Open Access Article
Open Access ArticleConformational stability, spectroscopic signatures and biological interactions of proton pump inhibitor drug lansoprazole based on structural motifs†
Megha Agrawala,
Amit Kumarab and
Archana Gupta *a
*a
aDepartment of Applied Physics, Faculty of Engineering & Technology, M. J. P. Rohilkhand University, Bareilly, India. E-mail: drarchana.physics@gmail.com
bDepartment of Physics, University of Lucknow, Lucknow, India
First published on 25th August 2017
Abstract
Structure based biological and chemical properties of Lansoprazole (LSP) have been studied by spectroscopic and quantum chemical methods. The geometrical parameters of the title compound obtained by DFT calculation are compared with single crystal XRD data. The conformational flexibility of the title compound has been discussed on the basis of the potential energy profile achieved from the rotation of various groups present in the molecule and the minimum energy conformer has been determined. The spectroscopic fingerprints are studied by variety of experiments (IR, Raman, UV, and NMR). Normal mode analysis is performed to assign the vibrational frequencies according to the potential energy distribution (PED). Simulation of infrared and Raman spectra has led to an excellent overall agreement with the observed spectral patterns by refinement of scale factors. TD-DFT approach is used to investigate the excited states of molecule and prediction of electronic absorption spectra. The 1H nuclear magnetic resonance (NMR) chemical shifts of the molecule are calculated by the gauge independent atomic orbital (GIAO) method and compared with experimental results. Prediction of Activity Spectra analysis (PASS) of the title compound has been done to explore several biological and toxic effects with high probability. The lipophilicity and aqueous solubility have been calculated to get insight into cell membrane penetration and drug absorption processes in biological systems. The molecular docking is performed to identify the binding energy of the ligand with the active site of protein. These studies show that several sites in the molecule are crucial for bonding and these results lead us to the conclusion that the compound might be metabolized by human protein. Electron density-based local reactivity descriptors such as Fukui functions have been calculated to describe the chemical reactivity sites within the molecule.
1. Introduction
Drug design is the inventive process of finding new medications based on the knowledge of a biological target. The ever increasing use of drugs has prompted pharmaceutical scientists to explore various approaches in finding and developing bioactive compounds suitable for the treatment of diseases and often for the maintenance of human health. During the pre-formulation stage of drug development, a great number of characterization methodologies can be employed and each has its associated utility and function. However when a large amount of information is to be gleaned from a small amount of sample, spectroscopic methods of analysis can be of great use. Our aim is to study the biological and chemical properties of the pharmaceutical sample lansoprazole using different spectroscopic techniques. Lansoprazole, a substituted benzimidazole belongs to a group of drugs called proton pump inhibitors. The benzimidazole scaffold is an accepted pharmacophore and represents an important synthetic precursor in new drug discovery.1–3 Lansoprazole suppresses gastric acid secretion by specific inhibition of the H+/K+ ATPase enzyme system at the secretary surface of the gastric parietal cell.4 An increased dose of lansoprazole is to be recommended for the extensive metabolizer (particularly homEM) patients who are refractory to the usual dose of a proton pump inhibitor.5 Literature survey reveals that no experimental and theoretical spectral analyses for LSP molecule have been performed yet. A study on spectral characterization, conformational distribution, chemical reactivity and biological interactions analysis will be useful in predicting several properties of compound. The spectroscopic techniques used here are FT-IR, Raman, UV-vis and NMR spectroscopy. Experimental spectroscopic features have been correlated with the theoretical results obtained from density functional theory (DFT) computations. The thermodynamically most stable configuration of the compound has been analyzed and the structure based properties are determined. The prospective biological activities and toxicity are evaluated by the predictions made by PASS software which predicts pharmacological effects on the basis of structural formula. As the biological activity of a drug compound is affected by the ability of the drug to be transported and absorbed, the lipophilicity and aqueous solubility were calculated. The study of drug–target interactions is important for the understanding of biological processes and for the efforts to develop clinical performance of drug molecules. Docking simulations have been done to determine the aspects of drug–target interactions. The global reactivity indices are calculated for interpreting and predicting diverse aspects of chemical bonding and reaction mechanism whereas local reactivity indices are employed to probe site selectivity of molecule.2. Experimental details
The compound under investigation is purchased from Sigma-Aldrich chemicals, USA with spectroscopic grade and it is used as such without any further purification for recording the spectra.2.1 Fourier transform infrared spectroscopy
The Infrared spectrum was recorded in the solid phase in the region 4000–400 cm−1 on Bruker FT-IR spectrometer, model ALPHA equipped with Diamond ATR module by averaging 24 scans with spectral resolution of 4 cm−1.2.2 Fourier transform Raman spectroscopy
The Raman spectrum was recorded in the region 4000–100 cm−1 on a BRUKER RFS-100/S FT-Raman spectrometer. A Nd:YAG laser operating at 1064 nm with an output of 30 mW was used as the exciting source. The detector was a Ge diode cooled to liquid nitrogen temperature. 64 scans were accumulated at 4 cm−1 resolution.2.3 UV-vis spectroscopy
The ultraviolet absorption spectrum of LSP was examined using a Shimadzu UV 1800 Spectrophotometer. The UV pattern is recorded from a 10−5 M solution prepared by dissolving LSP in ethanol solvent. The spectrum is shown in Fig. S1 of the ESI.†2.4 NMR spectroscopy
The 1H NMR spectrum was recorded on a Bruker DRX-300 spectrometer operating at 300 MHz. The measurements were done in CDCl3 solution. The solution was prepared by dissolving about 2 mg of the sample in 0.6 mL of CDCl3. The spectrum has following experimental parameters: number of scans 32; spectral width 9615.385 Hz; acquisition time 3.40 s. Chemical shifts were reported in ppm relative to TMS. The spectrum is shown in Fig. S2 of the ESI.†3. Computational details
The molecular geometry of the compound was optimized by DFT method with a hybrid functional B3LYP (Becke's three parameter hybrid functional using the LYP correlation functional) supplemented with standard 6-311++G(d,p) basis set. The way the functional is derived and the lack of empirical parameters fitted to specific properties make this functional widely applicable, both to quantum chemistry as well as to condensed matter physics. It has shown to predict a wide range of molecular properties successfully.6–8The equilibrium geometry corresponding to the true minima on the Potential Energy Surface (PES) has been obtained by solving self consistent field equation effectively. The optimized structural parameters were used in the vibrational frequency calculations. The entire calculations were performed using Gaussian 09W9 program package, invoking gradient geometry optimization.10 Due to systematic errors caused by basis set incompleteness, neglect of electron correlation and vibrational anharmonicity effects, the calculated harmonic vibrational frequencies were scaled by the wavenumber linear scaling (WLS) procedure of Yoshida et al.11
| νobs = (1.0087 − 0.0000163νcal)νcal cm−1 | (1) |
The WLS method employs essentially only one parameter to scale wavenumbers in the whole region and is applicable to a variety of compounds, except the ones for which the effect of dispersion forces is significant.11 It has excellently reproduced the experimental wavenumbers of pharmaceutical compounds studied by authors previously.12–14 Time dependent density functional theory (TD-DFT)15 is used to find the low-lying excited states and their nature within molecule. The solvent effects are taken care of by means of the polarizable continuum model (PCM)16 as there is no specific interaction, such as hydrogen bonds, between the solute and the solvent.17 The Gaussian software generated output file for NMR calculations using guage-independent atomic orbital (GIAO) approach18 provides the computation of absolute chemical shielding due to the electronic environment of the individual nuclei. In order to obtain the calculated results comparable with the experimental data, the absolute shielding returned by the program is transformed to chemical shift by subtracting it from the absolute shielding of tetramethylsilane (TMS).
