Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Metal–organic framework incorporated monolithic capillary for selective enrichment of phosphopeptides
Daojin Li *a and
Zijun Bieb
*a and
Zijun Bieb
aCollege of Chemistry and Chemical Engineering, Henan Key Laboratory of Fuction-Oriented Porous Materials, Luoyang Normal University, Luoyang 471022, P. R. China. E-mail: lidaojin7910@126.com
bDepartment of Chemistry, Bengbu Medical College, 233030, China
First published on 10th March 2017
Abstract
Protein phosphorylation is a major post-translational modification, which plays a central role in the cellular signaling of numerous biological processes. Mass spectrometry (MS) has been an essential tool for the analysis of protein phosphorylation, for which it is a key step to selectively enrich phosphopeptides from complex biological samples. In this study, a metal–organic framework (MOF) incorporated monolithic capillary has been successfully prepared as an effective sorbent for the selective enrichment of phosphopeptides and has been off-line coupled with matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) for efficient analysis of phosphopeptides. Using β-casein as a representative phosphoprotein, efficient phosphorylation analysis by this off-line platform was verified. Phosphorylation analysis of a nonfat milk sample was also well demonstrated. Due to the combination of MOFs with large surface areas and highly ordered pores and the monolithic column, the MOF incorporated monolithic capillary exhibited several significant advantages, such as excellent selectivity toward phosphopeptides, superb tolerance to interference and a simple operation procedure, making the MOF-based monolithic capillary an ideal sorbent for the selective enrichment of phosphopeptides. Because of these highly desirable properties, the MOF-based monolithic capillary could be a useful tool for protein phosphorylation analysis.
Introduction
Protein phosphorylation is one of the most common and most important post-translational modifications (PTMs) in mammalian species, which plays a vital and important role in many biological processes, such as cell growth, migration, division, and intercellular communication.1,2 Mass spectrometry (MS) has been an indispensable technique for proteome-wide phosphorylation analysis due to its high sensitivity, high-throughput, feasibility for identification of phosphorylation sites and quantification of changes in phosphorylation states. However, it often fails to identify phosphopeptides in complex peptide mixtures generated by phosphoprotein digestion because it often suffers from low abundance of phosphopeptides, poor ionization efficiency, and signal suppression by high-abundance non-phosphopeptides. Therefore, selective enrichment is usually a critical step for the analysis of phosphopeptides from a complex peptide mixture.3,4To date, a number of approaches, including immunoprecipitation,5–7 chemical coupling,8,9 ion exchange chromatography,10–12 metal oxide affinity chromatography (MOAC),13–17 immobilized metal ion affinity chromatography (IMAC)18–26 and molecularly-imprinted polymers (MIPs),27–29 have been developed for the selective enrichment of phosphoproteins/phosphopeptides. Among them, IMAC is one of the most commonly used techniques for selective isolation of phosphorylated peptides from predominantly non-phosphorylated peptides due to fast response, low cost and easy handling. The principle of IMAC is that metal ions (e.g. Fe3+ and Ti4+) immobilized on supporting materials form coordination with phosphate ion at certain pH environment while the coordination is disrupted when an elution solution of ammonia solution is applied. The selectivity of IMAC toward phosphopeptides is based on the high affinity of the phosphate groups with the metal ions bound on sorbents through iminodiacetic acid (IDA), nitrilotriacetic acid (NTA), polydopamine or phosphate groups.18,22–25,30–43
In recent years, porous IMAC materials also have been widely applied in phosphopeptides researches due to its merits of large surface area, unique pore volume, and regular porous structure.37,40,44–46 Metal–organic frameworks (MOFs), as a type of ordered porous IMAC materials, have been applied in storage,47 separation,48–51 sensing,52 drug delivery53 and catalysis54,55 because of their super high porosity, well-defined porous structure and super large surface areas. Recently, it has been reported that MOFs could function as sorbents for the separation and enrichment of phosphopeptides owing to the presence of plenty of Lewis acid sites.56,57 However, MOFs in its native format are not ideal materials since they usually require centrifugations at high speed during sample separation, which not only makes the enrichment process laborious and inconvenient, but also causes the undesirable loss of low-abundance phosphopeptides. MOFs-containing magnetic materials have been a solution to this issue. Magnetic microspheres modified with zirconium-based MOFs58 and magnetic nanoparticles with iron-based MOFs59 have been developed for phosphopeptides enrichment. However, MOFs in other formats for phosphopeptides enrichment have not been well explored.
Monolithic columns,60–63 which are defined as “continuous stationary phases that form as a homogeneous column in a single piece”, have been efficient media for chromatographic separation and sample pretreatment. Monolithic capillary can provide several significant advantages, such as ease of preparation, low cost, low back pressure, fast convective mass transfer, lower sample consumption, convenient online coupling to MS and easy post-modification. Particularly, monolithic capillary has been demonstrated to be an efficient and convenient platform for coupling with MS for proteomic analysis.12,64 Therefore, MOFs in the format of monolithic column would be efficient media for separation and sample pretreatment. Recently, several MOFs-based monolithic columns have been reported.65–67 However, they only focused on the separation of small molecules. To the best of our knowledge, there has been no report on phosphopeptides enrichment using the MOFs-based monolithic columns. Therefore, it is very necessary to develop MOFs-based monolithic columns for enrichment of phosphopeptides.
