 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Comparative effect of cationic gemini surfactant and its monomeric counterpart on the conformational stability and activity of lysozyme†
Taruna Sharmaa,
Neeraj Doharea,
Meena Kumaria,
Upendra Kumar Singha,
Abbul Bashar Khana,
Mahendra S. Borseb and
Rajan Patel *a
*a
aBiophysical Chemistry Laboratory, Centre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia (A Central University), New Delhi, India. E-mail: rpatel@jmi.ac.in; rajanpatelpcy@gmail.com; Fax: +91 11 26983409; Tel: +91 8860634100
bDepartment of Chemistry, Uttamrao Patil College Dahivel Taluka-sakri, Dhule, Maharashtra, India
First published on 17th March 2017
Abstract
Protein interactions with surfactants are dependent on their physiochemical properties. The effect of cationic gemini surfactant hexanediyl-α,ω-bis-(N-(2-hydroxyethyl)-N-methylhexadecylammonium dibromide) on the stability and activity of hen egg white lysozyme was compared with its monomeric counterpart N-(2-hyroxyethyl)-N,N-dimethylhexadecylammonium bromide at pre and post micellar concentrations. This study utilizes circular dichroism (CD), steady-state fluorescence spectroscopy, extrinsic fluorescence spectroscopy, time-resolved fluorescence spectroscopy, UV-visible spectroscopy, molecular docking and turbidity assays to resolve the conformational stability and antibacterial activity of lysozyme in the presence of surfactants. Micelles of both cationic surfactants were observed to stabilize the conformation of the protein, however, gemini was found to stabilize it in a much higher micellar concentration range. Detailed analysis of the time-resolved fluorescence spectroscopy results suggests contribution of the lifetime values of Trp62 and Trp108 to the overall conformation change of lysozyme with the increase in concentration of the respective surfactants, which is further correlated with the steady-state fluorescence and CD spectroscopy results. Furthermore, from the CD analysis it was found that the cationic single chain surfactant strongly perturbs the secondary and tertiary structure of the protein as compared to the gemini surfactant. Through docking results, it was found that the gemini surfactant binds weakly with lysozyme as compared to the single chain surfactant. Specifically, the antibacterial activity of lysozyme was found to be increased in the presence of cationic gemini surfactant, which extrapolates the use of these surfactants in pharmaceutics and industries.
1. Introduction
Protein–surfactant studies have been under thorough investigation since the late 18th century1 and there are a plethora of well-established facts related to this domain. The effect of surfactant on protein stability depends on the type of surfactant forming specific interactions.2 Protein conformation dynamics majorly involve hydrophobic effects3 and the degree of attraction for water.4 It has already been reported that monomeric detergents bind to the native state as conventional ligands, that is, they bind to a small number of sites in a saturable manner5,6 (through hydrophobic and electrostatic interactions mainly) and generally micelles act as denaturants.7 The process of denaturation depends on the effect of denaturants on the water structure and on the hydrophobic interactions in the tertiary structure of the protein8,9 which leads to nonspecific cooperative interactions. Unlike anionic surfactants, cationic surfactants bind weakly to the protein and are less efficient then anionic surfactants in promoting the cooperative transition. They are also known to maintain the three dimensional structure of the protein at considerable high concentrations2 even above their cmc values.10 This difference in binding pattern of cationic surfactants is attributed due to smaller relevance of electrostatic interactions at the pH's of interest,11 side chains involved in the basic protein–surfactant interaction (hydrophobic contribution and electrostatic interaction of charged residues) at monomeric concentrations and due to the difference in micellar structure of these surfactants at their micellar concentrations as explained by Otzen.7In addition, the structural and functional stability of some proteins can be more efficiently determined by analyzing their catalytic activity. It has already been mentioned that aqueous micelles can act as catalysts or inhibitors for enzymes, depending on the nature of the surfactants (cationic, anionic or neutral) forming the micelles.12 Cationic surfactants are known to increase or stabilize the activity of certain enzymes. In contrast to anionic surfactants,13 Lu et al.14 and Guzman et al.15 found catalytic activity of surface active enzymes like lysozyme and pepsin respectively and observed them to be reactivated and stabilized by interaction with cationic surfactants probably because of the lesser involvement of above mentioned electrostatic interactions.
Some recent reports on new generation surface active agents called as “Gemini surfactant” also suggests their potential role in protein stability and activity.16,17 Interaction of gemini surfactants with proteins suggest that they have stronger binding with proteins as compared to conventional surfactants.18–20 This difference in binding is majorly governed due to dissimilarity in their physiochemical properties they possess due to the presence of two long hydrocarbon chains and two ionic groups attached covalently to a spacer group. They have better solubility, lower cmc value and Krafft temperature.21 Also, they have unique binding properties due to their strong dependence on spacer length,22 special aggregate morphology, and strong hydrophobic microdomain.21
Recently we have reported the possible role of gemini surfactants in refolding of globular protein23 and stability of membrane bound peptide.24 Present communication is an attempt to get much deeper insight on the comparative effect of biscationic methyl ethanol amine gemini surfactant C16H33(CH3)2C2H4OHN+–(CH2)6–N+C2H4OH(CH3)2C16H332Br− abbreviated as G16 with spacer length –(CH2)6– and its monomeric counterpart C16H33(CH3)2C2H4OHN+Br− abbreviated as M16 on the structural stability and antibacterial activity of hen egg white lysozyme (HEWL) using various spectroscopic techniques and molecular docking method. The structure of M16 and G16 has been depicted in Scheme 1.