| δrel = δTMSabs − δabs | (2) |
To predict biological activity of LSP, an online structure activity relationship (SAR) tool ‘PASS’ (Prediction of Activity Spectra)19 has been used. Lipophilicity and aqueous solubility of LSP has been calculated using ALGOPS 2.1 program.20 This program combines electronic and topological characters to predict the lipophilicity and aqueous solubility of the analyzed molecule. In order to get an insight into ligand–receptor interactions, molecular docking calculations were performed using Auto Dock Vina software21 and Discover studio 4.0 Software.22
4. Results and discussion
4.1 Structural analysis
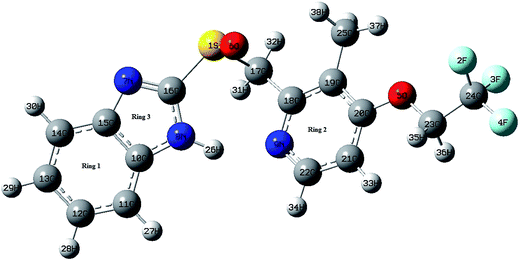
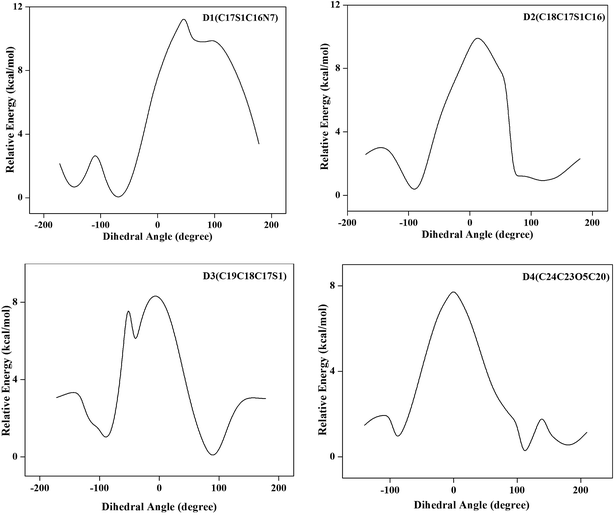
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) bond lengths. Similar observations are also found for C–N bond lengths in the benzimidazole ring. The replacement of C by N in pyridine ring distorts the hexagonal symmetry, and the ring angle C18–N9–C22 becomes 118.389° rather than 120°. Also the ring carbon atom exerts a large attraction on valence electron cloud of the nitrogen atom resulting in an increase in C–N force constant and a decrease in the corresponding bond length. Trifluoroethoxy group is almost coplanar with pyridine ring with dihedral angles C19–C20–O5–C23, C20–O5–C23–C24 and O5–C23–C24–F4 having values 178.154°, 179.617°, and 179.743° respectively. The DFT calculations give shortening of angle C19–C20–O5 by 3.734° and an enhancement of angle C21–C20–O5 by 3.474° from 120°; this asymmetry of exocyclic angles reveals the attraction between the methyl group and O5 atom of trifluoroethoxy group. The shortening of C20–O5 bond length (1.363 Å) is mainly due to the conjugation interaction between the pyridine ring and the strong electron withdrawing trifluoroethoxy group. The bond length of S1–C17 (1.907 Å) is greater than the bond length S1–C16. Elongation of S1–C17 bond is due to the induction effect of attachment of electron donating methylene group. In benzimidazole ring, the bond angle N7–C16–N8 is the largest (114.415°). This may be due to the electron donating substitution at C16 atom. Closer examination of the bond lengths relative to the XRD values shows the larger deviation in the bond length of N8–H26 and S1–O6 because of intramolecular N–H⋯N hydrogen bonding between the benzimidazole and pyridine ring of same molecule and intermolecular N–H⋯O hydrogen bonding between benzimidazole ring and S–O group of adjacent molecules. These intra and intermolecular bondings are shown in Fig. S4 of ESI.†
N) bond lengths. Similar observations are also found for C–N bond lengths in the benzimidazole ring. The replacement of C by N in pyridine ring distorts the hexagonal symmetry, and the ring angle C18–N9–C22 becomes 118.389° rather than 120°. Also the ring carbon atom exerts a large attraction on valence electron cloud of the nitrogen atom resulting in an increase in C–N force constant and a decrease in the corresponding bond length. Trifluoroethoxy group is almost coplanar with pyridine ring with dihedral angles C19–C20–O5–C23, C20–O5–C23–C24 and O5–C23–C24–F4 having values 178.154°, 179.617°, and 179.743° respectively. The DFT calculations give shortening of angle C19–C20–O5 by 3.734° and an enhancement of angle C21–C20–O5 by 3.474° from 120°; this asymmetry of exocyclic angles reveals the attraction between the methyl group and O5 atom of trifluoroethoxy group. The shortening of C20–O5 bond length (1.363 Å) is mainly due to the conjugation interaction between the pyridine ring and the strong electron withdrawing trifluoroethoxy group. The bond length of S1–C17 (1.907 Å) is greater than the bond length S1–C16. Elongation of S1–C17 bond is due to the induction effect of attachment of electron donating methylene group. In benzimidazole ring, the bond angle N7–C16–N8 is the largest (114.415°). This may be due to the electron donating substitution at C16 atom. Closer examination of the bond lengths relative to the XRD values shows the larger deviation in the bond length of N8–H26 and S1–O6 because of intramolecular N–H⋯N hydrogen bonding between the benzimidazole and pyridine ring of same molecule and intermolecular N–H⋯O hydrogen bonding between benzimidazole ring and S–O group of adjacent molecules. These intra and intermolecular bondings are shown in Fig. S4 of ESI.†
In the benzene ring the bond lengths C10–C15 (1.417 Å), C12–C13 (1.411 Å), C14–C15 (1.400 Å) are appreciably greater than other C–C bonds (C10–C11/C11–C12/C13–C14 = 1.397/1.388/1.387 Å). The internal bond angles C10–C11–C12 = 116.756° and C13–C14–C15 = 117.847° are consistently smaller than the normal bond angle of the phenyl ring. These are due to the intramolecular charge transfer from N7 and N8 to the ring system.