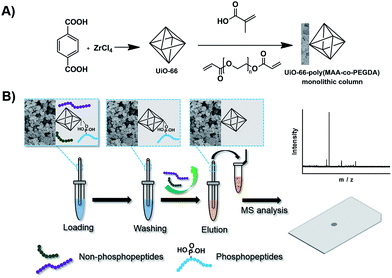
Herein, we synthesized a new MOF incorporated monolithic capillary and established a hyphenated approach of MOFs-monolithic capillary-based selective enrichment with matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) MS for highly efficient phosphorylation analysis. The synthetic procedure of the MOFs-based monolithic capillary is illustrated in Fig. 1A. It included two simple steps: (1) synthesis of MOFs (UiO-66); (2) preparation of MOFs-based monolithic capillary, in which UiO-66 was incorporated into a monolithic capillary. The porous structure of MOFs and the large amount of coordinated Zr(IV) endow the material with numerous functional spots for large enrichment capacity and high selectivity for phosphopeptides. In addition, the material structure format of monolithic capillary can provide nanoliter-scaled sample processing capability, online operation possibility and good reproducibility. Thus, the MOFs-based monolithic capillary could show its great potential in enrichment of phosphopeptides. By coupling this monolithic capillary with MALDI-TOF MS, a novel platform was established for efficient protein phosphorylation analysis. As depicted in Fig. 1B, the procedure of the approach included four steps: sample loading, washing, eluting, and MS analysis. The MOFs-based monolithic capillary exhibited excellent selectivity, superb tolerance to interference and simple operation procedure. Highly efficient phosphorylation analysis was demonstrated by the analysis of tryptic digestion of standard protein and nonfat milk.
Experimental
Materials
Trypsin, β-casein, bovine serum albumin (BSA), acetonitrile (ACN) and 2,5-dihydroxybenzoic acid (DHB) were purchased from Sigma-Aldrich (St. Louis, MO, USA). γ-Methacryloxypropyltrimethoxysilane (γ-MAPS), sodium hydroxide, poly(ethylene glycol)diacrylate (PEGDA, average molecular weight 256), poly (ethylene glycol) (PEG, Mn = 200), trifluoroacetic acid (TFA), ammonium hydroxide (25–28%), zirconium chloride (ZrCl4), 2-amino-1,4-benzenedicarboxylic acid, N,N-dimethylformamide (DMF) and HCl were purchased from J&K Scientific (Shanghai, China). Methanol, ethanol, 2,2-azobisisobutyronitrile (AIBN, recrystallized in methanol before use) were purchased from Nanjing Chemical Reagent (Nanjing, China). Fused-silica capillaries with 150 μm I.D. and 375 μm O.D. were purchased from Yongnian Optic Fiber Plant (Hebei, China). Human serum was purchased from Shuangliu Zhenglong Chemical and Biological Research Laboratory (Sichuan, China). Ultrapure water, purified with a Milli-Q Advantage A10 (Millipore, Milford, MA, USA), was used to prepare all solutions. Other chemical reagents were of analytical grade.Characterization
The chromatographic separations were performed on a TriSep 2100 system (Unimicro Technologies, Pleasanton, CA, USA). Scanning-electron microscopic (SEM) characterization was performed on a Hitachi FE-SEM S-4800 (Tokyo, Japan). Transmission electron microscopy (TEM) characterization was performed on a JEM-2100 system (JEOL, Tokyo, Japan). Nitrogen adsorption–desorption measurements were conducted at 77 K on an ASAP2020 instrument (Micromerities, Norcross, GA, USA). The surface area and the micropore volume were calculated by data the Brunauer–Emmett–Teller (BET) method using adsorption. The X-ray diffraction (XRD) patterns of samples were acquired on an D8-ADVANCEX diffractometer (BRUKER, German) with Cu Kα radiation in the 2θ range of 5–50°. MALDI-TOF MS analyses were carried out on a MALDI TOF/TOF Analyzer (Shimadzu/Kratos, Manches-ter, UK) with a pulsed nitrogen laser operated at 337 nm. The laser energy was adjusted to slightly above the threshold to obtain good resolution and signal-to-noise ratio (S/N). All mass spectra reported were obtained in the positive ion mode. The instrument was operated in reflectron mode for peptide detection. A typical spectrum was obtained by averaging 3000 laser shots from 30 positions within the sample well. The accelerating voltage was 20 kV. The whole process was controlled by the 4000 series Explorer Software V3.7.0. Data were processed using Data Explorer Software version 3.7 (Applied Biosystems, Framingham, MA). The matrixes for MALDI-TOF MS were 20 mg mL−1 DHB dissolved in 50% ACN containing 1% (v/v) H3PO4. The lyophilized powder of analytes were first dissolved in 2 μL of matrix solution and dropped onto the MALDI plate for MALDI-TOF MS analysis.Synthesis of UiO-66
UiO-66 was prepared according to the procedures reported before68–70 with minor modification. Briefly, ZrCl4 (0.080 g, 0.343 mmol), terephthalic acid (0.057 g, 0.343 mmol) and 0.6 mL acetic acid were mixed with 20 mL DMF in a Teflon-lined bomb. The bomb was sealed and placed in an oven at 120 °C for 24 h. After cooling down to room temperature, the white precipitates were obtained by centrifugation. The obtained powders were isolated by centrifugation and washed first with DMF (3 × 30 mL) and then with ethanol (3 × 30 mL). Finally, the product was dried under vacuum at 100 °C overnight.Preparation of UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column
The preparation procedure is also illustrated in Fig. 1A. Prior to the polymerization, the capillary was successively washed with NaOH (0.1 M), water, HCl (0.1 M), water and methanol respectively and then dried by passage of nitrogen gas. A solution of γ-MAPS (50% v/v in methanol) was injected into the capillary, and then the capillary was sealed with rubber at both ends and submerged in a water bath at 50 °C for 12 h. Finally, the capillary was washed with methanol to flush out the residual reagent and dried by passage of nitrogen gas overnight. The preparation of UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column was carried out through free radical reaction. Briefly, a mixture containing UiO-66 (25 mg), MAA (40 mg), PEGDA (51 mg), PEG 200 (320 mg) and AIBN (1 mg) was vortexed for 5 min and sonicated for 20 min to obtain a homogeneous solution. After that, the mixture was manually introduced into the pretreated capillary of an appropriate length by a syringe. After both ends of the capillary were sealed with two pieces of rubbers, the polymerization reaction was carried out at 60 °C for 12 h in the water bath. The resulting UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column was then flushed with 200 μL water and 100 μL methanol to remove the PEG and other residuals, respectively. For comparison, the blank monolithic column was prepared in parallel by adding PEG 200 instead of UiO-66 dispersion into polymerization mixture.The preparation of large-sized monolith for characterization
A UiO-66 incorporated poly(MAA-co-PEGDA) monolith was prepared in 1.5 mL centrifuge under otherwise identical conditions and cut into thin pieces. Then the pieces were rinsed with methanol using a Soxhlet extractor for 24 h and dried in vacuum at 60 °C for 12 h. The prepared UiO-66 incorporated poly(MAA-co-PEGDA) monolith was used for nitrogen adsorption–desorption measurements and XRD.Tryptic digestion of standard protein
The procedure for the tryptic digest of proteins (β-casein and BSA) used in the experiment was prepared as follows: 1 mg of protein was added to a centrifuge tube and dissolved in 1 mL of 100 mM NH4HCO3 buffer solution (pH 8.2). After being heated in a water bath of 100 °C for 10 min, the tube was cooled to room temperature. Then the protein solution was digested with trypsin for 24 h at 37 °C (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, w/w). Finally, the resulting mixture was heated again to 100 °C and kept for 10 min. The prepared protein digest solution was store at −20 °C.