 | ||
| Scheme 1 Structure of N-(2-hydroxyethyl)-N,N-dimethylhexadecyl ammonium bromide (M16) and hexanediyl-α,ω-bis-N-(2-hydroxyethyl)-N-methylhexadecyl ammonium dibromide (G16). | ||
Lysozyme is a well studied 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 603 Da globular protein. It is involved in protein misfolding disease, known as non-neuropathic systemic amyloidoses25 hence, understanding the effect of different ligands on structural and functional stability of this enzyme is highly significant. Lysozyme has two domains; α-domain having four α-helices and a C-terminal 310 helix and β-domain which contain triple-stranded antiparallel β-sheets, a 310 helix, and a long loop.26 The two domains are linked through short double stranded antiparallel β-sheet. It has total 129 amino acids with four disulphide bonds in its tertiary structure.27 In aqueous solution, lysozyme (Isoelectric point, IP 11.35) has a pH of 6.5 with a net positive charge of 8.28
603 Da globular protein. It is involved in protein misfolding disease, known as non-neuropathic systemic amyloidoses25 hence, understanding the effect of different ligands on structural and functional stability of this enzyme is highly significant. Lysozyme has two domains; α-domain having four α-helices and a C-terminal 310 helix and β-domain which contain triple-stranded antiparallel β-sheets, a 310 helix, and a long loop.26 The two domains are linked through short double stranded antiparallel β-sheet. It has total 129 amino acids with four disulphide bonds in its tertiary structure.27 In aqueous solution, lysozyme (Isoelectric point, IP 11.35) has a pH of 6.5 with a net positive charge of 8.28
Interaction of lysozyme with anionic surfactant like SDS has been studied in detail29 and is known to strongly perturb its three dimensional structure due to the presence of strong electrostatic interactions. Cationic surfactants possess comparatively weaker interaction with lysozyme because of the obvious electrostatic repulsion at physiological pH. However, they form weak electrostatic interactions with negatively charged amino acids (Asp and Glu)30 present in lysozyme. Chatterjee et al.31 using micro-calorimetry found that hydrophobic interaction dominates over electrostatic repulsion resulting in lysozyme-CTAB aggregate formation. The effect of gemini surfactant on lysozyme have also been reported previously,19,32,33 however relatively less interaction studies have been made till now. The aim of the present study is to decipher the binding interaction of M16 and G16 on the conformational stability and antibacterial activity of HEWL. The interaction pattern of M16 and G16 with lysozyme before and after cmc has been shown in Scheme 2. We hope that this study will endow greater landscape on the difference in binding of respective surfactants specifically with HEWL which will prove to be helpful from medicinal and pharmaceutical point of view. In addition, we hope that it will provide wide understanding on the structural and functional properties HEWL for augmenting the application of this enzyme in food preservation and as an effective antibiotic.
2. Experimental section
2.1 Materials
The details of all chemicals employed were summarized in Table 1. M16 and G16 surfactants were synthesized in our lab using protocol mentioned elsewhere.34,35 All the chemicals and reagents used were of analytical grade. Millipore water of specific conductance 2–2.5 μS was used throughout experiments. The pH measurements were carried out with a Khera Microprocessor pH meter (India) using a combined electrode consisting of glass and reference electrodes as a single entity.| Chemical name | Source | Purity | Purification method | Analysis method |
|---|---|---|---|---|
| a Sodium phosphate monobasic.b Sodium phosphate dibasic. | ||||
| HEWL | Sigma-Aldrich | ≥98.0% | Dialyzed and filtered | |
| Micrococcus lysodickticus | Sigma-Aldrich | — | — | |
| NaH2PO4a | Sigma-Aldrich | 98.0% | — | |
| Na2HPO4b | Sigma-Aldrich | 98.0% | — | |
| Pyrene | Sigma-Aldrich | 98.0% | — | |
| Ethanol | Merck | 99.5% | — | |
| Acetone | Merck | 99.0% | — | |
| M16 | Synthesized | 99.0% | Recrystallization | NMR41 |
| G16 | Synthesized | 99.0% | Recrystallization | NMR41 |
2.2 Sample preparation
The stock solution of protein was prepared in 10 mM phosphate buffer. It was then extensively dialyzed and filtered using 0.22 μM millipore syringe filter. The concentration of the stock solution was monitored by UV-visible spectrophotometer using molar extinction coefficient of HEWL ε280 = 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 932 M−1 cm−1.36 Protein working solution of 0.3 mg ml−1 (∼20 μM) was used in far-UV CD, UV-visible, steady-state and extrinsic fluorescence and time resolved fluorescence spectroscopy measurements. The stock solutions of surfactants were made in analytical grade millipore water. The cmc values M16 and G16 were measured by conductivity experiment and they were found to be ∼2.10 × 10−4 M and ∼3.63 × 10−6 M which is in good agreement with the reported values.34,37 The respective surfactants do not show any absorption in the protein absorbing region in all the concentrations used. All the working concentrations of samples were prepared by adding an appropriate amount of surfactant from its stock solution into the HEWL solution. All the samples of desired concentrations were incubated overnight at room temperature in all the experiments. Incubating surfactant-protein samples at higher concentration is necessary for maintaining equilibration of protein/surfactant ratio12 which otherwise decreases.
932 M−1 cm−1.36 Protein working solution of 0.3 mg ml−1 (∼20 μM) was used in far-UV CD, UV-visible, steady-state and extrinsic fluorescence and time resolved fluorescence spectroscopy measurements. The stock solutions of surfactants were made in analytical grade millipore water. The cmc values M16 and G16 were measured by conductivity experiment and they were found to be ∼2.10 × 10−4 M and ∼3.63 × 10−6 M which is in good agreement with the reported values.34,37 The respective surfactants do not show any absorption in the protein absorbing region in all the concentrations used. All the working concentrations of samples were prepared by adding an appropriate amount of surfactant from its stock solution into the HEWL solution. All the samples of desired concentrations were incubated overnight at room temperature in all the experiments. Incubating surfactant-protein samples at higher concentration is necessary for maintaining equilibration of protein/surfactant ratio12 which otherwise decreases.