4.2 Spectral analysis
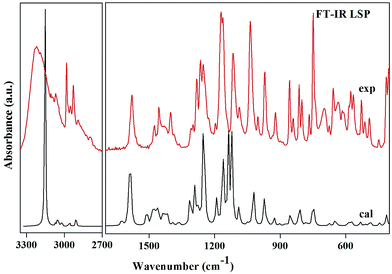
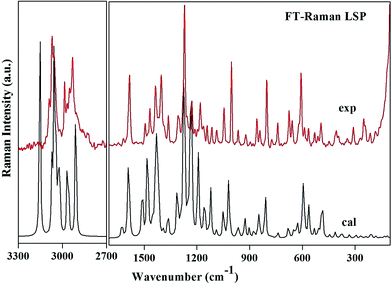
Region 3500–3000 cm−1. This is the usual range for appearance of N–H and C–H stretching vibrations. The position of absorption in this region depends upon the degree of hydrogen bonding and the physical state of the sample.27 The band observed at 3224 cm−1 in the IR spectrum corresponds to N–H stretching mode. This vibration is calculated at 3154 cm−1 and shows deviation from the observed band. The mismatch between the observed and the calculated wavenumbers is due to the involvement of this group in intra N–H⋯N as well as intermolecular hydrogen bonding N–H⋯O. The bands observed at 3071 in the IR and at 3068 in the Raman spectra are assigned to C–H stretching vibrations of the pyridine ring. These bands computed at 3072 cm−1, 3024 cm−1 and 3023 cm−1 shows good agreement with observed bands as well as with the literature.28 The bands in this region are not much affected due to the nature and position of the substitution.29 The methyl group C–H stretching vibrations occur at lower wavenumbers than those of aromatic ring.30 The asymmetric and symmetric C–H stretching modes are expected in the regions around 2980 cm−1, 2870 cm−1 respectively.30 In the present investigation, the medium bands at 2986 cm−1 in both FT-IR and FT-Raman and a weak band at 2932 cm−1 in FT-Raman corresponds to asymmetric stretching mode. A very weak FT-IR band at 2929 cm−1 and a medium FT-Raman band at 2932 cm−1 are assigned to CH3 symmetric stretching vibrations. The methylene asymmetric and symmetric C–H stretching vibrations are generally observed in regions 3000–2900 cm−1 and 2900–2800 cm−1 respectively. The methylene vibrational band positions are most consistent, when this group is attached to other carbon atoms. The spectral analysis reveals that the CH2 symmetric stretching modes are shifted to higher wavenumbers and appear at 2909 cm−1 and 2957 cm−1. As both the methylene C17H2 and C23H2 groups are attached to sulphur and oxygen respectively, so the bands shift their position due to electronic effects.
1650–1500 cm−1. The C–C stretching modes of benzimidazole and pyridine rings appear as sharp bands with medium to strong intensity in both FT-IR and FT-Raman spectra in the region 1635–1100 cm−1, which is the allowed region for these vibrations.27,29 These vibrations are mixed with C–H in plane bending vibrations of the corresponding aromatic ring. The actual positions are determined not so much by the nature of the substituents but by the form of substitution around the ring.31 The pyridine ring shows a strong absorption band in the region 1600–1500 cm−1 due to the C–N stretching vibrations.29 In the title compound a strong band observed at 1593 cm−1 is assigned to this mode. The wavenumber of this mode at the top end of the allowed region of the spectrum may be attributed to the substitution in the ring. Other C–N and C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N vibrations appear in lower range and are discussed below.
N vibrations appear in lower range and are discussed below.