1, w/w). Finally, the resulting mixture was heated again to 100 °C and kept for 10 min. The prepared protein digest solution was store at −20 °C.
Selective enrichment of phosphopeptides
To prove the selectivity of the prepared UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column, the tryptic digestion of β-casein and two mixtures of tryptic digestion of β-casein and BSA (w/w, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 1
100 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) were used, respectively. A typical experimental procedure is shown in Fig. 1B. Monolithic column (150 μm I.D.) was first equilibrated with 100 μL loading buffer (50% ACN/0.1% TFA) under the nitrogen pressure. Then the diluted protein digest solution of a certain concentration was continuously pumped through the column. Thereafter, the column was washed with 50 μL loading buffer in order to flush out the unbounded peptides. Then, the bound phosphopeptides were eluted with 5% ammonium hydroxide and collected in several tubes (5 μL each). Finally, the eluted phosphopeptides were vacuum freeze-dried and redissolved in 3 μL of DHB matrix solution before MALDI-TOF MS analysis. UiO-66 was first ultrasonically dispread into the loading buffer (50% ACN/0.1% TFA), then 10 μL peptide solution was added to 100 μL of the above-mentioned solution. The tubes were shaken on a rotator for 20 min at room temperature. Other procedures were the same as those described above.
1000) were used, respectively. A typical experimental procedure is shown in Fig. 1B. Monolithic column (150 μm I.D.) was first equilibrated with 100 μL loading buffer (50% ACN/0.1% TFA) under the nitrogen pressure. Then the diluted protein digest solution of a certain concentration was continuously pumped through the column. Thereafter, the column was washed with 50 μL loading buffer in order to flush out the unbounded peptides. Then, the bound phosphopeptides were eluted with 5% ammonium hydroxide and collected in several tubes (5 μL each). Finally, the eluted phosphopeptides were vacuum freeze-dried and redissolved in 3 μL of DHB matrix solution before MALDI-TOF MS analysis. UiO-66 was first ultrasonically dispread into the loading buffer (50% ACN/0.1% TFA), then 10 μL peptide solution was added to 100 μL of the above-mentioned solution. The tubes were shaken on a rotator for 20 min at room temperature. Other procedures were the same as those described above.
Tryptic digestion of proteins from nonfat milk
A volume of 30 mL of nonfat milk (Mengniu, China) was added into NH4HCO3 (1 mL, 25 mM), and this solution was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min. The supernatant was collected and then denatured at 100 °C for 10 min. The supernatant was digested with trypsin (40 mg) at 37 °C for 24 h. The prepared digest solution was stored at −20 °C.
000 rpm for 20 min. The supernatant was collected and then denatured at 100 °C for 10 min. The supernatant was digested with trypsin (40 mg) at 37 °C for 24 h. The prepared digest solution was stored at −20 °C.
Selective enrichment of digested phosphopeptides from nonfat milk
The monolithic column (150 μm I.D.) was first equilibrated with 100 μL loading buffer (50% ACN/0.1% TFA) under the nitrogen pressure. Then the diluted nonfat milk digest solution was continuously pumped through the column. Thereafter, the column was washed with loading buffer in order to flush out the unbounded peptides. Then, the bound phosphopeptides were eluted with 5% ammonium hydroxide and collected in several tubes (5 μL each). Finally, the eluted phosphopeptides were vacuum freeze-dried and redissolved in 4 μL of DHB matrix solution before MALDI-TOF MS analysis.Selective enrichment of phosphopeptides from human serum
20 mL of human serum was diluted with pure water (120 mL) and denatured for 5 min in boiled water, then the diluted human serum (5 mL) was continuously pumped through UiO-66 incorporated poly(MAA-co-PEGDA). After the column was washed with buffer 1 (ACN–H2O–TFA, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 34
34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, v/v/v, 200 mmol L−1 NaCl) and buffer 2 (ACN–H2O–TFA, 30
6, v/v/v, 200 mmol L−1 NaCl) and buffer 2 (ACN–H2O–TFA, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 69.9
69.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1, v/v/v), the bound phosphopeptides were eluted with 5% ammonium hydroxide and collected in several tubes (5 μL each) and the eluate was analyzed by MALDI-TOF MS.
0.1, v/v/v), the bound phosphopeptides were eluted with 5% ammonium hydroxide and collected in several tubes (5 μL each) and the eluate was analyzed by MALDI-TOF MS.