2.3 Methods
F = Fobs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) antilog[(Aex + Aem)/2] antilog[(Aex + Aem)/2]
| (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 counts in the peak unless otherwise indicated. The instrumental response function was recorded sequentially using a scattering solution and a time calibration of 114 ps per channel. Data were analyzed using a sum of exponentials, employing a nonlinear least squares reconvolution analysis of the form:42
000 counts in the peak unless otherwise indicated. The instrumental response function was recorded sequentially using a scattering solution and a time calibration of 114 ps per channel. Data were analyzed using a sum of exponentials, employing a nonlinear least squares reconvolution analysis of the form:42
 | (2) |
The goodness of fit was judged in terms of both a chi-squared (χ2) value and weighted residuals. Time-resolved fluorescence decays were analyzed making use of the impulse response function (IBH DAS6 software). The average fluorescence lifetimes (〈τ〉) of HEWL for the decay curves were calculated from the decay times and the relative contribution of the components using the following equation:38
 | (3) |
A fit was considered acceptable when plots of the weighted residuals and the autocorrelation function showed random deviation about zero with a minimum χ2 value approaching 1.38
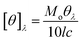 | (4) |
 | (5) |
The value 4000 is the MRE of the β-sheet and random coil conformation cross at 208 nm and 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 is the MRE value of a pure α-helix at 208 nm.44
000 is the MRE value of a pure α-helix at 208 nm.44
 | (6) |
3. Results and discussion
3.1 Circular dichroism spectroscopy
CD spectroscopy is an absorption-based technique and gives information about the secondary and tertiary structure of the protein. In the far-UV region of the spectrum (240–180 nm), different forms of regular secondary structure found in proteins give rise to characteristic CD spectra. The CD spectrum in this region can be analyzed in terms of the content of α-helix, β-sheet, β-turn, etc.47 Where, in the near-UV region (320–260 nm) CD signals principally arises from the aromatic side chains Phe, Tyr and Trp and it provides a detailed fingerprint of the tertiary structure of the protein.47In the present study, far-UV CD spectroscopy was used in order to monitor the changes induced by M16 and G16 on the secondary structure of HEWL. From the spectrum shown in Fig. 3, it can be clearly seen that HEWL untreated with M16 and G16 gives a characteristic negative peak at 208 nm and 222 nm signifies the presence of α-helix in HEWL. The percentage of α-helix content at 208 nm was calculated by using eqn (5) and values were reported in Table S1.† α-Helical content of pure HEWL was found to be ∼42% which is in good agreement as mentioned by Sethuraman et al.26
It was observed that addition of M16 (Fig. 1(a)) in the concentration range from 8.26 × 10−8 M to 1.50 × 10−3 M, the α-helical content of HEWL marginally increased from 42% to 45%. Signal at M16 concentration 3.20 × 10−3 M there was a decrease in α-helical content observed from 45% to 39%. In case of G16 (Fig. 1(b)), the α-helical content was found be increased from 42% to 45% in the same way as in case of M16, however, in the concentration range from 8.26 × 10−8 M to 2.10 × 10−4 M. At concentration 3.0 × 10−4 M an overlapping spectrum just like that of native HEWL was observed with a subtle blue shift of 206 nm. Signal at 1.5 × 10−3M G16 was observed to get shifted in upward direction which signifies destabilizing effect of surfactant on the secondary structure of the protein. The variation in signals at 208 nm can be more clearly analyzed in Fig. 2 at various concentrations of both the surfactants. Owing to the limitation of far-UV CD spectroscopy, the stability effect by M16 and G16 at the concentration above 3.20 × 10−3 M and 1.5 × 10−3 M respectively, could not be measured as far-UV CD signals above these concentrations could not be deconvoluted due to high noise/signal ratio, as also observed in some others protein–surfactant studies at higher surfactant concentrations.12 However, due to the shift of spectrum in an upward direction (Fig. 2) it can be assumed that after these concentrations of M16 and G16 the helical content will decrease.
 | ||
| Fig. 1 Far-UV CD spectra of HEWL as a function of increase in concentration of (a) M16 and (b) G16 at 298 K. | ||
Thus, it can be concluded from the results that monomeric, as well as micellar forms of both the surfactants are involved in stabilization of secondary structure of HEWL up to certain concentration range. Cationic surfactant can bind to the positively charged HEWL through weak electrostatic (with negatively charged residues like Glu and Asp) and strong hydrophobic interactions. However, its stabilizing effect on the secondary structure of HEWL is imparted may be due to the positive charge associated with it, as elucidated in a study by Gospodarczyk et al.32 The increase in α-helical content can be elucidated due to the formation of the hydrophobic linkage between the hydrophobic chains and non-polar residues present on HEWL causes a local increase in the α-helical content of the protein as also observed in a study by Gull et al.20 and Wang et al.48 In the case of G16, the stabilizing effect on HEWL cannot be attributed to the positive charge present on the two head groups. The reason for this discrepancy can be explained due to the presence of longer spacer group17 which causes lower charge density on the surfactant and thus possibly it binds weekly with HEWL as compared to M16 unlike of some reported observations with much smaller spacer group.18,20 Therefore, increase in helical content, in this case is governed by stronger hydrophobic microdomains which not only limit the unfolding but also compress or increase the α-helical content of the protein20 by interacting with the non-polar residues of the protein. These results also suggested that G16 stabilizes the secondary structure of protein up to considerable high concentration range even beyond its cmc value (3.63 × 10−6 M to 3.00 × 10−4 M). Whereas, a decrease in the α-helical content was observed in the case of M16 at 3.20 × 10−3 M thus micelles of single chain surfactant stabilize the protein to considerable low concentration range (2.10 × 10−4 M to 1.50 × 10−3 M) as compared to its gemini counterpart. Nevertheless, at very high surfactant concentration (M16 and G16 respectively) excess micellar structures form intrachain hydrophobic interactions in the protein which are replaced by the electrostatic repulsive forces between the micellar aggregates, which is possibly the reason for decrease in the α-helical content of HEWL20 as explained by Gull et al.20 in a study with gemini surfactant and HSA.
For understanding, the conformational changes imparted by both the surfactants more clearly, near-UV CD experiments were done. The typical near-UV CD spectra (Fig. 3) of untreated HEWL gives positive triplet like signal from 280 to 300 nm range. This signal is indicative of lysozyme's active conformation.49 In addition, the weak negative band at 295 nm has been attributed to Trp108, which is associated with the active site.50 This gives direct insight for monitoring the environment of the active site of the enzyme.
 | ||
| Fig. 3 Near UV-CD spectra of HEWL as a function of increase in concentration of (a) M16 and (b) G16 at 298 K. | ||
It can be observed from the near-UV CD spectra shown in Fig. 3(a) that addition of M16 from concentration range 8.26 × 10−8 M to 1.50 × 10−3 M stabilizes the tertiary structure of the protein with intact active conformation as also observed by far-UV CD results. After this, a significant fall in the native conformation with diminished peaks in 280–300 nm regions was observed. This indicates that at these concentrations the micellar structure of M16 strongly interferes with the tertiary structure of HEWL.50 The weak peak at 295 nm got almost diminished suggesting that the active site of the enzyme got significantly affected at higher micellar concentration and such disruption by different surfactants have been observed earlier.51 On the other hand, it can be clearly observed from Fig. 3(b) that no change in the tertiary structure of HEWL was monitored even at a very high concentration of G16 (from 8.26 × 10−8 to 2.11 × 10−2 M) and found to be resistant to its addition. These observations suggest that G16 at very high concentration do not interferes with the tertiary structure of the protein, though it affects the local secondary structure of HEWL as observed in far-UV CD results. This type of unusual protein conformation has also been reported elsewhere.52,53 Some other gemini's on interaction with proteins like HSA, are also found to increase their tertiary structure.20
3.2 Steady-state fluorescence measurements
Steady-state fluorescence spectroscopy is sensitive technique to evaluate the changes in the microenvironment around the fluorophore and to monitor the tertiary structure of the protein. HEWL has six tryptophan molecules out of which steady state fluorescence data have shown that 80% of the fluorescence in the native protein comes from Trp62 and Trp108.54 These two residues reside in the hydrophobic core (Trp62 is most exposed) of HEWL and their intensity for fluorescence is highly dependent on the microenvironment. The emission spectrum was monitored by exciting HEWL at 280 nm and monitoring the emission at 342 nm. This gives an overall picture of the globular protein folding.20With the addition of M16 concentration from 8.26 × 10−8 to 1.20 × 10−4 M in HEWL, no change (Fig. 4(a)) in the intensity was measured and the spectra overlap on the native HEWL, suggesting native like microenvironment around the fluorophores. At M16 concentration 2.10 × 10−4 M increase in the intensity was monitored, the maximum increase was observed at concentration 1.50 × 10−3 M. This may suggest that M16 micelles providing a structural moiety to HEWL where fluorophores may suppress the internal quenching which otherwise interferes with their fluorescence. Thus, from the results, it can be interpreted that HEWL conformational changes observed in far-UV CD results lead to the exposure of Trp and Tyr molecules to a more flexible environment by M16 micelles which can also be related to HEWL compact structure.55 Such results have also been observed by Celej et al.12 At concentrations 3.2 × 10−3 M to 2.11 × 10−2 M the spectra showed a sharp decrease (Fig. 4(a)). This observation suggests that at these mentioned concentration the M16 micelles strongly interfere with the tertiary structure of HEWL that leads to its unfolding which is also confirmed by near-UV CD results at these concentrations.