1500–1200 cm−1. The asymmetric and symmetric deformations of the methyl group are expected in the regions 1480–1390 cm−1 and 1390–1340 cm−1 (ref. 32) respectively. The asymmetric deformation vibrations of LSP are observed in the accepted region and are in agreement with the calculated values as well as the literature values.32 The intense band at 1402 cm−1 in both IR and Raman spectra can be attributed to the symmetrical umbrella mode of the methyl group which is calculated at 1391 cm−1 (with PED 85%) by the DFT calculations. This mode may contribute to the linear electro-optic effect in LSP. The influence of the CH3 umbrella vibration can be understood in terms of in-phase displacement of the three H atoms which changes the electronic environment for the donor and the acceptor of the chromophore and results in modification in the polarizability. Molecular polarizability of a molecule characterizes the capability of the electronic system to be distorted by the external field, and plays an important role in modeling many molecular properties and biological activities.33 Highly polarizable molecules may be expected to have strong attractions with other molecules. The polarizability of a molecule can also enhance aqueous solubility. The relatively large values of IR and Raman intensities of this mode suggest a large positive charge localized on hydrogen, which further supports presence of hyperconjugation. The identification of wavenumber for C–N and C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching vibrations is rather difficult since the mixing of vibrations is possible in this region; so there are problems in differentiating those wavenumbers from others. Pinchas et al.34 assigned the C–N stretching band at 1368 cm−1 in benzamide. Kahovec and Kohlreusch35 identified the stretching wavenumber of the C
N stretching vibrations is rather difficult since the mixing of vibrations is possible in this region; so there are problems in differentiating those wavenumbers from others. Pinchas et al.34 assigned the C–N stretching band at 1368 cm−1 in benzamide. Kahovec and Kohlreusch35 identified the stretching wavenumber of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N band in salicylic aldoxime at 1617 cm−1. Referring to the above workers, C
N band in salicylic aldoxime at 1617 cm−1. Referring to the above workers, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C–N stretching modes of both the rings are identified in the range 1480–1440 cm−1 and 1300–1190 cm−1 respectively. These modes are mixed with methyl asymmetric deformation and methylene out of plane bending modes respectively. The CF3 group possesses as many normal vibrations as methyl. However, the halogen atoms are much heavier than hydrogen; hence CF3 modes appear at considerably lower values than those of methyl. Usually symmetric and antisymmetric C–F stretching vibrations are in the ranges 1290–1235 and 1226–1200 cm−1 respectively. The bands of the C–F symmetric and antisymmetric stretching vibrations have been observed to give rise to extremely intense FT-IR absorption and rather weak Raman bands and these appear in the range 1300–850 cm−1. These vibrations are coupled with the several other vibrations.
N and C–N stretching modes of both the rings are identified in the range 1480–1440 cm−1 and 1300–1190 cm−1 respectively. These modes are mixed with methyl asymmetric deformation and methylene out of plane bending modes respectively. The CF3 group possesses as many normal vibrations as methyl. However, the halogen atoms are much heavier than hydrogen; hence CF3 modes appear at considerably lower values than those of methyl. Usually symmetric and antisymmetric C–F stretching vibrations are in the ranges 1290–1235 and 1226–1200 cm−1 respectively. The bands of the C–F symmetric and antisymmetric stretching vibrations have been observed to give rise to extremely intense FT-IR absorption and rather weak Raman bands and these appear in the range 1300–850 cm−1. These vibrations are coupled with the several other vibrations.
Below 1200 cm−1. In this region a large mixing of different types of vibrations appears. Most prominent vibrations appearing in this region are twisting, wagging and rocking vibrations of methylene group; trigonal deformation, puckering, asymmetric deformation and asymmetric torsion vibrations of six membered rings; deformation and torsion vibrations of five membered ring. Absorption due to methylene twisting and wagging vibrations, are observed in 1240–1050 cm−1 region. These bands are appreciably weaker than those resulting from methylene scissoring. Methyl rocking frequencies are observed in the range 1120–1050 cm−1 and 900–800 cm−1.32 These vibrations are mass sensitive and variable in position due to the interaction with skeletal stretching modes. In the present study the modes involving methyl rocking vibrations fall within the commonly accepted region of these modes. A Raman band observed at 1046 cm−1 is assigned to S–O stretching mode which is calculated at 1022 cm−1. S–O stretching vibrations usually occur at about 1080–1120 cm−1.36 This large deviation in wavenumbers is due to the involvement of oxygen atom of S–O in intermolecular hydrogen bonding N–H⋯O. The characteristic vibration of the pyridine ring, the ring breathing (ring pulsation), has been calculated at 739 cm−1 and matches well with the Raman band observed at the same frequency. This mode is observed at 995 cm−1 in 2-amino-5-methylpyridine37 and at 1022 cm−1 in 2-acetylamino-5-bromo-6-methylpyridine.32 The variation in wavenumbers of ring breathing modes shows that these vibrations are sensitive to the mass and position of the substituent. A Raman active band at 250 cm−1 with medium intensity is assigned to benzimidazole ring butterfly mode and is calculated at 259 cm−1. It matches with the value reported by T. S. Xavier et al. at 225 cm−1 in the Raman spectrum in 2-(4-bromophenyl)-1H-benzimidazole.38
The scaled wavenumbers obtained showed respectable agreement with the observed FT-IR and FT-Raman spectra and also with the literature values. The deviation of the scaled wavenumbers from the experimental ones was less than 10 cm−1, with exception in few modes where intra and intermolecular interactions come into play. This reveals that B3LYP functional is suitable for the study of pharmaceutical compounds.
4.3 Biological activities
If for any particular activity Pa > 0.7 the substance is very likely to exhibit that activity in experiment. Activities for title molecule analysed by Pharma Expert are listed in Table 1 with Pa > 0.7. The highest probability for LSP is to show the Mucositis treatment effect with Pa value 0.784. We also predicted the toxic effects of the compound. The prediction for toxicity shows that the title compound has high probability to show toxic/side effect bronchoconstrictor with Pa value 0.772. It means that toxic effects of LSP should be scrutinized in further investigation.