Results and discussion
Characterization of UiO-66
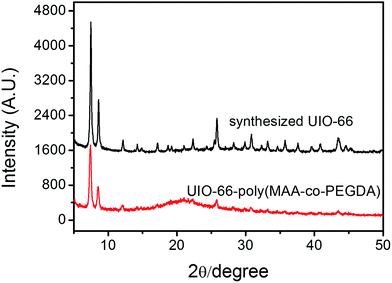
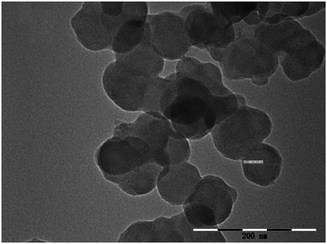
The XRD data of the synthesized UiO-66 is shown in Fig. 2. It can be seen from the XRD pattern of UiO-66 that all diffraction peaks match well with the reported data in the ref. 70, which suggest characteristic close packed structure. This result indicates that the UiO-66 has been successfully prepared. The TEM images shown in Fig. 3 reveal that the UiO-66 crystals had satisfactory dispersibility and relatively homogeneous size distribution. Based on the TEM images, the average diameter of UiO-66 was estimated to be about 60 nm. Clearly, the particle size of UiO-66 is relative small, being particularly advantageous for the preparation of MOFs-based monolithic column. The BET surface areas of UiO-66 was calculated to be 845 m2 g−1. The pore size of the obtained UiO-66 was determined to be 1.2 nm, corresponding to the tetrahedral and octahedral cages.Characterization of UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column
In general, porogenic solvent served as a dual functional agent during the synthetic process, not only working as a porogen, but also inducing the phase separation. In this work, PEG was used as porogenic solvent because not only PEGDA can be dissolved in PEG but also UiO-66 can be well dispersed into PEG. The morphology of the prepared UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column was characterized by SEM. As shown in Fig. 4, this MOFs-based monolith attached tightly with the inner wall of capillary and exhibited a well distributed open channel network. The existence of the characteristic signal of UiO-66 at 2θ = 5–10° in the XRD pattern of UiO-66 incorporated poly(MAA-co-PEGDA) monolith (Fig. 2) confirmed the successful preparation of MOFs-based monolithic column. In addition, the BET surface areas and the pore size of UiO-66 incorporated poly(MAA-co-PEGDA) monolith were calculated to be 375 m2 g−1, and 1.3 nm, which also showed the successful incorporation of UiO-66 into the monolith.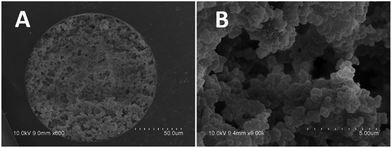 | ||
| Fig. 4 The SEM images of the UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column. (A) 600×; (B) 9000×. | ||
Selective enrichment of phosphopeptides from the tryptic digest of standard phosphoproteins
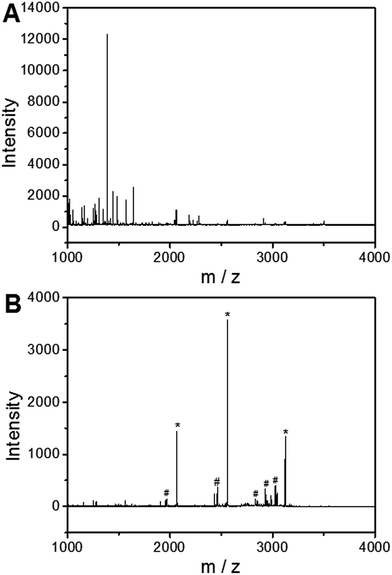
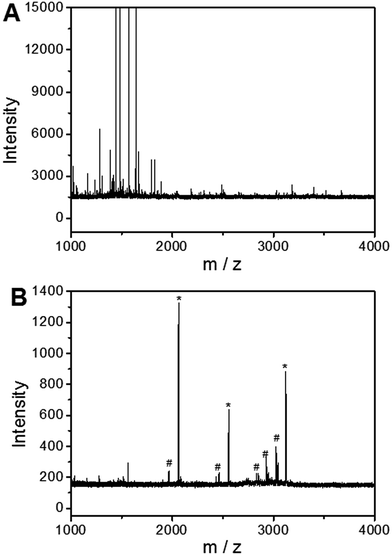
To investigate the applicability of the UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column for the selective enrichment of phosphopeptides, tryptic digest of a standard phosphoprotein (e.g. bovine β-casein) was used as representative sample. Phosphopeptides from tryptic digest of β-casein was first extracted using the UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column. The extracted compounds were then eluted and the resulting solutions as well as the original sample were subject to MALDI-TOF MS analysis. As shown in Fig. 5A, for the direct analysis of β-casein tryptic digest, only one phosphopeptide with poor signal-to-noise (S/N) ratio was detected due to the relatively low concentration of phosphopeptides and severe signal suppression by the abundant non-phosphopeptides. In contrast, after enrichment by the monolithic column, the signals of non-phosphopeptides were eliminated and three expected phosphopeptides (β1, β2, and β3) were detected with strong MS signal intensities, along with their four dephosphorylated peptides (Fig. 5B). The dephosphorylated peptides were likely formed during the MALDI ionization process. The structural details of the detected phosphopeptides from β-casein are given in Table 1. These results well demonstrate the excellent selectivity of the UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column towards phosphopeptides. In order to confirm the fact that the good selectivity stems from the UiO-66 rather than poly(MAA-co-PEGDA) monolith, MALDI-TOF mass spectra of the tryptic digest of β-casein after enrichment by UiO-66 and poly(MAA-co-PEGDA) monolith were investigated, respectively. As shown in Fig. 6A, after enrichment by UiO-66, the signals of non-phosphopeptides were eliminated and three expected phosphopeptides were detected with strong MS signal intensities, along with their four dephosphorylated peptides, which were in good agreement with those obtained by enrichment using UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column. In contrast, after exaction by poly(MAA-co-PEGDA) monolith without UiO-66, no phosphopeptide was detected (Fig. 6B). The results indicated that the good selectivity results from UiO-66 rather than poly(MAA-co-PEGDA) monolith.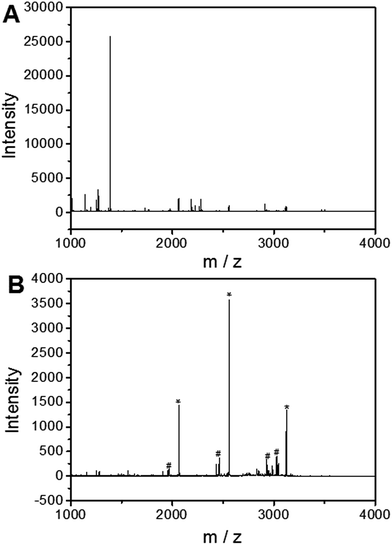 | ||
