD+ biosynthesis. Notably, LASPO distinguishes from the family members of the amino acid oxidases, most closely resembling the succinate dehydrogenase/fumarate reductase family of flavoproteins; indeed, LASPO is not evolutionarily related to the L-amino acid oxidase members.2In past years, we focused on LASPO from the thermophilic archaea Sulfolobus tokodaii (StLASPO), a monomeric protein (comprising 472 residues, 52 kDa) which folds into three distinct domains: a FAD-binding domain, a capping domain, and a helical domain.3 StLASPO is produced as a recombinant protein in E. coli, while classical L-amino acid oxidases with a broad substrate specificity are not. It is produced as active holoenzyme (the flavin cofactor is tightly, but non-covalently, bound to the protein moiety) reaching up to 9% of the total proteins in the crude extract and 13.5 mg L−1 in the fermentation broth.3,4
We focused on this enzyme since it possesses distinctive features that make it attractive for biotechnological applications.5 Indeed, LASPO possesses high thermal stability (it is fully stable up to 80 °C), high temperature optimum, stable activity in a broad range of pH (7.0–10.0), weak inhibition by the product and by the D-isomer of aspartate, and a quite low Km for dioxygen (0.3 mM). These properties make StLASPO a potential useful novel tool in biocatalysis where it can be used in applications resembling those developed for D-amino acid oxidase of opposite enantioselectivity.6 In particular, it can be used for the production of D-aspartate, a molecule employed in the pharmaceutical industry, for parenteral nutrition, as food additive and in sweetener manufacturing.5 On this side, when applied in the free form, the resolution of a 50 mM racemic mixture of D,L-aspartate was obtained in one day using 9 U of StLASPO (final e.e. > 99.5%).4
To facilitate industrial applications by improving reusability and, hence, by reducing the costs, StLASPO was immobilized on various matrices. The best results in terms of immobilization yield and volumetric activity have been obtained through the free amino groups of the enzyme by using the supports Relizyme™ HA403/S R and SEPABEADS® EC-EP/S or when prepared as cross-linked enzyme aggregates. The Relizyme-StLASPO immobilized preparation was reused for three cycles keeping full oxidation of L-aspartate, in ≤2 hours starting from a 50 mM racemic mixture: in a semi-preparative scale reaction, 66 mg of D-aspartate per day were produced using 20 U of StLASPO.7
With the development of nanotechnology, magnetic nanoparticles (NPs) have become available for the immobilization of enzymes. Magnetic NPs, when small enough, show superparamagnetic behaviors with a fast response to applied magnetic fields and with negligible residual magnetism and coercitivity. This means that these NPs can be magnetized with an external magnetic field and immediately redispersed once the magnet is removed.8 We have recently functionalized iron oxide NPs with D-amino acid oxidase for therapeutic purposes and obtained a magnetic nano-enzymatic system capable of producing, in presence of its substrate, reactive oxygen species.9,10 This system, which possesses relatively low toxicity,11 might be driven to the target area by an external magnetic field. Magnetic nano-enzymes can be used in several other fields such as biosensors for environmental pollutants and clinical sensors for biomolecules. A caveat, when using NPs, is always a concern about their safety.12 Iron NPs, however, have been shown to exert a relatively low toxicity.11,13
The application of StLASPO has few limitations due to the high cost per enzymatic unit and inability of separation. Among the approaches useful to improve the enzyme stability, the immobilization has proven particularly valuable. On this side, significant improvement has been made by enzyme immobilization onto magnetic nanocarriers. They can be advantageously employed as industrial catalysts since they have high surface area, large surface-to-volume ratio, lower hindrance, allow to regulate the orientation of the enzyme on the support and to easily remove the enzyme by applying a magnetic field.14,15
To generate an immobilized StLASPO system differing in mechanical, physical and diffusional properties, we focused on the setup of magnetic NP-LASPO that can be easily recovered from the reaction mixture and, potentially, applied under different practical conditions as compared to classical resin-immobilized enzyme preparations. Following the optimization of the immobilization procedure, the NP-LASPO system was characterized for the main physical–chemical properties and then used to convert L-aspartate into oxaloacetate and ammonia. The flavoenzyme system can be reused for 4 cycles with no loss of activity. This result paves the way to the set-up of innovative biocatalytic processes.