 | ||
| Fig. 4 Steady-state fluorescence spectra of HEWL with increase in concentration (a) M16 and (b) G16 from 8.26 × 10−8 to 2.11 × 10−2 M at 298 K. | ||
On the other hand, the addition of various concentrations of G16 didn't produce a much pronounced effect on the intensity of HEWL unlike of M16 (Fig. 4(b)). Similar kind of result was also observed by Amiri et al.17 in which G16 treated RNase A showed minor changes in absorbance as well as fluorescence spectra. They attributed this effect due to the low charge density on G16 as also observed in the present study. Li et al.56 also experienced such observations with much decreased fluorescence intensity with increase in spacer length of gemini surfactants. The intensity of the spectra remained almost constant from 8.26 × 10−8 M to 3.4 × 10−4 M G16 in a similar way as observed with M16 at its lower concentrations. A subtle increase in intensity was observed at concentration 6.07 × 10−5 M to 2.10 × 10−4 M. As mentioned above this subtle increase in the intensity can be attributed to the compact tertiary structure as well as the increase in the α-helical content of HEWL as observed through far and near-UV CD experiments. The spectrum slightly decreased at concentration 3.0 × 10−4 M and this can be attributed due to the destabilization of α-helix as observed in far-UV CD results. The decrease in fluorescence with a subtle blue shift of 4 nm (∼342 to ∼338 nm) was observed at concentration 2.11 × 10−2 M suggesting movement of the fluorophore to a more hydrophobic environment may be because of some loss of α-helical content at high concentration of G16 micelles. The difference in the change of the fluorescence intensity of both the ligands can be more clearly visualized from the graph plotted between maximum fluorescence intensity (Fmax) as a function of increase in the concentration of M16 and G16 (Fig. 5).
These observations give the possible binding mechanism of both the surfactants with HEWL. Apart from the difference in charge density on the surface of M16 and G16 which governs the binding with HEWL, other possible reason for discrepancy in the intensity change of HEWL in presence of respective surfactants lies on the fact that possibly the micellar shapes of both the surfactants have different structure which produces different effect on the secondary and tertiary structure of the protein.7 Thus, more research is needed to confirm these possibilities.
Synchronous fluorescence spectroscopy was also done in order to decipher the involvement of Trp and Tyr molecule during the interaction of M16 and G16 with HEWL respectively. It was observed (Fig. 1S and 2S†) that in both the cases the Tyr molecule marginally got affected and the major effect of both the ligands was observed on Trp molecule. Effect of G16 on Trp microenvironment (Fig. 2S(a)†) at higher concentration depicts a blue shift which is same as observed in steady-state fluorescence experiment. This observation suggests the movement of Trp residues to a less solvent exposed environment at high G16 concentrations.
3.3 Time resolved spectroscopy
Structural and conformational dynamics of proteins can be thoroughly analyzed by measuring steady-state fluorescence spectroscopy in combination with studying the fluorescence lifetime of these molecules.55,57 In case of HEWL, it has been reported that Trp62 has the longest lifetime of ∼2.5–2.8 ns (τ value) as it is most exposed to the solvent where as Trp108 (buried in the hydrophobic core) has the lifetime of ∼1.5–1.7 ns,54,58 also the lifetime decay values are pH dependent.58 Solvent exposure is not only the reason for Trp62 longer lifetime. The fluorescence lifetime of proteins is highly affected by the quenching effect of the nearby sulfur containing groups. These groups are effective collision quenchers of indole fluorescence. Trp62 does not reside near cystine (involved in disulfide bond formation) or methionine residues; this increases its lifetime.58 Trp108 also has very less contact with the cystine residues. Trp108 and Trp28 are associated with methionine residues however methionine is not involved in fluorescence quenching. It's mainly Trp62 which decreases the fluorescence lifetime of Trp108 through energy transfer process.54 Other three Trp63, Trp111 and Trp123 have high contact with cystine residues which contributes to their shorter lifetimes that is ∼0.5 ns.58The presented life time values (Tables S2 and S3†) in the present study are markers of the microenvironment of the fluorophore, where τ3, τ2 and τ1 values are designated to Trp62, Trp108, and life time of other Trp as discussed above. The relative amplitude here is regarded as the relative population of Trp62, Trp108 and other remaining Trp in the excited states. The value of τ is thus system dependent. Mandal et al.57 illustrated that when quenching favorably occurs by different influencing interactions the value of τ can decrease, when ligand does not affect the fluorophore the τ value remains unaffected, and on removing the quenching factors the τ value even can increase. Increase/decrease in fluorescence lifetime was also observed in the unfolding/refolding studies of proteins as reported by Anand et al.55
The obtained τ3, τ2 and τ1 values of untreated HEWL (Tables S2 and S3†) are in fair agreement with the above mentioned values. This difference in the τavg of HEWL with increased concentration of M16 and G16 can be more clearly visualized through Fig. 7. In the case of M16, from the spectra represented in Fig. 6(a), it can be clearly analyzed that calculated τavg values remained almost constant from M16 concentrations between 8.26 × 10−8 M to 6.07 × 10−5 M indicating compactness maintained by the surfactant. A subtle increase in τavg value was observed at concentration 3.00 × 10−4 M to 1.50 × 10−3 M. These observations are in accordance with our previous results suggesting structural stability provided by the M16 at these micellar concentrations. The τavg values (Table S2† and Fig. 6(a)) start decreasing at concentration 3.20 × 10−3 M to 2.11 × 10−2 M. This observation can be attributed to the quenching effect which is mainly exhibited due to the presence of highly hydrophobic micellar aggregates of M16 at very high micellar concentrations which changes the conformation of HEWL and perturbs its secondary and tertiary structure. The quenching effect can be explained in accordance to the explanations given by Anand et al.59 They explained the role of planarity of Trp indole ring at the ground and excited states. As the substrate interact with the Trp residue, it leads to distortion of the planarity of the indole ring which results in the reduction of fluorescence lifetime.59 Also, out of three lifetime values, Trp62 (most exposed) was found to be most effected in the present data. This suggests that lifetime of Trp residues is highly dependent upon its native position in a folded protein. Thus, the substrate in the present study is M16 micelles which at higher micellar concentrations distort the planarity of the Trp62 residue due to change in the overall conformation of HEWL which causes a change in the local environment around it and decrease in its lifetime as well as relative amplitude. In the case of G16, the τavg value (Table S3† and Fig. 6(b)) was found to be in the increasing order which can be attributed to the attainment of HEWL overall conformation where the fluorophores are possibly not being affected by the internal quenchers (relative amplitude of Trp residues other then Trp62). The relative contribution by longer component (Trp62) at very high G16 concentration was found to be decreased considerably (Table S3†), this happened because of the decreased α-helical content, suggesting its movement to a less solvent exposed environment as observed in far-UV CD as well as in steady state fluorescence spectroscopy results (confirmed by blue shift).