| Fig. 5 MALDI-TOF mass spectra of the tryptic digest of β-casein. (A) Direct analysis and (B) after enrichment by UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column. | ||
| No. | Peptide sequence | Number of phosphoryl groups | Observed m/z |
|---|---|---|---|
| a [pS]: phosphorylated site. | |||
| β1 | FQ[pS]EEQQQTEDELQDK | 1 | 2062.8 |
| β2 | FQ[pS]EEQQQTEDELQDKIHPF | 1 | 2555.9 |
| β3 | RELEELNVPGEIVE[pS]L[pS][pS][pS]EESITR | 4 | 3121.7 |
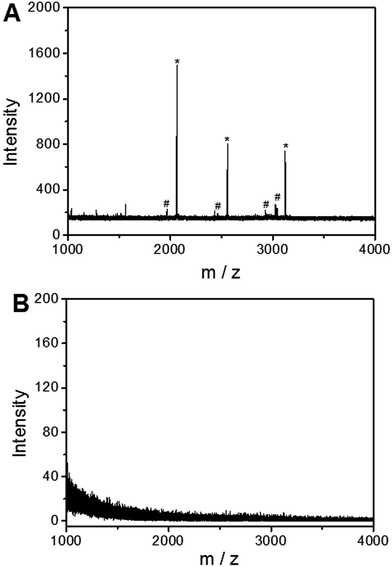 | ||
| Fig. 6 MALDI-TOF mass spectra of the tryptic digest of β-casein after enrichment by UiO-66 (A) and poly(MAA-co-PEGDA) monolithic column (B). | ||
To further evaluate the selectivity of the monolithic column for phosphopeptides under severe interference of non-phosphopeptides, a mixture of the tryptic digests of β-casein and BSA was used as the test sample. As depicted in Fig. 7A and 8A, the molar ratio of β-casein and BSA was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 1
100 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, respectively, nearly no phosphopeptide MS peak was detected and non-phosphopeptide peaks with high intensity dominated the spectrum through direct MS detection due to the serious signal suppression of the non-phosphorylated peptide peaks. As a comparison, after extraction by the monolithic column, the S/N ratio of phosphopeptides was significantly improved, leading to the identification of three phosphopeptides and their five dephosphorylated peptides (Fig. 7B and 8B). These results confirm the ability of the UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column for the selective enrichment of phosphopeptides from complex samples.
1000, respectively, nearly no phosphopeptide MS peak was detected and non-phosphopeptide peaks with high intensity dominated the spectrum through direct MS detection due to the serious signal suppression of the non-phosphorylated peptide peaks. As a comparison, after extraction by the monolithic column, the S/N ratio of phosphopeptides was significantly improved, leading to the identification of three phosphopeptides and their five dephosphorylated peptides (Fig. 7B and 8B). These results confirm the ability of the UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column for the selective enrichment of phosphopeptides from complex samples.
Application to real samples
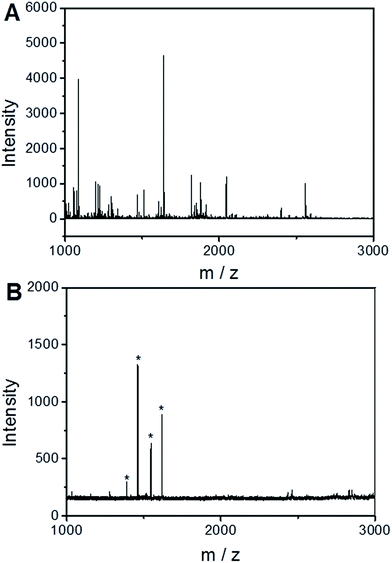
To demonstrate the feasibility of the UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column for the selective enrichment of low-abundance phosphopeptides from real samples, nonfat milk was used as a test sample. For direct analysis, only two peaks with low intensities for phosphopeptides were detected and non-phosphopeptides dominated the spectrum (Fig. 9A). After enrichment with the monolithic column, six peaks of phosphopeptides could be distinctly observed with a clean background (Fig. 9B). The detailed information of the detected phosphopeptides from the tryptic digests of nonfat milk is listed in Table 2. The results indicate that the UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column is capable of highly selective enrichment for phosphopeptides from complex real samples. In order to further evaluate the performance of the UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column in selective enrichment of low-abundance phosphopeptides from a more complex sample, human serum was applied as real sample. For diluted human serum, as shown in Fig. 10A, only one phosphopeptide with weak MS signal intensity appeared owing to the low-abundance phosphopeptides and high salt content. After treatment with the UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column, four peaks of phosphopeptides with higher MS intensities and a clean background can be clearly detected (Fig. 10B). The detailed information of the four phosphopeptides from human serum is given in Table 3. The results indicated that the UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column is capable of highly selective trapping of phosphopeptides from a naturally obtained complex biological sample.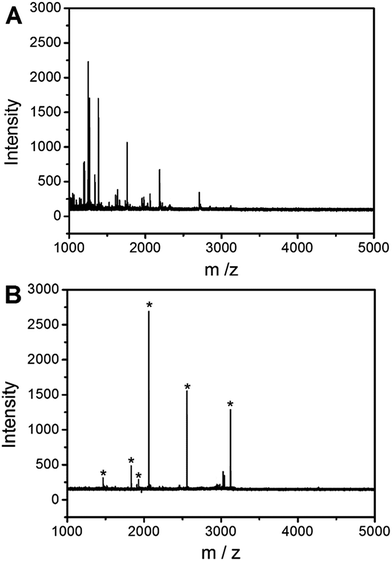 | ||