Experimental
Chemicals
Iron oxide nanoparticles (Fe3O4NPs, <50 nm), (3-aminopropyl)triethoxylsilane (APTES), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), hydroxysuccinimide (NHS), 4-amino antipyrine (4-AAP), L-aspartic acid, glycine, phenol: all purchased from Sigma Aldrich (Milan, Italy). Horseradish peroxidase (POD) was purchased from Roche (Milan, Italy).Enzyme preparation
StLASPO was overexpressed in E. coli cells and was purified to >95% purity by the procedure described by (ref. 4). The purified StLASPO was stored in 20 mM Tris–HCl buffer at pH 7.5 and 10% (v/v) glycerol; its specific activity was ∼0.43 U mg−1 of protein on L-aspartate as substrate.Functionalization of NPs with APTES
Fe3O4NPs were functionalized according to the protocol of (ref. 16). Briefly, 5 mL of 2% (v/v) APTES in milliQ water were added to a suspension of Fe3O4NPs (150 mg) and the reaction was maintained under mechanical stirring for 5 h at 50 °C. Amino-functionalized nanoparticles (NP-APTES) were then separated from unbound APTES by a commercial parallelepiped neodymium magnet (Webcraft GmbH, Uster, Switzerland; Ni–Cu–Ni plated; magnetization: N45; size: 30 × 30 × 15 mm), washed several times with water and anhydrified with ethanol overnight. NPs were suspended in water, isolated and dried at 50 °C overnight.Synthesis of NP-LASPO
16 mg of NP-APTES were sonicated in 5 mM sodium pyrophosphate buffer at pH 8.5. Then a solution of EDC (65 mM final concentration) and NHS (13 mM final concentration) were added under sonication in a final volume of 2 mL. Finally, 200 μg of pure StLASPO were added and the reaction was carried out for 2 h at room temperature on a rotating plate tube stirrer. Subsequently, StLASPO conjugated NPs (NP-LASPO) were collected by a magnet and washed twice with 2 mL of 5 mM sodium pyrophosphate buffer, pH 8.5. The resulting conjugated enzyme was stored at 4 °C until use.Characterization
Shape, size, and size distribution of Fe3O4NPs, NP-APTES and NP-LASPO were investigated by transmission electron microscopy (TEM) with JEOL 1010 electron microscope (JEOL, Tokyo, Japan). Samples were dispersed in milliQ water on carbon coated copper grids and dried at room temperature.Activity assay
Activity of StLASPO and NP-LASPO was assayed by measuring the initial rate of production of hydrogen peroxide with a coupled peroxidase/dye assay at different temperatures. The standard assay mixture contained 10 mM L-aspartate in 50 mM sodium pyrophosphate buffer, pH 8.5, 1.5 mM 4-aminoantipyrine, 2 mM phenol, 20 μM FAD, 2.5 U of peroxidase, and 30 μg of StLASPO in a total volume of 1 mL. The colored product generated by peroxidase from H2O2 and 4-aminoantipyrine was detected spectrophotometrically at 505 nm (ε = 6.58 mM−1 cm−1).4 One unit (U) is defined as the amount of enzyme that catalyzes the degradation of 1 μmol of L-aspartate per min.Oxidation of L-aspartate by conjugated StLASPO
The oxidation of L-aspartate was carried out by adding 50 μL of 50 mM amino acid solution (in water, prepared and adjusted to the desired pH with 0.5 M NaOH) and 150 μL of water to the immobilized enzyme (4 mg of support corresponding to ≈ 0.104 U of StLASPO). All reactions were performed at 70 °C, in a thermomixer set to 600 rpm and at different pH values (in the 8–11 range). The dependence of the residual activity after 60 min of incubation on pH was fitted using an equation for a single ionization according to Harris and Colleagues.17To investigate the stability of NP-LASPO on temperature, the same reaction was performed at pH 10.0 in the 25–80 °C range: samples were collected at different time intervals for measurements. For the recycling of the NP-LASPO, at the end of each cycle NPs were collected with a magnet, washed several times and stored in the storage buffer. In all cases, the oxidation of L-aspartate was assayed by HPLC separation (see below).