 | ||
| Fig. 6 Variation in fluorescence lifetime of HEWL as a function of increase in concentration of (a) M16 and (b) G16 at 298 K. | ||
3.4 UV-visible spectroscopy
HEWL shows two absorption bands. One strong band arises due to the π–π* electronic transition of the peptide backbone C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. It lies in between 200–230 nm and gives information about the scaffold of HEWL.60 Second absorption band can be seen at 280 nm involving n–π* transitions which arise mainly due to the presence of aromatic amino acids (largely because of Trp). UV-visible spectroscopy was employed to monitor the effect of M16 and G16 on HEWL.
O. It lies in between 200–230 nm and gives information about the scaffold of HEWL.60 Second absorption band can be seen at 280 nm involving n–π* transitions which arise mainly due to the presence of aromatic amino acids (largely because of Trp). UV-visible spectroscopy was employed to monitor the effect of M16 and G16 on HEWL.
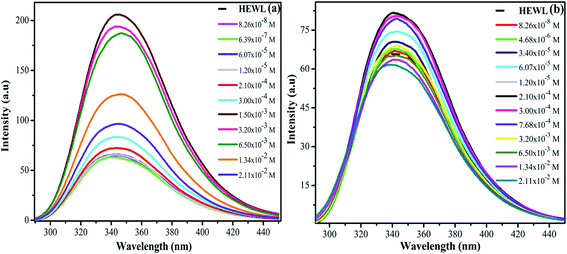
From Fig. 8(a) and (b), it can be clearly seen that the absorbance at 280 nm of HEWL substantially increases in both the cases. The variation in absorbance intensities can be more clearly analyzed by plotting a graph between absorbance values obtained at 280 nm as a function of increased concentration of M16 and G16 as shown in Fig. 9. In the case of M16, it can be clearly seen through the plot that the absorbance values remained almost constant from 8.26 × 10−8 M to 3.20 × 10−3 M. At higher concentrations (1.34 × 10−2 to 2.11 × 10−2 M) absorbance values sharply increased. On the other hand, the same trend was observed for G16 with not much changes in the absorbance value were observed at various concentrations. Additionally, more increase in absorbance values were observed at higher micellar concentrations of G16 (1.34 × 10−2 M and 2.11 × 10−2 M).
 | ||
| Fig. 8 UV-visible spectra of HEWL as a function of increase in concentration of (a) M16 and (b) G16 at 298 K. | ||
 | ||
| Fig. 9 Difference in variation of absorbance (at 280 nm) of HEWL with increased concentration of M16 and G16. | ||
The small rise in absorbance is indicative that the addition of G16 and M16 hinders the penetration of light due to the compaction of HEWL and are in line with the observations reported by Mandal et al.57 on interaction of SDDS with HEWL. The hyperchromic effect observed at a higher micellar concentration (more pronounced in M16) with no shift is mainly attributed due to the conformational changes in the protein as observed in our previous results also. This indicates that at higher micellar concentration, due to strong electrostatic and hydrophobic interaction of M16 and G16 with HEWL causing exposure of chromophores to the bulk solvent and this result in increased observed absorbance values.
3.5 Extrinsic fluorescence spectroscopy
The pyrene probe method is highly sensitive to even minor changes in the polarity of the medium and thus used for measuring cmc of surfactants in different mediums and ligand–protein interactions.61 Out of the five characteristic peaks (∼375 (I), 379 (II), 385 (III), 395 (IV) and 410 (V) nm) peak I is most sensitive to the non-polar environment and show a sharp decrease with increase in hydrophobicity.62 The ratio between the intensity of the emission peak I (I1) and III (I3) gives information about the polarity of the medium.63 The I1/I3 ratio decreases with increase in hydrophobicity of the medium.63 The reason behind this fact relies on the low solubility of pyrene in water and as the hydrophobicity of the medium increases, this ratio starts decreasing due to the interaction of pyrene molecules with the micellar aggregates.64 Direct and exact determination of cmc is not possible by using pyrene fluorescence probe method64 however its sensitiveness gives sufficiently good results.63 In a study by Akram et al.,33 pyrene probe method has also been used to study the micropolarity of HEWL in the presence of different concentration of ligand. In the present study, pyrene probe method was used to analyze the alteration in the cmc value of respective surfactants in presence and absence of HEWL. It was also used to monitor the interaction of M16 and G16 in presence and absence of HEWL respectively.It can be clearly seen from Fig. 10, in the presence of HEWL more decrease in the I1/I3 ratio curve was observed as compared to the decrease observed in an aqueous medium for both the surfactants. This observed decrease was comprehended to the increase in the hydrophobicity of the medium as discussed above. Non-polar pyrene molecule binds at the hydrophobic core of the HEWL through various weak interactions. The increase in concentration of M16 and G16 in the medium causes dislodging of pyrene molecules from the hydrophobic core of HEWL. This dislodging of pyrene molecule indicates interaction of M16 and G16 to the hydrophobic core of HEWL. After this stage, the dislodged pyrene starts residing within the micelle like aggregates of M16 and G16 as detailed by De et al.65 This can be clearly seen by the sharp decrease in the I1/I3 ratio. With further increase in M16 and G16 concentration, the I1/I3 ratio reached a constant value which indicates strong interaction of M16 and G16 with HEWL as compared to pyrene which is present in a constant environment. This observation suggests the complex formation of M16 and G16 with HEWL respectively and such results have also been reported earlier.61
 | ||
| Fig. 10 Variation in I1/I3 of pyrene as a function of (a) M16 concentration (1.98 × 10−2 to 3.30 × 10−1 mM) and (b) G16 (0.34–5.44 μM) in absence and presence of HEWL. | ||
In the absence of HEWL, pyrene senses the polar environment around it which can be observed through higher I1/I3 ratio. The decrease in the I1/I3 ratio is attributed to the solubilization of pyrene molecule in the hydrophobic part of the surfactant micelle as mentioned above.