| Fig. 9 MALDI-TOF mass spectra of tryptic digests of the nonfat milk (A) before and (B) after enrichment by UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column. * indicates phosphopeptides. | ||
| No. | Protein | Peptide sequence | Number of phosphoryl groups | Observed m/z |
|---|---|---|---|---|
| a [pS]: phosphorylated site. | ||||
| 1 | αS2-Casein | TVDME[pS]TEVFTK | 1 | 1466.8 |
| 2 | αS1-Casein | YLGEYLIVPN[pS]AEER | 1 | 1832.5 |
| 3 | αS1-Casein | DIG[pS]E[pS]TEDQAMEDIK | 2 | 1927.3 |
| 4 | β-Casein | FQ[pS]EEQQQTEDELQDK | 1 | 2062.9 |
| 5 | β-Casein | FQ[pS]EEQQQTEDELQDKIHPF | 1 | 2555.9 |
| 6 | β-Casein | RELEELNVPGEIVE[pS]L[pS][pS] [pS]EESITR | 4 | 3121.8 |
 | ||
| Fig. 10 MALDI-TOF mass spectra of human serum (A) before and (B) after enrichment by UiO-66 incorporated poly(MAA-co-PEGDA) monolithic column. * indicates phosphopeptides. | ||
| No. | Peptide sequence | Number of phosphoryl groups | Observed m/z |
|---|---|---|---|
| 1 | D[pS]GEGDFLAEGGGV | 1 | 1388.91 |
| 2 | AD[pS]GEGDFLAEGGGV | 1 | 1460.16 |
| 3 | D[pS]GEGDFLAEGGGVR | 1 | 1544.43 |
| 4 | AD[pS]GEGDFLAEGGGVR | 1 | 1616.27 |
Conclusions
In this study, a UiO-66 incorporated poly(MAA-co-PEGDA) monolithic capillary was successfully prepared. The MOFs-based monolithic capillary has been successfully exploited as an efficient extraction sorbent for phosphopeptide enrichment owing to the significant advantages, including excellent selectivity toward phosphopeptides and tolerance to interference of nonphosphopeptides. Off-line combination of UiO-66 incorporated poly(MAA-co-PEGDA) monolithic capillary-based enrichment with MALDI-TOF MS has been developed for the efficient phosphorylation analysis. The efficiency of this off-line platform has been demonstrated with the phosphorylation analysis of tryptic digestion of standard phosphoprotein and nonfat milk. We foresee such MOFs-containing monolithic capillaries and off-line platform could find more promising applications in proteomic analysis.Acknowledgements
We are grateful to the National Natural Science Foundation of China (No. 21372112) for financial support of this work.References
- M. J. Hubbard and P. Cohen, Trends Biochem. Sci., 1993, 18, 172–177 CrossRef CAS PubMed.
- T. Hunter, Cell, 2000, 100, 113–127 CrossRef CAS PubMed.
- J. Reinders and A. Sickmann, Proteomics, 2005, 5, 4052–4061 CrossRef CAS PubMed.
- C. Temporini, E. Callerli, G. Massolini and G. Caccialanza, Mass Spectrom. Rev., 2008, 27, 207–236 CrossRef CAS PubMed.
- J. Rush, A. Moritz, K. A. Lee, A. Guo, V. L. Goss, E. J. Spek, H. Zhang, X. M. Zha, R. D. Polakiewicz and M. J. Comb, Nat. Biotechnol., 2005, 23, 94–101 CrossRef CAS PubMed.
- T. P. Conrads and T. D. Veenstra, Nat. Biotechnol., 2005, 23, 36–37 CrossRef CAS PubMed.
- K. Rikova, A. Guo, Q. Zeng, A. Possemato, J. Yu, H. Haack, J. Nardone, K. Lee, C. Reeves, Y. Li, Y. Hu, Z. Tan, M. Stokes, L. Sullivan, J. Mitchell, R. Wetzel, J. Macneill, J. M. Ren, J. Yuan, C. E. Bakalarski, J. Villen, J. M. Kornhauser, B. Smith, D. Li, X. Zhou, S. P. Gygi, T. L. Gu, R. D. Polakiewicz, J. Rush and M. J. Comb, Cell, 2007, 131, 1190–1203 CrossRef CAS PubMed.
- Y. Oda, T. Nagasu and B. T. Chait, Nat. Biotechnol., 2001, 19, 379–382 CrossRef CAS PubMed.
- G. Arrigoni, S. Resjo, F. Levander, R. Nilsson, E. Degerman, M. Quadroni, L. A. Pinna and P. James, Proteomics, 2006, 6, 757–766 CrossRef CAS PubMed.
- S. A. Beausoleil, M. Jedrychowski, D. Schwartz, J. E. Elias, J. Villen, J. Li, M. A. Cohn, L. C. Cantley and S. P. Gygi, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 12130–12135 CrossRef CAS PubMed.
- S. A. Beausoleil, J. Villen, S. A. Gerber, J. Rush and S. P. A. Gygi, Nat. Biotechnol., 2006, 24, 1285–1292 CrossRef CAS PubMed.
- M. M. Dong, M. H. Wu, F. J. Wang, H. Q. Qin, G. H. Han, J. Dong, R. A. Wu, M. L. Ye, Z. Liu and H. F. Zou, Anal. Chem., 2010, 82, 2907–2915 CrossRef CAS PubMed.
- M. W. Pinkse, P. M. Uitto, M. J. Hilhorst, B. Ooms and A. J. Heck, Anal. Chem., 2004, 76, 3935–3943 CrossRef CAS PubMed.