| Effect | Pa | Pi |
|---|---|---|
| Mucositis treatment | 0.784 | 0.020 |
| Ankylosing spondylitis treatment | 0.757 | 0.017 |
| Phobic disorders treatment | 0.736 | 0.070 |
| Alopecia treatment | 0.726 | 0.009 |
| Kidney function stimulant | 0.718 | 0.020 |
| Antimutagenic | 0.712 | 0.004 |
| Proliferative diseases treatment | 0.701 | 0.027 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P and log
P and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S respectively are the key determinants of pharmacokinetic properties used in the quantitative structure–activity relationship (QSAR) studies and drug design, since these properties are related to drug absorption, metabolism, bioavailability, and toxicity. Many of the processes of drug disposition depend on the capability to cross membranes and hence these have a high correlation with measures of lipophilicity. On the basis of review of the relevant literature41 high lipophilicity (>5) frequently leads to compounds with rapid metabolic turnover, low solubility, and poor absorption however if lipophilicity is too low, a drug will generally display poor ADMET (absorption, distribution, metabolism, elimination and toxicology) properties. The average lipophilicity value (between ∼1 and 3) ensures optimal physiochemical properties. The calculated value of log
S respectively are the key determinants of pharmacokinetic properties used in the quantitative structure–activity relationship (QSAR) studies and drug design, since these properties are related to drug absorption, metabolism, bioavailability, and toxicity. Many of the processes of drug disposition depend on the capability to cross membranes and hence these have a high correlation with measures of lipophilicity. On the basis of review of the relevant literature41 high lipophilicity (>5) frequently leads to compounds with rapid metabolic turnover, low solubility, and poor absorption however if lipophilicity is too low, a drug will generally display poor ADMET (absorption, distribution, metabolism, elimination and toxicology) properties. The average lipophilicity value (between ∼1 and 3) ensures optimal physiochemical properties. The calculated value of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P for LSP is 2.84. This suggests that the drug LSP may penetrate through the apolar cell membranes so that it is distributed well within the body after adsorption and is rapidly metabolized and excreted. This value of log
P for LSP is 2.84. This suggests that the drug LSP may penetrate through the apolar cell membranes so that it is distributed well within the body after adsorption and is rapidly metabolized and excreted. This value of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P is in the range of other benzimidazole derivatives.42,43 The high value of lipophilicity ensures the high plasma level that provides the good gastrointestinal absorption. Aqueous solubility limits the amount of compound that can be present in solution in the gut and in the circulation. Solubility can limit the bioavailability of a compound and lead to high patient-to-patient and fed-to-fasted variation. Solubility of a compound must be accurately determined to assess the concentrations that the drug will achieve in the target area, to establish the therapeutic level, and to prevent toxicity. The value of log
P is in the range of other benzimidazole derivatives.42,43 The high value of lipophilicity ensures the high plasma level that provides the good gastrointestinal absorption. Aqueous solubility limits the amount of compound that can be present in solution in the gut and in the circulation. Solubility can limit the bioavailability of a compound and lead to high patient-to-patient and fed-to-fasted variation. Solubility of a compound must be accurately determined to assess the concentrations that the drug will achieve in the target area, to establish the therapeutic level, and to prevent toxicity. The value of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S for LSP is −3.2, which is in the range of good solubility describing the balanced pharmacokinetic properties of LSP. Most of the marketed drugs are found in this category.44 High solubility of LSP leads to high hydrophilicity that supports absorption across the biological membrane and distribution to reach the target organ or tissue. Lower solubility can hinder the absorption and distribution of compound.
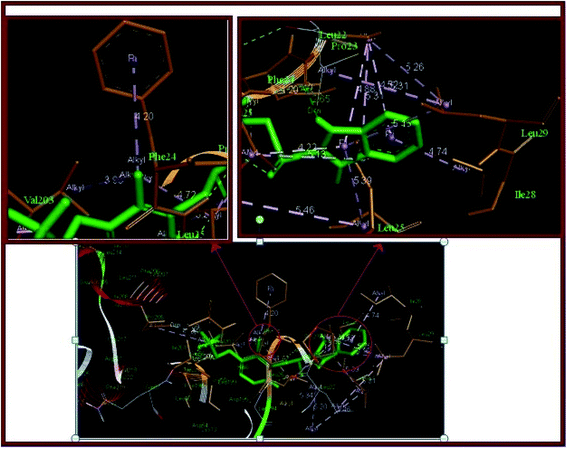
S for LSP is −3.2, which is in the range of good solubility describing the balanced pharmacokinetic properties of LSP. Most of the marketed drugs are found in this category.44 High solubility of LSP leads to high hydrophilicity that supports absorption across the biological membrane and distribution to reach the target organ or tissue. Lower solubility can hinder the absorption and distribution of compound.| S. No. | Affinity (kcal mol−1) | Distance from the best mode | |
|---|---|---|---|
| RMSD (lower bound) | RMSD (upper bound) | ||
| 1 | −7.8 | 0 | 0 |
| 2 | −7.2 | 2.512 | 1.726 |
| 3 | −7.1 | 2.199 | 1.409 |
| 4 | −7.1 | 8.391 | 4.393 |
| 5 | −6.9 | 20.648 | 19.238 |
| 6 | −6.9 | 18.425 | 17.183 |
| 7 | −6.8 | 9.203 | 4.871 |
| 8 | −6.8 | 14.061 | 12.976 |
| 9 | −6.6 | 19.924 | 18.407 |
4.4 Reactivity predictions
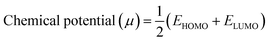 | (3) |
 | (4) |
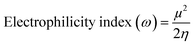 | (5) |
The chemical potential expresses the tendency of a substance to react with other substances, to transform into another state and to redistribute in space. The greater the chemical potential, the more active or “driven” the substance is. The smaller the chemical potential, the more passive or “phlegmatic” it is. The chemical potential calculated for LSP is −4.1689 eV. So the compound is likely to be active. The chemical hardness for LSP is 2.3718 eV. There is a correspondence between the chemical hardness and the stability of the respective reagent so that the molecular system becomes more stable when its chemical hardness increases. The increase of the chemical hardness may induce an increase of the covalent energy interaction and stabilize the structure. Electrophilicity index measures the stabilization in energy when the system acquires an additional electronic charge from the environment. It demonstrates the toxicity of various pollutants in terms of their reactivity and site selectivity.49 This reactivity index for LSP is 3.6683 eV; this is positive and definite quantity. Where two molecules react, the one that acts as nucleophile/electrophile will depend on which has a higher/lower electrophilicity index.