HPLC analysis
The OPA-NAc reagent was prepared by dissolving 4 mg of ortho-phthalaldehyde (OPA) in 300 μL of methanol, followed by the addition of 250 μL of 0.4 M borate buffer at pH 9.4.18 To this mixture, 15 μL of 1 M N-acetylcysteine (NAc) in 1 M NaOH were added, and then the solution was diluted with 435 μL of distilled water. At fixed intervals, 10 μL aliquots were withdrawn from the reaction mixture and diluted with 40 μL of distilled water: 10 μL of this solution were derivatized with 25 μL of OPA-NAc reagent, diluted with 25 μL of HPLC eluent and analyzed by HPLC chromatography on a symmetry C8 column (100 Å, 5 μm, 3.9 mm × 150 mm; Waters, Milano, Italy). Eluent: 50 mM sodium acetate buffer, pH 5.2/acetonitrile 9/1; isocratic flux ofz 1 mL min−1; absorbance detection at 340 nm.7 The separation allows to easily distinguish and quantify L-aspartate from glycine (used as internal standard) and thus to calculate the bioconversion yield; Rt (glycine): 10.5 min, Rt (L-aspartate) 4.5 min.Results and discussion
Optimization of StLASPO conjugation
StLASPO was conjugated to NP-APTES using EDC/NHS reaction by a two-step process. The scheme of NP activation and enzyme conjugation is illustrated in Fig. 1. With this two steps reaction, different conjugations can occur (Fig. 1): an enzyme molecule could interact with just one NP-APTES (product A), an enzyme molecule could react with amino groups of different NPs (product B) and an enzyme molecule could react with more than one amino groups of the same NP (product C).19,20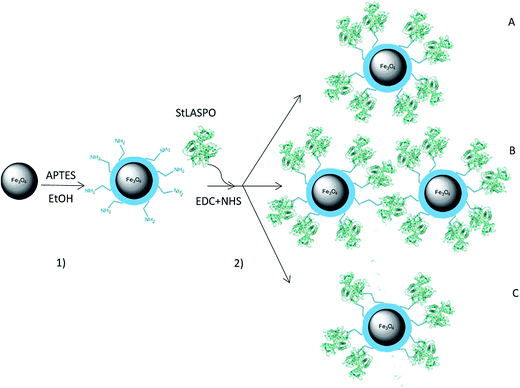 | ||
| Fig. 1 Synthetic route for StLASPO conjugation on Fe3O4NPs (not in scale). The protein structure corresponds to PDB code 2E5V. (1) Functionalization of Fe3O4NPs with APTES; (2) conjugation of StLASPO through covalent linkage via EDC/NHS. The enzyme can interact with just one NP-APTES (product A) or with amino groups of different NPs (product B) or with more than one amino groups of the same NP (product C). | ||
In order to identify the best conditions for the enzyme conjugation, several factors have been taken into consideration: EDC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NHS ratio, NPs: enzyme ratio, the presence of FAD (the enzyme cofactor), substrate or product (Table 1). At each condition, the immobilization yield and the enzyme activity bound to NP-APTES were determined.