- M. Mazanek, G. Mituloviae, F. Herzog, C. Stingl, J. R. A. Hutchins, J. M. Peters and K. Mechtler, Nat. Protoc., 2007, 2, 1059–1061 CrossRef CAS PubMed.
- S. B. Ficarro, J. R. Parikh, N. C. Blank and J. A. Marto, Anal. Chem., 2008, 80, 4606–4613 CrossRef CAS PubMed.
- Z. D. Lu, M. M. Ye, N. W. Li, W. W. Zhong and Y. D. Yin, Angew. Chem., Int. Ed., 2010, 49, 1862–1866 CrossRef CAS PubMed.
- Q. H. Min, X. X. Zhang, H. Y. Zhang, H. F. Zhou and J. J. Zhu, Chem. Commun., 2011, 47, 11709–11711 RSC.
- T. S. Nuhse, A. Stensballe, O. N. Jensen and S. C. Peck, Mol. Cell. Proteomics, 2003, 2, 1234–1243 Search PubMed.
- X. Q. Xu, C. H. Deng, M. X. Gao, W. J. Yu, P. Y. Yang and X. M. Zhang, Adv. Mater., 2006, 18, 3289–3293 CrossRef CAS.
- T. E. Thingholm, O. N. Jensen, P. J. Robinson and M. R. Larsen, Mol. Cell. Proteomics, 2007, 7, 661–671 Search PubMed.
- S. Feng, M. L. Ye, H. J. Zhou, X. G. Jiang, X. N. Jiang, H. F. Zou and B. L. Gong, Mol. Cell. Proteomics, 2007, 6, 1656–1665 CAS.
- H. J. Zhou, M. L. Ye, J. Dong, G. H. Han, X. N. Jiang, R. A. Wu and H. F. Zou, J. Proteome Res., 2008, 7, 3957–3967 CrossRef CAS PubMed.
- J. Y. Ye, X. M. Zhang, C. Young, X. L. Zhao, Q. Hao, L. Cheng and O. N. Jensen, J. Proteome Res., 2010, 9, 3561–3573 CrossRef CAS PubMed.
- H. J. Zhou, M. L. Ye, J. Dong, E. Corradini, A. Cristobal, A. J. Heck, H. F. Zou and S. Mohammed, Nat. Protoc., 2013, 8, 461–480 CrossRef CAS PubMed.
- Y. H. Yan, Z. F. Zheng, C. H. Deng, Y. Li, X. M. Zhang and P. Y. Yang, Anal. Chem., 2013, 85, 8483–8487 CrossRef CAS PubMed.
- F. Wang, Y. T. Zhang, P. Yang, S. Jin, M. Yu, J. Guo and C. C. Wang, J. Mater. Chem. B, 2014, 2, 2575–2582 RSC.
- M. Emgenbroich, C. Borrelli, S. Shinde, I. Lazraq, F. Vilela, A. J. Hall, J. Oxelbark, E. De Lorenzi, J. Courtois, A. Simanova, J. Verhage, K. Irgum, K. Karim and B. Sellergren, Chem.–Eur. J., 2008, 14, 9516–9529 CrossRef CAS PubMed.
- S. Helling, S. Shinde, F. Brosseron, A. Schnabel, T. Muller, H. E. Meyer, K. Marcus and B. Sellergren, Anal. Chem., 2011, 83, 1862–1865 CrossRef CAS PubMed.
- Y. Chen, D. J. Li, Z. J. Bie, X. P. He and Z. Liu, Anal. Chem., 2016, 88, 1447–1454 CrossRef CAS PubMed.
- L. Trojer, G. Stecher, I. Feuerstein, S. Lubbad and G. K. Bonn, J. Chromatogr. A, 2005, 1079, 197–207 CrossRef CAS PubMed.
- M. C. Posewitz and P. Tempst, Anal. Chem., 1999, 71, 2883–2892 CrossRef CAS PubMed.
- Y. Zhang, X. Yu, X. Wang, W. Shan, P. Yang and Y. Tang, Chem. Commun., 2004, 24, 2882–2883 RSC.
- Y. H. Yan, Z. F. Zheng, C. H. Deng, X. M. Zhang and P. Y. Yang, Chem. Commun., 2013, 49, 5055–5057 RSC.
- M. M. Dong, Y. Y. Bian, J. Dong, K. Y. Wang, Z. Y. Liu, H. Q. Qin, M. L. Ye and H. F. Zou, J. Proteome Res., 2015, 14, 5341–5347 CrossRef CAS PubMed.
- M. M. Dong, M. L. Ye, K. Cheng, J. Dong, J. Zhu, H. Q. Qin, Y. Y. Bian and H. F. Zou, Proteomics, 2014, 14, 2410–2416 CrossRef CAS PubMed.
- Z. C. Xiong, L. Y. Zhang, C. L. Fang, Q. Q. Zhang, Y. S. Ji, Z. Zhang, W. B. Zhang and H. F. Zou, J. Mater. Chem. B, 2014, 2, 4473–4480 RSC.
- H. Q. Qin, F. J. Wang, P. Y. Wang, L. Zhao, J. Zhu, Q. H. Yang, R. A. Wu, M. L. Ye and H. F. Zou, Chem. Commun., 2012, 48, 961–963 RSC.
- L. Zhao, H. Q. Qin, Z. Y. Hu, Y. Zhang, R. A. Wu and H. F. Zou, Chem. Sci., 2012, 3, 2828–2838 RSC.
- B. Xu, L. P. Zhou, F. J. Wang, H. Q. Qin, J. Zhua and H. F. Zou, Chem. Commun., 2012, 48, 1802–1804 RSC.
- Y. Zhang, C. Chen, H. Q. Qin, R. A. Wu and H. F. Zou, Chem. Commun., 2010, 12, 2271–2273 RSC.