The cmc values of M16 and G16 in presence and absence of HEWL were summarized in Table S4.† The cmc value of M16 and G16 in the absence of HEWL was found to be 2.40 × 10−4 M and 3.63 × 10−6 M which are in good agreement with the reported values.34,37 The cmc value of G16 was found to be much lower as compared to M16. The low cmc value of G16 as compared to its single chain counterpart can be attributed to the fact that it has two C2H4OH group as compared to the single C2H4OH group in M16. This causes more hydrogen bond formation with water and with oxygen atom present in the head group. This results in the screening of repulsive forces between the head groups, which imposes the aggregate formation between them.34 Also, due to the characteristic properties of gemini surfactants, that is, two long hydrophobic chains and a spacer groups contributes to G16 lower cmc value. In the presence of HEWL, the cmc of M16 and G16 increased to 2.60 × 10−4 M and 3.91 × 10−6 M. The delayed in cmc for both the cases in the presence of HEWL can be attributed to the obvious prior interaction of surfactants with the HEWL, after which they start forming micelles.
3.6 Antibacterial activity assay of HEWL
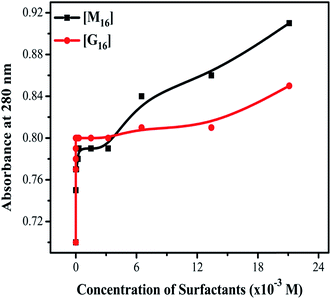
Activity profile of HEWL was determined by measuring the decrease in turbidity of Micrococcus luteus cells (cell lysis) in the presence of HEWL at 450 nm using UV-visible spectroscopy. The main residues which are involved in HEWL catalytic mechanism are Asp52 and Glu35. Glu35 execute proton donation and Asp52 work as a nucleophile that produces glycosyl intermediates. Trp62, Trp63 and Trp108 are also present in the substrate binding site and are sensitive to the microenvironment around them. This increases the significance of studying these residues residing in the active site of HEWL for monitoring the changes around the active site at different conditions.The % RA of HEWL at different concentration of M16 and G16 was calculated by using eqn (6) and the values thus obtained were reported in Table S5.† In the case of the interaction of HEWL with M16 (Fig. 11(a)), with an increase in M16 concentration from 8.26 × 10−8 M to 3.63 × 10−6 M % RA (relative activity) of HEWL was decreased by ∼20%. This remained almost constant till 2.10 × 10−4 M and then further decreased by ∼25% at 3.00 × 10−4 M concentration of M16. The % RA drastically decreased by 64% at 3.20 × 10−3 M concentration. In the case of G16 (Fig. 11(b)), ∼61% activity was observed by HEWL in comparison to untreated HEWL at 8.26 × 10−8 M G16 concentration. It gradually increases from ∼66% to 117% in the presence of 3.0 × 10−4 M G16 concentration. After the above mentioned concentrations of respective surfactants, no antibacterial activity was observed.
 | ||
| Fig. 11 Relative activity profile of HEWL as a function of increased concentrations of (a) M16 and (b) G16 (error bars represent 5% relative error). | ||
In reviewing the literature it has been mentioned that activity of the surface active enzyme is dependent upon the surface per area of the enzyme with the surface of contact (in the present case it is bacterial membrane). HEWL is a surface active protein which acts on the surface of the bacterial membrane. It has been reported that HEWL even shows antibacterial activity at denatured conditions.66 Ibrahim et al.67 have illustrated the antibacterial activity of mutant and heat denatured lysozyme and confirmed that it is independent of its catalytic function. This in turn also proves that the antibacterial activity to some extent dependent on the structural components of this enzyme.
In the case of M16, its binding at the hydrophobic core of HEWL significantly affects its antibacterial activity in its monomeric and micellar concentrations, however, more pronounced effect was observed at micellar concentrations. Whereas, G16 substantially decreases the activity of HEWL at its monomeric concentrations, however, the activity was found to get gradually increased at its higher micellar concentration. Celej et al.12 reported that micelles of cationic surfactant like CTABr substantially increases the enzymatic activity of α-chymotrypsin by stabilizing its secondary and tertiary structure. As by looking at the far-UV CD results (at concentration 3.00 × 10−4 M) it can be concluded that increase in the antibacterial activity by G16 is not due to its stabilization effect on the secondary structure of HEWL. Apparently, this suggests that increased HEWL activity by G16 as compared to M16, is probably due to increased surface activity of HEWL by G16, nevertheless, these possibilities deserves further investigations. Also, such explanations have been previously mentioned by Dasgupta et al.68 and Mitra et al.69 As already mentioned in the introductory section, lysozyme has majorly α-helix in its secondary structure present in the α-domain (Fig. 12: a-domain). The active site of the protein lies in between the two domains26 (Fig. 12). It seems that, at very high concentration these micellar structures strongly interfere with the overall integrity of the two domains and due to this no activity were observed at very high G16 and M16 concentrations. Thus, decrease in secondary structure; considerably affect the activity of HEWL. Such results has also been reported by Li et al.13 and are further confirmed from our study.
3.7 Molecular docking studies
Docking studies were done to visualize and identify the possible binding sites in the complex formation between M16 and G16 with HEWL respectively. The docked conformation with lowest binding energy was selected. In the case of M16, it was found to be −20 kJ mol−1 and in G16 it was −17.99 kJ mol−1, where, Akram et al.33 reported docking of gemini surfactants with very high binding energy. The negative sign shows spontaneity of the interaction. Through the binding energies it can be concluded that G16 binds slightly weaker with HEWL, as also inferred in the above discussions. Binding of G16 and M16 with HEWL was found to occur in the HEWL principle binding region lies between the two domains as can be seen in Fig. 12 (a (α) and b (β) domains) and also reported by Akram et al.33Leu75, Asn103, Ile98, Ala107, Asp52, Gln57, Ile58, Asn59, Trp62, Trp63 and Asp101 strongly influencing the M16-HEWL binding complex majorly through van der Waal's interactions and various electrostatic and hydrophobic interactions (Fig. 13). The weak hydrophobic pi–alkyl interaction was found between methyl group in the tail region of M16 and Trp63 residue (6.86 Å distance). Trp62 and Trp63 were found to strongly influence the complex through van der Waal's and hydrophobic interaction as the hydrophobic tail was also found to be in close proximity with these residues. One carbon hydrogen bond (5.73 Å distance) was also observed to form between Ala107 and methyl group present in the head region of the M16.