NHS ratio, NPs: enzyme ratio, the presence of FAD (the enzyme cofactor), substrate or product (Table 1). At each condition, the immobilization yield and the enzyme activity bound to NP-APTES were determined.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NHS molar ratios are reported in the second columna,b,c
NHS molar ratios are reported in the second columna,b,c
| NP-APTES (mg) | EDC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NHS ratio NHS ratio |
Other components | Vf (mL) | Yield (%) | Relative activity (%) | Specific activity (U mg−1 enzyme) |
|---|---|---|---|---|---|---|
| a Reaction conditions: during StLASPO conjugation, 100 μM FAD 1 mM L-aspartate and/or 200 μM succinate were added.b Yield has been calculated from units present in the supernatant of the immobilization reaction relatively to the total units.c Relative activity is reported as the ratio between the activity assayed for the NP-LASPO and the activity of the free enzyme. | ||||||
| 4 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
— | 1 | 75 | 4 | 0.32 |
| 4 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
FAD | 1 | 100 | 14 | 0.43 |
| 4 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
L-Aspartate | 1 | 100 | 15 | 0.43 |
| 4 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
Succinate | 1 | 77 | 5 | 0.33 |
| 4 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
L-Aspartate + FAD | 1 | 86 | 5 | 0.37 |
| 4 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
Succinate + FAD | 1 | 77 | 7 | 0.33 |
| 8 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
— | 2 | 84 | 4 | 0.36 |
| 8 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
FAD | 2 | 100 | 15 | 0.43 |
| 8 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
L-Aspartate | 2 | 100 | 12 | 0.43 |
| 8 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
Succinate | 2 | 92 | 5 | 0.40 |
| 8 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
L-Aspartate + FAD | 2 | 85 | 6 | 0.37 |
| 8 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
Succinate + FAD | 2 | 88 | 7 | 0.38 |
| 8 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 2 | 91 | 8 | 0.39 |
| 8 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
— | 2 | 84 | 4 | 0.36 |
| 8 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 2 | 85 | 8 | 0.37 |
| 8 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
— | 2 | 88 | 9 | 0.38 |
| 8 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
— | 2 | 75 | 10 | 0.32 |
| 8 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
— | 2 | 74 | 10 | 0.32 |
| 8 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 2 | 67 | 13 | 0.29 |
| 8 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
— | 2 | 79 | 8 | 0.34 |
| 8 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 2 | 86 | 11 | 0.37 |
| 8 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
— | 2 | 80 | 8 | 0.34 |
| 4 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 2 | 39 | 6 | 0.17 |
| 8 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 2 | 95 | 11 | 0.41 |
| 16 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 2 | 100 | 15 | 0.43 |
| 24 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 2 | 100 | 12 | 0.43 |
| 4 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
FAD | 2 | 37 | 8 | 0.16 |
| 8 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
FAD | 2 | 52 | 10 | 0.22 |
| 16 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
FAD | 2 | 94 | 10 | 0.41 |
| 24 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
FAD | 2 | 95 | 12 | 0.41 |
The concentrations of EDC and NHS have to be carefully chosen to yield complete activation of the binding sites and, at the same time, to prevent formation of unwanted surface by-products,21 such as the N acetyl-substituted, derivative of the unstable intermediate of the EDC. Indeed, during the reaction in aqueous solution, O-acetyl urea is formed and, if it fails to react with an amine, undergoes to hydrolysis.22 Furthermore, an excess of EDC may promote unwanted polymerization due to the abundance of both amines and carboxylates on protein molecules leading to a protein-to-protein cross-linking.20 There is also the risk that the NHS esters formed on the protein molecule may then couple to other protein molecules to give poorly defined polymers.23 As a general rule, the amount of StLASPO activity bound to the NPs increases at higher EDC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NHS ratios (reaching the maximum at a 5
NHS ratios (reaching the maximum at a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 value) and using increasing NP-APTES amounts.
1 value) and using increasing NP-APTES amounts.
The presence of FAD significantly increases both the units and the amount of the enzyme bound to NPs at low (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) EDC
2) EDC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NHS ratio. The positive effect of the flavin cofactor is less evident at higher EDC
NHS ratio. The positive effect of the flavin cofactor is less evident at higher EDC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NHS ratios (e.g., its presence resulted in a 1.3 fold increase in immobilized StLASPO activity at 5
NHS ratios (e.g., its presence resulted in a 1.3 fold increase in immobilized StLASPO activity at 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 EDC
1 EDC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NHS ratio and using 4 mg of NP-APTES).
NHS ratio and using 4 mg of NP-APTES).
Similarly, the substrate L-aspartate also positively affects StLASPO immobilization in an active form, while the product analogue succinate does not. Notably, the use of the FAD and L-aspartate together results in an enzyme conformational change that does not favor its immobilization.