| f−k = ρ(N) − ρ(N − 1) | (6) |
| f+k = ρ(N + 1) − ρ(N) | (7) |
| f+k = qk(N + 1) − qk(N), for a nucleophilic attack | (8) |
| f−k = qk(N) − qk(N − 1), for an electrophilic attack | (9) |
The atom with the largest f value is the most reactive during reaction: f+k for nucleophilic attack, f−k for electrophilic attack. From Table 3 it has been found that the most prone site for electrophilic attack is C17 having highest f−k value. The reactivity order for the electrophilic case is C17 > C16 > C15 > C19 > C21 > C23 > C10 > C12 > N9. The possible sites for nucleophilic attack are predicted by calculating f+k values and the reactivity order for nucleophilic case is C16, C19, C20, C23, C15, C13 and C24. It is seen that sites C16, C19, C23 and C15 are involved in both electrophilic and nucleophilic attack. To overcome this difficulty Morell et al.50 proposed a local reactivity descriptor called the dual descriptor f(2)(k) ≡ Δf(k) to reveal nucleophilic and electrophilic sites on a molecular system at the same time. The general working equation to obtain dual descriptor is given by the arithmetic difference between nucleophilic and electrophilic Fukui functions.
| f(2)(k) = f+k − f−k | (10) |
| kth atom | f+k | f−k | f(2)(k) | s(2)(k) |
|---|---|---|---|---|
| 1 S | −0.2798 | −0.0893 | −0.1905 | −0.0339 |
| 2 F | −0.0117 | −0.0059 | −0.0058 | −0.0010 |
| 3 F | −0.0133 | −0.0086 | −0.0047 | −0.0008 |
| 4 F | −0.0185 | −0.0125 | −0.0059 | −0.0011 |
| 5 O | −0.0149 | −0.0077 | −0.0072 | −0.0013 |
| 6 O | −0.0035 | −0.1609 | 0.1573 | 0.0279 |
| 7 N | −0.0529 | −0.0771 | 0.0241 | 0.0043 |
| 8 N | −0.0173 | −0.0764 | 0.0591 | 0.0105 |
| 9 N | −0.0451 | 0.0037 | −0.0488 | −0.0087 |
| 10 C | −0.0298 | 0.0129 | −0.0426 | −0.0076 |
| 11 C | −0.0306 | −0.0827 | 0.0521 | 0.0093 |
| 12 C | −0.0085 | 0.0044 | −0.013 | −0.0023 |
| 13 C | 0.01318 | −0.0268 | 0.0400 | 0.0071 |
| 14 C | −0.0319 | −0.0557 | 0.0238 | 0.0042 |
| 15 C | 0.01399 | 0.0165 | −0.0025 | −0.0005 |
| 16 C | 0.14010 | 0.0234 | 0.1167 | 0.0207 |
| 17 C | −0.0343 | 0.0617 | −0.0959 | −0.0171 |
| 18 C | −0.0332 | −0.0437 | 0.0105 | 0.0019 |
| 19 C | 0.05399 | 0.0163 | 0.0377 | 0.0067 |
| 20 C | 0.02920 | −0.0089 | 0.03822 | 0.0068 |
| 21 C | −0.1097 | 0.0157 | −0.1255 | −0.0223 |
| 22 C | −0.0343 | −0.0215 | −0.0128 | −0.0023 |
| 23 C | 0.02368 | 0.0140 | 0.0096 | 0.0017 |
| 24 C | 0.00018 | −0.0059 | 0.0062 | 0.0011 |
| 25 C | −0.0046 | −0.0259 | 0.0213 | 0.0038 |
When f(2)(k) > 0, the process is driven by a nucleophilic attack on atom k and then that atom acts as an electrophilic species; conversely, when f(2)(k) < 0, the process is driven by an electrophilic attack over atom k and therefore atom k acts as a nucleophilic species. However, dual descriptor is a sub intensive property; its condensed values become insignificant as the size of the molecule increases. To overcome the intrinsic behavior of this local reactivity descriptor, another local reactivity descriptor has been defined which permits one to measure local reactivity according to the molecular size. Such a descriptor is the local hyper softness which is expressed as follows:
| s(2)(k) = f(2)(k)S2 | (11) |
The results for the calculation of the condensed electrophilic Fukui function f−(r), nucleophilic Fukui function f+(r), and dual descriptor f(2)(r) along with the local hyper softness s(2)(r) over the atoms (with the exception of hydrogen atoms) of the LSP molecule are presented in Table 3. On the basis of dual descriptor the order for the nucleophilic species is as follows O6 > C16 > N8 > C11 > C13 > C20 > C19 > N7 > C14 > C25 > C18 > C23 > C24, evidently O6 is more likely to undergo a nucleophilic attack, whereas electrophilic attacks will take place preferentially on atom S1 followed by atoms in order given as S1 > C21 > C17 > N9 > C10 > C12 > C22 > O5 > F4 > F2 > C15 > F3.
The dual descriptor is positive at S1 and negative at O6. This shows that LSP undergoes both electrophilic substitution (at S1) and nucleophilic substitution (at O6) at different sites. Therefore it is a dual molecule. Fukui functions are important in deciphering the initial stage of covalent bonding. As S1 and O6 are the most reactive atoms for receiving electrophilic and nucleophilic attacks, respectively and they are pretty close to each other in the middle of the molecule, so the most intense covalent interactions can be concentrated in this part of molecule. For isolated molecules, f(2)(k) and s(2)(k) should give similar reactivity descriptions. The observation of the trend of the reactive sites in LSP by s(2)(k) is found identical to f(2)(k).