As can be seen in Fig. 14, binding of G16 with HEWL includes various weak interactions with Thr47, Asn46, Asp52, Trp108, Gln57, Asn59, Ile98, Ile58, Trp62, val109, Ala107, Asn103, Asp101, Trp63, Arg112 and Asn106 residues out of which van der Waal's interactions playing a significant role. The nitrogen group of G16 is making electrostatic attractive interaction with the negatively charge Asp52 residues with a bond distance of 7.24 Å. As already mentioned, Asp52 is the major residue which acts as a nucleophile and forms glycosyl intermediate during the catalytic process. Due to this electrostatic interaction HEWL antibacterial activity tremendously affected when it interacts with the monomeric form of G16. Also, it can be clearly seen that one hydrogen bond was observed to form between Asn46 (at a distance of 4.06 Å) and hydroxyl group (shown as a red stick) present in the head region of G16 molecule. One other carbon hydrogen bond is also forming between Asn106 (at a distance of 4.99 Å) and methyl group present at the head region of G16 molecule. The hydrophobic tail of the G16 was found to be in close proximity with Trp63 and distant from the two major fluorophores Trp62 and Trp108. This may be attributed to low changes in absorbance as well as emission intensities observed in the spectroscopic experiments of the G16–HEWL complex.
4. Conclusions
We have brought in the difference between the interaction mechanism of cationic gemini surfactant and its monomeric counterpart with HEWL using standard spectroscopic techniques and molecular docking method. From the study, it can be concluded that surfactants used in the work have considerable effect on the structural stability of HEWL. The G16 was more evident in stabilizing the secondary (at higher micellar concentrations) as well as tertiary structure of HEWL at considerably high cmc value. At high G16 concentration apparently, the native HEWL becomes less solvent exposed may be due to aggregation and it can be said that protein might have adopted an unusual structural moiety where it has a compact tertiary structure with less pronounced helical content. On the other hand, M16 at higher concentrations affects the secondary as well as tertiary structure of the protein. This discrepancy in the effect of respective surfactant on HEWL conformation is attributed to the difference in the physiochemical properties of both the surfactants. These distinguished properties of such surfactants can prove to be useful in their applicability in different industries (cosmetics, food, feed). The detailed account on the time-resolved analysis of different τ values of fluorophores in HEWL on interaction with surfactants is new addition to the literature, nonetheless strewed information and associations are available in some reports. The antibacterial activity was also found to increase in the presence of gemini surfactant where single chain surfactant was found to decrease it substantially. Thus, this study can provide a better insight for the usage of this enzyme in enhancing its role in food preservation as well as a more efficient antibiotic. For future prospective more in vitro as well as in vivo studies are required for the above mentioned possible significance of this study.Acknowledgements
Dr Rajan Patel greatly acknowledges the financial supports from Science and Engineering Research Board (EEQ/2016/000339 and SB/EMEQ-097/2013) and University Grant Commission New Delhi, India (F. No. 39-841/2010 (SR)). Dr Abbul Bashar Khan is thankful to Science and Engineering Research Board (SERB), New Delhi for providing research grant with Sanction Order No. SB/FT/CS- 031/2013. Authors also thank DST for providing the FIST grant with Sanction Order No. (SR/FIST/LS-541/2012).References
- D. E. Otzen, Biochim. Biophys. Acta, 2011, 1814, 562–591 CrossRef CAS PubMed.
- C. Tanford, Y. Nozaki, J. A. Reynolds and S. Makino, Biochemistry, 1974, 13, 2369–2376 Search PubMed.
- T. E. Creighton, Curr. Opin. Struct. Biol., 1991, 1, 5–16 CrossRef CAS.
- A. Hvidt and P. Westh, J. Solution Chem., 1998, 27, 395–402 CrossRef CAS.
- A. Yonath, A. Podjarny, B. Honig, A. Sielecki and W. Traub, Biochemistry, 1977, 16, 1418–1424 CrossRef CAS PubMed.
- J. A. Reynolds and C. Tanford, J. Biol. Chem., 1970, 245, 5161–5165 CAS.
- D. E. Otzen, Biophys. J., 2002, 83, 2219–2230 CrossRef CAS PubMed.
- N. Gull, P. Sen and R. H. Khan, J. Biochem., 2007, 141, 261–268 CrossRef CAS PubMed.
- N. Gull, S. Kumar, B. Ahmad and R. H. Khan, Colloids Surf., B, 2006, 51, 10–15 CrossRef CAS PubMed.
- M. N. Jones, H. A. Skinner, E. Tipping and A. Wilkinson, Biochem. J., 1973, 135, 231–236 CrossRef CAS PubMed.
- A. Few, R. Ottewill and H. Parreira, Biochim. Biophys. Acta, 1955, 18, 136–137 CrossRef CAS.
- M. S. Celej, M. G. D. Andrea, P. T. Campana, G. D. Fidelio and M. L. Bianconi, Biochem. J., 2004, 378, 1059–1066 CrossRef CAS PubMed.
- Q. Li, T. Zhai, K. Du, Y. Li and W. Feng, Colloids Surf., B, 2013, 112, 315–321 CrossRef CAS PubMed.
- R. C. Lu, J. X. Xiao, A. N. Cao, L. H. Lai, B. Y. Zhu and G. X. Zhao, Biochim. Biophys. Acta, 2005, 1722, 271–281 CrossRef CAS PubMed.
- M. L. Guzman, M. R. Marques, M. E. O. Me and E. S. Stippler, Results Pharma Sci., 2016, 6, 15–19 CrossRef PubMed.
- R. Amiri, A. K. Bordbar and D. V. Laurents, J. Phys. Chem. B, 2014, 118, 10633–10642 CrossRef CAS PubMed.
- R. Amiri, A. K. Bordbar, D. V. Laurents, A. R. Khosropour and I. Mohammadpoor-Baltork, Int. J. Biol. Macromol., 2012, 50, 1151–1157 CrossRef CAS PubMed.
- N. Gull, P. Sen and R. H. Khan, Langmuir, 2009, 25, 11686–11691 CrossRef CAS PubMed.
- Y. Li, M. Cao and Y. Wang, J. Phys. Chem. B, 2006, 110, 18040–18045 CrossRef CAS PubMed.
- N. Gull, P. Sen and R. H. Khan, J. Biochem., 2009, 145, 67–77 CrossRef CAS PubMed.
- Y. Han and Y. Wang, Phys. Chem. Chem. Phys., 2011, 13, 1939–1956 RSC.
- M. A. Mir, J. M. Khan, R. H. Khan, G. M. Rather and A. A. Dar, Colloids Surf., B, 2010, 77, 54–59 CrossRef CAS PubMed.
- R. Patel, M. U. H. Mir, U. K. Singh, I. Beg, A. Islam and A. B. Khan, J. Colloid Interface Sci., 2016, 484, 205–212 CrossRef CAS PubMed.
- R. Patel, M. ud din Parray, U. K. Singh, A. Islam, P. Venkatesu, S. Singh and H. B. Bohidar, Colloids Surf., A, 2016, 508, 150–158 CrossRef CAS.