Under the best experimental conditions – i.e. 16 mg of NP-APTES in 5 mM sodium pyrophosphate buffer, pH 8.5, 65 mM EDC, 13 mM NHS, 200 μg of enzyme, final volume 2 mL – the amount of enzyme bound to NPs was approximately 100%, with an enzymatic activity of 0.026 U mg−1 of NPs and a relative activity of 15% (Table 1). Relative activity values strongly depend on the enzyme and the conditions used. Indeed, relative activities ranging from less than 10% to more than 80% in relation to the diameter of the NP, its functionalization and the immobilized enzyme are reported in the literature.24–26 Furthermore, the reduction of relative activity might be due also to the chemistry of EDC/NHS conjugation that determines non-specific bindings between NP-APTES and the enzyme molecules. Actually, when the covalent bonds are formed close to the active site, an activity loss due to conformational changes can occur.27,28 Under the best conjugation conditions, the specific activity of the immobilized enzyme for the L-aspartate is 0.43 U mg of enzyme (Table 1), comparable to the specific activity of the free enzyme. As for the relative activity, specific activity depends on the enzyme and the conditions of the immobilization process. Therefore, the conjugation can affect also the specific activity.29–31
NP-LASPO properties
TEM images indicate that Fe3O4NPs and NP-APTES show a spherical shape, with an average diameter of 14.6 ± 3.8 nm and 17.2 ± 3.2 nm respectively, and with a good dispersion (Fig. 2A and B). On the other end, NP-LASPO show the tendency to aggregate and a more irregular shape, with an average diameter of 38.0 ± 9.8 nm (Fig. 2C). This slight tendency to aggregation can be explained by the possibility that several carboxylic groups located on the enzyme surface may react with the amine groups of different NP-APTES causing a cross-linking leading to aggregation.32 However, this phenomenon is not massive since no NPs precipitation was apparent.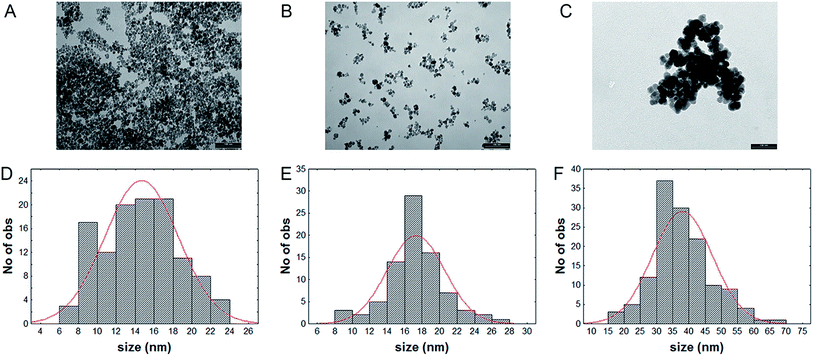 | ||
| Fig. 2 Transmission electron microscopy (TEM) and size distribution of Fe3O4NPs (A and D), NP-APTES (B and E), NP-LASPO (C and F). Bars in panel A–C indicate 100 nm. | ||
The immobilized NP-LASPO maintains the absolute stereo-selectivity of the free enzyme, i.e., the D-isomer of L-aspartate is not oxidized. The addition of exogenous, free FAD positively affect the activity of NP-LASPO: a 2-fold increase in the activity of the enzyme immobilized on the NPs was obtained in the presence of a large excess of exogenous cofactor in the HRP-coupled assay mixture, this indicating that half of StLASPO is immobilized in the apoprotein, inactive form.
To investigate the storage stability, the NP-LASPO system was stored in 5 mM sodium pyrophosphate buffer (pH 8.5) at 25 °C: 70% of the initial activity was maintained after 35 days.
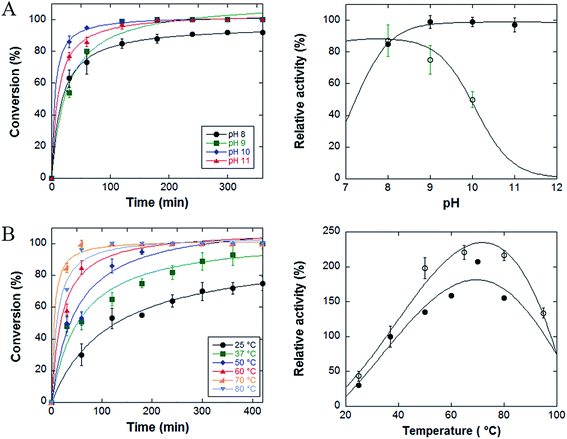
The effect of pH on the catalytic activity of NP-LASPO was investigated in the 8.0 to 11.0 range, by following the disappearance of the L-aspartate peak by HPLC separation (Fig. 3).