5. Conclusion
In the present work, we have analyzed the ground state geometric structure of LSP together with conformational stability, spectroscopic signatures, electronic properties, biological activities and chemical reactivity predictions using quantum chemical computations. The experimental spectral properties of LSP are successfully studied by FT-IR, FT-Raman, UV-vis, NMR spectroscopic techniques. The lowest energy conformer of LSP has been identified using PES scan as a function of the selected torsion angle. This conformer reproduces the experimentally reported structure. As spectroscopic methods probe the molecular level, changes in the molecular environment (e.g., H-bonding or π–π interactions) due to solid state changes are detected as a result of altered vibrations of functional groups involved in these interactions. The various modes of vibrations are explicitly assigned on the results of PED output obtained from the normal coordinate analysis. The mismatch between the observed and the calculated wavenumbers in NH and SO stretching vibrations is due to the involvement of these groups in hydrogen bonding. TD-DFT calculations predict a weak transition (oscillator strength = 0.0794) at 287.10 nm which can be correlated to the experimental band at 284.0 nm in ethanol. This is likely due to vanishing overlapping between the MOs involved in the corresponding electronic transition, which usually results in small oscillator strength. The calculated HOMO–LUMO energy gap is 4.7436 eV. It reveals the significant degree of charge transfer interactions taking place in the molecule and is responsible for the enhanced chemical as well as biological activity of the molecule. The nuclear magnetic resonance spectrum shows that the hydrogen atom attached to or near to an electronegative atom gives the most downfield region. In the present compound the largest chemical shift value 11.882 ppm is for the hydrogen atom H26 attached to the nitrogen atom N8. The hydrogen atom H34 in the pyridine ring is also near to the nitrogen atom N9 and has higher chemical shift value than other atom H33. Pass software predicts the mucositis treatment effect with high probability and some other effects as ankylosing spondylitis, phobic disorders, alopecia, proliferative diseases treatment, kidney function stimulant and antimutagenic with decreasing probabilities. Pass predicted effects of this drug are important and may promote quite a number of investigations in the health area. The values of lipophilicity and aqueous solubility are 2.84 and −3.2 respectively. These values indicate tendency of the compound to partition into lipid vs. aqueous environments. Such insights are useful for correlating important pharmaceutical processes such as gastrointestinal absorption, membrane permeation, solubility, volume of distribution, and protein binding. The results of molecular docking studies speculate that this biologically active molecule is metabolized by cytochrome CYP3A4 enzyme thereby indicating its possible pharmacological importance. Under reactivity predictions molecular electrostatic potential map clearly demonstrates that the possible sites for electrophilic attack are spread around the oxygen atom O6 and the nitrogen atom N7, while the nucleophilic sites are around the hydrogen atoms. This shows that the electrophilic ability strengthens and nucleophilic ability weakens as one move outwards from the inner core of the molecule. LSP has negative chemical potential (μ = −4.1689 eV). This implies that it has a weaker tendency to change into the elements it is produced from and does not decompose. It can be produced voluntarily from the elements. The chemical hardness is 2.3718 eV which advocates its relative kinetic stability. Global reactivity descriptors based on structural properties eventually obtained legitimacy within density functional theory. On the basis of Fukui function the possible molecular sites for the nucleophilic attack are O6, C16, N8, C11, C13, C20, C19, N7, C14, C25, C18, C23, C24 and for the electrophilic attack are S1, C21, C17, N9, C10, C12, C22, O5, F4, F2, C15, F3. Locally, a molecular site is more prone to electrophilic attack or another more prone to nucleophilic attack, but the overall philicity of the whole molecule remains conserved. Thus, the present investigation provides a complete and reliable structural, spectral, biological and structure–activity relation of the compound. This investigation not only presents these features of LSP for the first time, but also prepares the ground for future investigations.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors are thankful to Central Drug Research Institute, Lucknow for providing instrumentation facilities for recording NMR and UV-vis spectra of the sample. The authors are grateful to Dr Shreeram Oak, Bruker Optik GmbH, Mumbai for help in recording FT-IR spectra.References
- R. Marquis, J. Sheng, T. Nguyen, J. Baldeck and J. Olsson, Arch. Oral Biol., 2006, 51, 1015–1023 CrossRef PubMed.
- A. T. Mavrova, K. Anichina, D. Vuchev, J. Tsenov, P. Denkova, M. Kondeva and M. Micheva, Eur. J. Med. Chem., 2006, 41, 1412–1420 CrossRef CAS PubMed.
- M. Boiani and M. Gonzalez, Mini-Rev. Med. Chem., 2005, 5, 409–424 CrossRef CAS PubMed.
- X. Li, T. B. Andersson, M. Ahlstrom and L. Weidolf, Drug Metab. Dispos., 2004, 32, 821–827 CrossRef CAS.
- T. Furuta, N. Shirai, F. Watanabe, S. Honda, K. Takeuchi, T. Iida, Y. Sato, M. Kajimura, H. Futami, S. Takayanagi, M. Yamada, K. Ohashi, T. Ishizaki and H. Hanai, Clin. Pharmacol. Ther., 2002, 72, 453–460 CrossRef CAS PubMed.
- J. T. Rives and W. L. Jorgensen, J. Chem. Theory Comput., 2008, 4, 297–306 CrossRef PubMed.
- M. Orio, D. A. Pantazis and F. Neese, Photosynth. Res., 2009, 102, 443–453 CrossRef CAS PubMed.
- F. Neese, Coord. Chem. Rev., 2009, 253, 526–563 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski, and D. J. Fox, Gaussian 09, Revision B.01, Gaussian, Inc., Wallingford, CT, 2010 Search PubMed.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865 CrossRef CAS PubMed.
- H. Yoshida, K. Takeda, J. Okamura, A. Ehara and H. Matsurra, J. Phys. Chem. A, 2002, 106, 3580–3586 CrossRef CAS.
- N. Choudhary, S. Bee, A. Gupta and P. Tandon, Comput. Theor. Chem., 2013, 1016, 8–21 CrossRef CAS.
- V. Deval, A. Kumar, V. Gupta, A. Sharma, A. Gupta, P. Tandon and K. Kunimoto, Spectrochim. Acta, Part A, 2014, 132, 15–26 CrossRef CAS PubMed.
- M. Agrawal, V. Deval, A. Gupta, B. R. Sangala and S. S. Prabhu, Spectrochim. Acta, Part A, 2016, 167, 142–156 CrossRef CAS PubMed.
- G. Onida, L. Reining and A. Rubio, Rev. Mod. Phys., 2002, 74, 601–659 CrossRef CAS.