- F. Chiti and C. M. Dobson, Annu. Rev. Biochem., 2006, 75, 333–366 CrossRef CAS PubMed.
- A. Sethuraman and G. Belfort, Biophys. J., 2005, 88, 1322–1333 CrossRef CAS PubMed.
- N. A. Fazili, W. F. Bhat and A. Naeem, Int. J. Biol. Macromol., 2014, 64, 36–44 CrossRef CAS PubMed.
- B. Mandal, S. Mondal, A. Pan, S. P. Moulik and S. Ghosh, Colloids Surf., A, 2015, 484, 345–353 CrossRef CAS.
- M. D. Lad, V. M. Ledger, B. Briggs, R. J. Green and R. A. Frazier, Langmuir, 2003, 19, 5098–5103 CrossRef CAS.
- K. N. Vennila and D. Velmurugan, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2011, 67, 1662–1665 CrossRef CAS PubMed.
- A. Chatterjee, S. Moulik, P. Majhi and S. Sanyal, Biophys. Chem., 2002, 98, 313–327 CrossRef CAS PubMed.
- W. Gospodarczyk and M. Kozak, Colloid Polym. Sci., 2015, 293, 2855–2866 CAS.
- M. Akram, I. A. Bhat and D. Kabir ud, RSC Adv., 2015, 5, 102780–102794 RSC.
- D. Tikariha, N. Singh, M. L. Satnami, K. K. Ghosh, N. Barbero and P. Quagliotto, Colloids Surf., A, 2012, 411, 1–11 CrossRef CAS.
- M. Borse, V. Sharma, V. Aswal, N. K. Pokhriyal, J. V. Joshi, P. S. Goyal and S. Devi, Phys. Chem. Chem. Phys., 2004, 6, 3508–3514 RSC.
- R. Steiner, Biochim. Biophys. Acta, 1964, 79, 51–63 CAS.
- S. S. Borse and T. J. Patil, J. Chem. Pharm. Res., 2014, 6, 904–911 CAS.
- J. R. Lakowicz, Principles of fluorescence spectroscopy, Springer Science & Business Media, 2013 Search PubMed.
- Q. Yue, T. Shen, C. Wang, C. Gao and J. Liu, Int. J. Spectrosc., 2012, 2012, 1–9 CrossRef.
- K. Kalyanasundaram and J. K. Thomas, J. Am. Chem. Soc., 1977, 99, 2039–2044 CrossRef CAS.
- M. Yan, B. Li and X. Zhao, Food Chem., 2010, 122, 1333–1337 CrossRef CAS.
- J. Tian, Y. Zhao, X. Liu and S. Zhao, Luminescence, 2009, 24, 386–393 CAS.
- G. C. Chen and J. T. Yang, Anal. Lett., 1977, 10, 1195–1207 CrossRef CAS.
- N. J. Greenfield and G. D. Fasman, Biochemistry, 1969, 8, 4108–4116 CrossRef CAS PubMed.
- D. Shugar, Biochim. Biophys. Acta, 1952, 8, 302–309 CrossRef CAS.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS PubMed.
- N. C. Price, Biotechnol. Appl. Biochem., 2000, 31, 29–40 Search PubMed.
- Y. Wang, R. Guo and J. Xi, J. Colloid Interface Sci., 2009, 331, 470–475 CrossRef CAS PubMed.
- G. D. Fasman, in Circular Dichroism and the Conformational Analysis of Biomolecules, Springer, 1996, pp. 381–412 Search PubMed.
- T. Knubovets, J. J. Osterhout and A. M. Klibanov, Biotechnol. Bioeng., 1999, 63, 242–248 CrossRef CAS PubMed.
- W. Zhu and T. A. Keiderling, Biochim. Biophys. Acta, 2013, 1834, 593–600 CrossRef CAS PubMed.
- A. Ratnaparkhi, S. A. Muthu, S. M. Shiriskar, R. R. Pissurlenkar, S. Choudhary and B. Ahmad, J. Biomol. Struct. Dyn., 2015, 33, 1866–1879 CAS.
- S. Venkataramani, J. Truntzer and D. R. Coleman, J. Pharm. BioAllied Sci., 2013, 5, 148 CrossRef PubMed.
- J. M. Beechem and L. Brand, Annu. Rev. Biochem., 1985, 54, 43–71 CrossRef CAS PubMed.
- U. Anand and S. Mukherjee, Biochim. Biophys. Acta, 2013, 1830, 5394–5404 CrossRef CAS PubMed.
- Y. Li, X. Wang and Y. Wang, J. Phys. Chem. B, 2006, 110, 8499–8505 CrossRef CAS PubMed.
- B. Mandal, S. Ghosh and S. Moulik, New J. Chem., 2016, 40, 4617–4624 RSC.
- C. Rmoso and L. S. Forster, J. Biol. Chem., 1975, 250, 3738–3745 Search PubMed.
- U. Anand and S. Mukherjee, Phys. Chem. Chem. Phys., 2013, 15, 9375–9383 RSC.
- J. Wang, X. Yang, J. Wang, C. Xu, W. Zhang, R. Liu and W. Zong, New J. Chem., 2016, 40, 3738–3746 RSC.
- M. Kumari, J. K. Maurya, M. Tasleem, P. Singh and R. Patel, J. Photochem. Photobiol., B, 2014, 138, 27–35 CrossRef CAS PubMed.
- G. Bains, A. B. Patel and V. Narayanaswami, Molecules, 2011, 16, 7909–7935 CrossRef CAS PubMed.
- K. Ananthapadmanabhan, E. Goddard, N. Turro and P. Kuo, Langmuir, 1985, 1, 352–355 CrossRef PubMed.
- M. Vasilescu, D. Angelescu, M. Almgren and A. Valstar, Langmuir, 1999, 15, 2635–2643 CrossRef CAS.
- S. De, A. Girigoswami and S. Das, J. Colloid Interface Sci., 2005, 285, 562–573 CrossRef CAS PubMed.
- W. Carrillo, A. Garcia-Ruiz, I. Recio and M. V. Moreno-Arribas, J. Food Prot., 2014, 77, 1732–1739 CrossRef CAS PubMed.
- H. R. Ibrahim, T. Matsuzaki and T. Aoki, FEBS Lett., 2001, 506, 27–32 CrossRef CAS PubMed.
- A. Dasgupta, D. Das, R. N. Mitra and P. K. Das, J. Colloid Interface Sci., 2005, 289, 566–573 CrossRef CAS PubMed.
- R. N. Mitra, A. Dasgupta, D. Das, S. Roy, S. Debnath and P. K. Das, Langmuir, 2005, 21, 12115–12123 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra00172j |
| This journal is © The Royal Society of Chemistry 2017 |