Fig. 4, panel A left, displays the activity curves of the enzyme at different pH values. At pH 9, 10 and 11 the complete oxidation of L-aspartate is observed in 3 hours, while at pH 8 the reaction stops to approximately 90% of conversion. When compared to the free enzyme form, the immobilized enzyme shows a full stability at pH values 9–11 after 60 min of incubation, while for the free StLASPO the stability strongly decreases at pH > 8 (Fig. 4A, right). Similarly, earlier studies demonstrated that immobilized enzymes are frequently more stable than free enzymes in an alkaline environment.24,29
The effect of temperature on the catalytic activity of conjugated StLASPO was investigated in the 25–80 °C range (Fig. 4B). The NP-enzyme system shows a good activity in the range of 60–80 °C: the fastest L-aspartate oxidative deamination is apparent at 70 °C. Only at ≤37 °C a partial conversion of the 50 mM L-aspartate solution was obtained after 420 min. Fig. 4B right shows that the thermostability of NP-LASPO after 30 min of incubation parallel the behavior observed for the free enzyme.
The catalytic parameters of StLASPO immobilized on the Fe3O4NPs were determined by a HRP-coupled spectrophotometric assay at pH 10.0 at 25 °C. The apparent Vmax at air oxygen-saturation for the NP-LASPO system is 0.11 μmol per min per mg of protein and Km for L-aspartate is 4.3 mM. The corresponding values for the free enzyme form are 0.98 μmol per min per mg enzyme and 1.3 mM at pH 8.0 and 37 °C.4 The higher Km of NP-LASPO suggests a lower affinity for L-aspartate by the immobilized enzyme so a higher substrate concentration is needed to achieve a given enzyme activity. Diffusional limitations and steric effects may contribute to the increased apparent Km value due to a decrease in the accessibility of substrate to the enzyme active site. Similar effects are frequently reported for enzyme immobilized with EDC/NHS protocol, e.g., Tee and colleagues33 suggest that the high loading of the enzyme molecules using a zero-length crosslinker might restrict the access of the substrate to the active sites.
Bioconversion by NP-LASPO
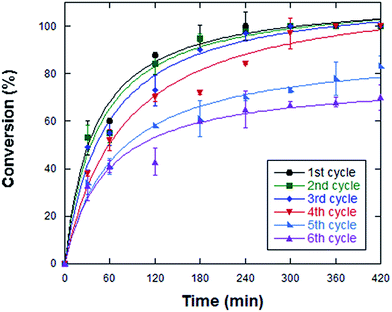
The enzyme immobilization on magnetic NPs allows a simple recovery of the biocatalyst and its reuse, making this system interesting for industrial applications. To determine the number of reaction cycles that immobilized StLASPO can carry out, the same batch of enzyme was reused for subsequent cycles. Full oxidative deamination of 50 μL of 50 mM L-aspartate was obtained in 240 min using optimal operational conditions, i.e., at pH 10, 50 °C and using 0.2 U of the NP-enzyme system. Under these conditions, a full conversion was observed for three sequential cycles, while a longer reaction time was required for the following cycles (Fig. 5): after 420 min of reaction, ≤70% of the substrate was converted in the fifth and sixth cycle. Notably, under optimized conditions, the Relizyme-StLASPO fully converted 25 mM L-aspartate for three sequential cycles of 4 hours onlyd,7 pointing to partial inactivation of the immobilized flavoenzyme. Taking into consideration the four cycles that resulted in full L-aspartate oxidation, 50 μmoles of substrate were converted per unit of NP-LASPO. As compared to the free enzyme form, which fully converted 30 mL of 25 mM L-aspartate in 24 h by using 9 U of StLASPO4 corresponding to 3.5 μmoles per h per unit, the NP-LASPO converted 3.1 μmole per h per unit at pH 10.0 and 50 °C. Indeed, the Relizyme-StLASPO – using 10 mL of 100 mM D,L-Asp, at pH 10.0 and 70 °C – converted 1 μmoles per h per unit (or ∼4.4 μmoles per h per unit using 0.5 mL of 142 mM D,L-Asp).7Conclusions
In this work, the stereo-selective flavoenzyme L-aspartate oxidase from the thermophilic archaea Sulfolobus tokodaii has been covalently immobilized on Fe3O4NPs using EDC and NHS as cross-linking agents. The optimal conditions for the enzyme immobilization were identified by investigating the interaction effect of different variables.The NP-LASPO showed a better stability than the free enzyme at pH ≥ 9.0 and was reused five times reaching full L-aspartate conversion, similarly to that previously obtained for the Relizyme-StLASPO preparation which employed a 2.5-fold higher amount of enzyme and longer times.7 L-Aspartate conversion by NP-LASPO yielded a productivity similar to that obtained using the free enzyme (that cannot be recycled) or the enzyme bound to the amino support Relizyme™ HA403/S R (3.1 vs. 1–4 μmol per h per unit of enzyme at 50 mM L-aspartate concentration). These results indicate that the prepared NPs are favorable for immobilization of LASPO and the immobilized enzyme has promising industrial applications.