- M. Cossi, G. Scalmani, N. Rega and V. Barone, J. Chem. Phys., 2002, 117, 43–54 CrossRef CAS.
- D. Jacquemin, V. Wathelet, E. A. Perpete and C. Adamo, J. Chem. Theory Comput., 2009, 5, 2420–2435 CrossRef CAS PubMed.
- R. Ditchfield, Mol. Phys., 1974, 27, 789–807 CrossRef CAS.
- D. A. Filimonov, V. V. Poroikov, E. I. Karaicheva, R. K. Kazaryan, A. P. Boudunova, E. M. Mikhailovsky, A. V. Rudnitskih, L. V. Goncharenko and Y. V. Burov, Exp. Clin. Psychopharmacol., 1995, 58, 56–62 CAS.
- I. V. Tetko, J. Gasteiger, R. Todeschini, A. Mauri, D. Livingstone, P. Ertl, V. A. Palyulin, E. V. Radchenko, N. S. Zefirov, A. S. Makarenko, V. Y. Tanchuk and V. V. Prokopenko, J. Comput.-Aided Mol. Des., 2005, 19, 453–463 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
- http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php.
- K. Vyas, A. Sivalakshmidevi and G. O. Reddy, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2000, 56, e572–e573 Search PubMed.
- J. M. L. Martin and C. V. Alsenoy, Gar2ped, University of Antwerp, Antwerp, 1995 Search PubMed.
- P. Pulay, G. Fogarasi, F. Pang and J. E. Boggs, J. Am. Chem. Soc., 1979, 101, 2550–2560 CrossRef CAS.
- G. Fogarasi, X. Zhou, P. W. Taylor and P. Pulay, J. Am. Chem. Soc., 1992, 114, 8191–8201 CrossRef CAS.
- S. Sudha, M. Karabacak, M. Kurt, M. Cinar and N. Sundaraganesan, Spectrochim. Acta, Part A, 2011, 84, 184–195 CrossRef CAS PubMed.
- L. Dyminska, A. Gagor, M. Maczka, Z. Weglinskia and J. Hanuza, J. Raman Spectrosc., 2010, 41, 1021–1029 CrossRef CAS.
- M. Karnan, V. Balachandran and M. Murugan, Spectrochim. Acta, Part A, 2012, 96, 51–62 CrossRef CAS PubMed.
- R. Meenakshi, RSC Adv., 2016, 6, 21822–21831 RSC.
- V. Arjunan, P. S. Balamourougane, S. T. Govindaraja and S. Mohan, J. Mol. Struct., 2012, 1018, 156–170 CrossRef CAS.
- S. Premkumar, T. N. Rekha, R. M. Asath, T. Mathavan, A. Milton and F. Benial, Eur. J. Pharm. Sci., 2016, 82, 115–125 CrossRef CAS PubMed.
- J. Wang, X. Q. Xie, T. J. Hou and X. Xu, J. Phys. Chem. A, 2007, 111(20), 4443–4448 CrossRef CAS PubMed.
- S. Pinchas, D. Samuel and M. Weiss-Broday, J. Chem. Soc., 1961, 1688 RSC.
- L. Kahovec and K. W. F. Kohlreusch, Monatsh. Chem., 1941, 74, 333 CrossRef.
- M. W. Wong, Y. Steudel and R. Steudel, Chem.–Eur. J., 2007, 13, 502–514 CrossRef CAS PubMed.
- N. Sundaraganesan, C. Meganathan and M. Kurt, J. Mol. Struct., 2008, 891(1–3), 284–291 CrossRef CAS.
- T. S. Xavier, N. Rashid and I. H. Joe, Spectrochim. Acta, Part A, 2011, 78(1), 319–326 CrossRef CAS PubMed.
- D. D. Eley, H. Pines and P. B. Weisz, Advances in Catalysis, Academic Press, United States, 1966, vol. 16 Search PubMed.
- V. V. Poroikov, D. A. Filimonov, W. Ihlenfeldt, T. A. Gloriozova, A. A. Lagunin, Y. V. Borodina, A. V. Stepanchikova and M. C. Nicklaus, J. Chem. Inf. Comput. Sci., 2003, 43, 228–236 CrossRef CAS PubMed.
- J. A. Arnott, R. Kumar and S. L. Planey, J. Appl. Biopharm. Pharmacokinet., 2013, 1, 31–36 Search PubMed.
- S. O. Podunavac-Kuzmanovic, D. D. Cvetkovic and D. J. Barna, J. Serb. Chem. Soc., 2008, 73(10), 967–978 CrossRef CAS.
- C. Sarbu, T. Djakovic-Sekulic and N. Perisic-Janjic, J. Pharm. Biomed. Anal., 2001, 30, 739–745 CrossRef.
- J. Huuskonen, M. Salo and J. Taskinen, J. Chem. Inf. Comput. Sci., 1998, 38, 450–456 CrossRef CAS PubMed.
- F. P. Guengerich, Chem. Res. Toxicol., 2008, 21(1), 70–83 CrossRef CAS PubMed.
- J. P. Perdew, R. G. Parr, M. Levy and J. L. Balduz, Phys. Rev. Lett., 1982, 49, 1691–1694 CrossRef CAS.
- M. Levy, J. P. Perdew and V. Sahni, Phys. Rev. A, 1984, 30, 2745–2748 CrossRef.
- J. P. Perdew and M. Levy, Phys. Rev. B: Condens. Matter Mater. Phys., 1997, 56(24), 16021–16028 CrossRef CAS.
- R. Parthasarathi, J. Padmanabhan, V. Subramanian, B. Maiti and P. K. Chattaraj, J. Phys. Chem. A, 2003, 107(48), 10346–10352 CrossRef CAS.
- C. Morell, A. Grand and A. Toro-Labbe, J. Phys. Chem. A, 2005, 109, 205–212 CrossRef CAS PubMed.
- A. M. Sapse, Molecular Orbital Calculations for Biological Systems, Oxford University Press, Oxford, New York, 1998 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra00130d |
| This journal is © The Royal Society of Chemistry 2017 |