- Z. Y. Yu, G. H. Han, S. T. Sun, X. N. Jiang, R. Chen, F. J. Wang, R. A. Wu, M. L. Ye and H. F. Zou, Anal. Chim. Acta, 2009, 636, 34–41 CrossRef CAS PubMed.
- L. Zhao, R. A. Wu, G. H. Han, H. J. Zhou, L. B. Ren, R. J. Tian and H. F. Zou, J. Am. Soc. Mass Spectrom., 2008, 19, 1176–1186 CrossRef CAS PubMed.
- S. Y. Xu, J. C. Whitin, T. T. S. Yu, H. J. Zhou, D. Z. Sun, H. J. Sue, H. F. Zou, H. J. Cohen and R. N. Zare, Anal. Chem., 2008, 80, 5542–5549 CrossRef CAS PubMed.
- S. Feng, C. S. Pan, X. G. Jiang, S. Y. Xu, H. J. Zhou, M. L. Ye and H. F. Zou, Proteomics, 2007, 7, 351–360 CrossRef CAS PubMed.
- C. S. Pan, M. L. Ye, Y. G. Liu, S. Feng, X. G. Jiang, G. H. Han, J. J. Zhu and H. F. Zou, J. Proteome Res., 2006, 5, 3114–3124 CrossRef CAS PubMed.
- Y. Zhang, W. F. Ma, C. Zhang, C. C. Wang and H. J. Lu, ACS Appl. Mater. Interfaces, 2014, 6, 6290–6299 CAS.
- L. J. Murray, M. Dinca and J. R. Long, Chem. Soc. Rev., 2009, 38, 1294–1314 RSC.
- N. Chang, Z. Y. Gu and X. P. Yan, J. Am. Chem. Soc., 2010, 132, 13645–13647 CrossRef CAS PubMed.
- Z. Y. Gu and X. P. Yan, Angew. Chem., Int. Ed., 2010, 49, 1477–1480 CrossRef CAS PubMed.
- Z. Y. Gu, C. X. Yang, N. Chang and X. P. Yan, Acc. Chem. Res., 2012, 45, 734–745 CrossRef CAS PubMed.
- C. X. Yang and X. P. Yan, Anal. Chem., 2011, 83, 7144–7150 CrossRef CAS PubMed.
- L. E. Kreno, K. Leong, O. K. Farha, M. Allendorf, R. P. Van Duyne and J. T. Hupp, Chem. Rev., 2012, 112, 1105–1125 CrossRef CAS PubMed.
- S. Rojas, P. S. Wheatley, E. Quartapelle-Procopio, B. Gil, B. Marszalek, R. E. Morris and E. Barea, CrystEngComm, 2013, 15, 9364–9367 RSC.
- S. Bhattacharjee, Y. R. Lee, P. Puthiaraj, S. M. Cho and W. S. Ahn, Catal. Surv. Asia, 2015, 19, 203–222 CrossRef CAS.
- J. W. Liu, L. F. Chen, H. Cui, J. Y. Zhang, L. Zhang and C. Y. Su, Chem. Soc. Rev., 2014, 43, 6011–6061 RSC.
- C. B. Messner, M. R. Mirza, M. Rainer, O. M. D. Lutz, Y. Guzel, T. S. Hofer, C. W. Huck, B. M. Rode and G. K. Bonn, Anal. Methods, 2013, 5, 2379–2383 RSC.
- X. Y. Zhu, J. L. Gu, J. Yang, Z. Wang, Y. S. Li, L. M. Zhao, W. R. Zhao and J. L. Shia, J. Mater. Chem. B, 2015, 3, 4242–4248 RSC.
- M. Zhao, C. H. Deng and X. M. Zhang, Chem. Commun., 2014, 50, 6228–6231 RSC.
- Y. J. Chen, Z. C. Xiong, L. Peng, Y. Y. Gan, Y. M. Zhao, J. Shen, J. H. Qian, L. Y. Zhang and W. B. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 16338–16347 CAS.
- G. Guiochon, J. Chromatogr. A, 2007, 1168, 101–168 CrossRef CAS PubMed.
- K. K. Unger, R. Skudas and M. M. Schulte, J. Chromatogr. A, 2007, 1184, 393–415 CrossRef PubMed.
- N. Tanaka, H. Kobayashi, K. Nakanishi, H. Minakuchi and N. Ishizuka, Anal. Chem., 2001, 73, 420A–429A CrossRef CAS PubMed.
- H. Y. Li and Z. Liu, Trends Anal. Chem., 2012, 37, 148–161 CrossRef CAS.
- Z. J. Bie, Y. Chen, H. Y. Li, R. H. Wu and Z. Liu, Anal. Chim. Acta, 2014, 834, 1–8 CrossRef CAS PubMed.
- A. Ahmed, M. Forster, R. Clowes, P. Myers and H. F. Zhang, Chem. Commun., 2014, 50, 14314–14316 RSC.
- Y. Y. Fu, C. X. Yang and X. P. Yan, Chem. Commun., 2013, 49, 7162–7164 RSC.
- S. C. Yang, F. G. Ye, Q. H. Lü, C. Zhang, S. F. Shen and S. L. Zhao, J. Chromatogr. A, 2014, 1360, 143–149 CrossRef CAS PubMed.
- M. J. Katz, Z. J. Brown, Y. J. Colon, P. W. Siu, K. A. Scheidt, R. Q. Snurr, J. T. Hupp and O. K. Farha, Chem. Commun., 2013, 49, 9449–9451 RSC.
- Y. Y. Zhang, X. Feng, H. W. Li, Y. F. Chen, J. S. Zhao, S. Wang, L. Wang and B. Wang, Angew. Chem., Int. Ed., 2015, 54, 4259–4263 CrossRef CAS PubMed.
- S. J. Garibay and S. M. Cohen, Chem. Commun., 2010, 46, 7700–7702 RSC.
| This journal is © The Royal Society of Chemistry 2017 |