Acknowledgements
Ilaria Armenia and Riccardo Balzaretti are PhD students of the “Biotechnology, Biosciences and Surgical Technology” course at Università degli studi dell'Insubria. Chiara Allegretti is a PhD student of the Research Doctorate Program in Chemical Engineering and Industrial Chemistry at Politecnico di Milano. We thank the support from Consorzio Interuniversitario per le Biotecnologie.Notes and references
- G. Tedeschi, A. Negri, M. Mortarino, F. Ceciliani, T. Simonic, L. Faotto and S. Ronchi, Eur. J. Biochem., 1996, 239, 427–433 CAS.
- P. Macheroux, O. Seth, C. Bollschweiler, M. Schwarz, M. Kurfürst, L. C. Au and S. Ghisla, Eur. J. Biochem., 2001, 268, 1679–1686 CrossRef CAS PubMed.
- H. Sakuraba, K. Yoneda, I. Asai, H. Tsuge, N. Katunuma and T. Ohshima, Biochim. Biophys. Acta, Proteins Proteomics, 2008, 1784, 563–571 CrossRef CAS PubMed.
- D. Bifulco, L. Pollegioni, D. Tessaro, S. Servi and G. Molla, Appl. Microbiol. Biotechnol., 2013, 97, 7285–7295 CrossRef CAS PubMed.
- L. Pollegioni, P. Motta and G. Molla, Appl. Microbiol. Biotechnol., 2013, 97, 9323–9341 CrossRef CAS PubMed.
- L. Pollegioni and G. Molla, Trends Biotechnol., 2011, 29, 276–283 CrossRef CAS PubMed.
- P. D'Arrigo, C. Allegretti, A. Fiorati, L. Piubelli, E. Rosini, D. Tessaro, M. Valentino and L. Pollegioni, Catal. Sci. Technol., 2015, 5, 1106–1114 Search PubMed.
- A. H. Lu, E. L. Salabas and F. Schüth, Angew. Chem., Int. Ed., 2007, 46, 1222–1244 CrossRef CAS PubMed.
- A. Bava, R. Gornati, F. Cappellini, L. Caldinelli, L. Pollegioni and G. Bernardini, Nanomedicine, 2013, 8, 1797–1806 CrossRef CAS PubMed.
- R. Balzaretti, F. Meder, M. P. Monopoli, L. Boselli, I. Armenia, L. Pollegioni, G. Bernardini and R. Gornati, RSC Adv., 2017, 7, 1439–1442 RSC.
- F. Cappellini, C. Recordati, M. De Maglie, L. Pollegioni, F. Rossi, M. Daturi, R. Gornati and G. Bernardini, Future Sci. OA, 2015, 1, 4 CrossRef PubMed.
- A. G. Cattaneo, R. Gornati, E. Sabbioni, M. Chiriva-Internati, E. Cobos, M. R. Jenkins and G. Bernardini, J. Appl. Toxicol., 2010, 30, 730–744 CrossRef CAS PubMed.
- R. Gornati, E. Pedretti, F. Rossi, F. Cappellini, M. Zanella, I. Olivato, E. Sabbioni and G. Bernardini, J. Appl. Toxicol., 2016, 36, 385–393 CrossRef CAS PubMed.
- C. G. C. M. Netto, H. E. Toma and L. H. Andrade, J. Mol. Catal. B: Enzym., 2013, 85–86, 71–92 CrossRef CAS.
- H. Vaghari, H. Jafarizadeh-Malmiri, M. Mohammadlou, A. Berenjian, N. Anarjan, N. Jafari and S. Nasiri, Biotechnol. Lett., 2016, 38, 223–233 CrossRef CAS PubMed.
- A. Bava, F. Cappellini, E. Pedretti, F. Rossi, E. Caruso, E. Vismara, M. Chiriva-Internati, G. Bernardini and R. Gornati, BioMed Res. Int., 2013, 2013, 314091 Search PubMed.
- C. M. Harris, L. Pollegioni and S. Ghisla, Eur. J. Biochem., 2001, 268, 5504–5520 CrossRef CAS PubMed.
- Y. Mutaguchi and T. Ohshima, Biochemistry, 2014, 4, 2–5 Search PubMed.
- H. Jędrzejewska and R. Ostaszewski, J. Mol. Catal. B: Enzym., 2013, 90, 12–16 CrossRef.
- G. T. Hermanson, Bioconjugate techniques, Academic Press, 3rd edn, 2013 Search PubMed.
- S. Sam, L. Touahir, J. Salvador Andresa, P. Allongue, J. N. Chazalviel, A. C. Gouget-Laemmel, C. H. De Villeneuve, A. Moraillon, F. Ozanam, N. Gabouze and S. Djebbar, Langmuir, 2010, 26, 809–814 CrossRef CAS PubMed.
- S. K. Vashist, Diagnostics, 2012, 2, 23–33 CrossRef CAS PubMed.
- L. S. Wong, F. Khan and J. Micklefield, Chem. Rev., 2009, 109, 4025–4053 CrossRef CAS PubMed.
- M. Jain, A. Mariya Sebatini, P. Radha, S. Kiruthika, C. Muthukumaran and K. Tamilarasan, J. Mol. Catal. B: Enzym., 2016, 128, 1–9 CrossRef CAS.
- F. Kazenwadel, H. Wagner, B. E. Rapp and M. Franzreb, Anal. Methods, 2015, 7, 10291–10298 RSC.
- K. Turcheniuk, A. V. Tarasevych, V. P. Kukhar, R. Boukherroub and S. Szunerits, Nanoscale, 2013, 5, 10729–10752 RSC.
- J. Xu, J. Sun, Y. Wang, J. Sheng, F. Wang and M. Sun, Molecules, 2014, 19, 11465–11486 CrossRef PubMed.
- H. J. Park, J. T. McConnell, S. Boddohi, M. J. Kipper and P. A. Johnson, Colloids Surf., B, 2011, 83, 198–203 CrossRef CAS PubMed.
- C. Yen, Y. Chuang, C. Ko, L. O. Chen, S. Chen, C. Lin, Y. Chou and J. Shaw, Molecules, 2016, 21, 972 CrossRef PubMed.
- B. Sahoo, S. K. Sahu and P. Pramanik, J. Mol. Catal. B: Enzym., 2011, 69, 95–102 CrossRef CAS.
- S. L. Hosseinipour, M. S. Khiabani, H. Hamishehkar and R. Salehi, J. Nanopart. Res., 2015, 17(9), 382 CrossRef.
- R. A. Sperling and W. J. Parak, Philos. Trans. R. Soc., A, 2010, 368, 1333–1383 CrossRef CAS PubMed.
- B. L. Tee and G. Kaletunç, Biotechnol. Prog., 2009, 25, 436–445 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |

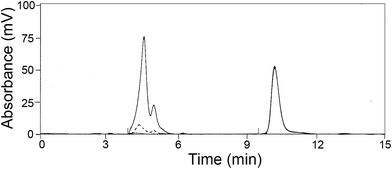
![[thick line, graph caption]](https://www.rsc.org/images/entities/char_e117.gif) ) before NP-LASPO addition; (
) before NP-LASPO addition; (![[dash dot, graph caption]](https://www.rsc.org/images/entities/char_e090.gif) ) 30 and (⋯) 60 min after NP-LASPO addition. Glycine, Rt 10.5 min, was used as internal standard.
) 30 and (⋯) 60 min after NP-LASPO addition. Glycine, Rt 10.5 min, was used as internal standard.