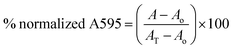Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
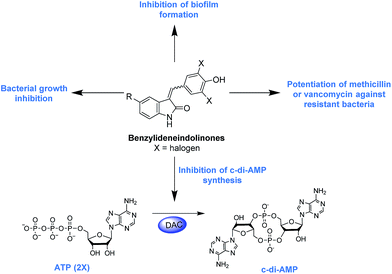
Hydroxybenzylidene-indolinones, c-di-AMP synthase inhibitors, have antibacterial and anti-biofilm activities and also re-sensitize resistant bacteria to methicillin and vancomycin†
Clement Opoku-Temengac,
Neetu Dayalab,
Jacob Millerb and
Herman O. Sintim *abd
*abd
aPurdue Institute for Drug Discovery, Purdue University, West Lafayette, IN 47907, USA. E-mail: hsintim@purdue.edu
bDepartment of Chemistry, Purdue University, West Lafayette, IN 47907, USA
cBiochemistry Graduate Program, University of Maryland, College Park, Maryland 20742, USA
dPurdue Institute of Inflammation, Immunology and Infectious Disease, West Lafayette, IN 47907, USA
First published on 24th January 2017
Abstract
c-di-AMP signaling regulates a myriad of physiological processes in Gram-positive bacteria and mycobacteria. c-di-AMP synthase (DAC) is essential in many human pathogens including Staphylococcus aureus, Listeria monocytogenes and Streptococcus pneumoniae and could become an important antibacterial drug target. In our continuing efforts to identify diverse DAC inhibitors, we uncovered hydroxybenzylidene-indolinones as new DAC inhibitors. Interestingly, these compounds also possess antibacterial activities and inhibit biofilm formation. Importantly, methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus faecalis could be re-sensitized to methicillin and vancomycin, respectively, by hydroxybenzylidene-indolinones.
Introduction
Each year millions of people become infected with drug-resistant bacteria and a significant number succumb to the pathogens. It has been estimated that if new antibacterial agents or adjuvants that re-sensitize resistant bacteria to traditional antibiotics are not developed/found, in the near future (by 2050) deaths due to bacterial infections will surpass 10 million.1 In light of this gloomy forecast, several groups have embarked on the search of essential pathways or proteins that could be targeted to develop a new generation of antibacterial agents.2 Recently, it was revealed that cyclic dimeric adenosine 3′,5′-monophosphate (c-di-AMP), initially discovered as a ligand bound to the DNA integrity scanning protein A (DisA) of Thermatoga maritima,3 is an important second messenger that is present in a myriad of clinically-relevant bacteria, including Staphylococcus aureus,4 Listeria monocytogenes,5 Streptococcus pyogenes,6 Mycobacterium tuberculosis,7 Chlamydia trachomatis8 among others. The physiological roles of c-di-AMP include peptidoglycan homeostasis,9,10 cell size regulation,4 fatty acid metabolism and transport,11 ion transport,12 biofilm formation13 and a host of other physiological processes (Fig. 1).c-di-AMP is synthesized by diadenylate cyclases (DACs) from two molecules of ATP (Fig. 1). The DAC gene has been shown to be essential in some Gram-positive bacteria such as L. monocytogenes, S. aureus and S. pneumoniae,5,10,14,15 and this raises the potential that new drugs against these problematic bacteria could be found by screening for inhibitors of c-di-AMP synthesis. Of note c-di-AMP regulates cell wall homeostasis9,10 and because many antibiotics in clinical use also target the cell wall,16 it is anticipated that inhibitors of c-di-AMP could potentiate the action of several cell wall-acting antibiotics. In a seminal study by Gründling, it was disclosed that depletion of intracellular c-di-AMP via the over-expression of a c-di-AMP PDE (GdpP) sensitized S. aureus to the β-lactams oxacillin and penicillin G.4 The gdpP mutant strain was observed to have increased biofilm formation relative to the wildtype.4 Recently, another important work from Peng et al. revealed that c-di-AMP regulates biofilm formation in S. pneumoniae. These insights regarding the role of c-di-AMP in biofilm formation is interesting (it is known that biofilm bacteria are several orders of magnitude more resistant to antibiotics than planktonic bacteria) and could lead to new treatment paradigms against Gram-positive bacteria.17
Motivated by the central role played by c-di-AMP in some bacteria, especially in S. aureus, our group has been pursuing inhibitors of c-di-AMP synthesis with the hope that some of these compounds could be developed into antibacterial agents. Previously we revealed that bromophenol-TH,18 suramin19 and theaflavin digallate20 inhibit the prototypical c-di-AMP synthase, DisA. The first generation c-di-AMP synthase inhibitors are however not drug-like. For example suramin,19 is polyanionic and suffers from poor cell penetration and the theaflavins are easily metabolized in vivo.21 3′-deoxyATP, a nucleotide analog was also identified as DAC inhibitor by Müller and others22 but this molecule also suffers from cell permeation issue (it is polyanionic as well). In our continuing efforts to identify cell-permeable compounds that also inhibit c-di-AMP synthesis, we identified a benzylidene-indolinone derivative as a cell permeable inhibitor of c-di-AMP synthesis. This molecule and analogs thereof demonstrate potent antibacterial properties and could synergize the action of other antibiotics.
Results and discussion
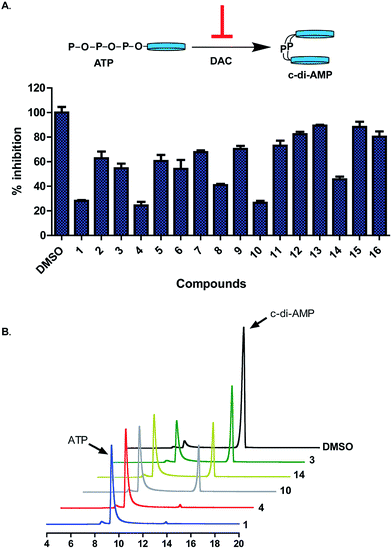
We previously used the coralyne assay23 (Fig. S1A†) to identify bromophenol-TH,18 suramin19 and theaflavin digallate20 as inhibitors of c-di-AMP synthase DisA from B. subtilis. Based on the success of the coralyne assay, we screened a 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compound library containing pharmacologically active compounds in order to identify cell permeable inhibitors of DAC. From the screen we identified a benzylidene-indolinone derivative, compound 1 (Fig. 2) as an inhibitor of c-di-AMP synthesis (Fig. S1B†). The DAC inhibition was confirmed using 32P-ATP assays (Fig. S1C†). Compound 1 (also called GW5074), was originally developed as a selective c-Ras inhibitor. It is non-toxic to mammalian cells and has been used in a few mouse studies without showing any adverse effects.24,25 Compound 1 possesses neuroprotective properties and in an in vivo model of Huntington's disease, it was shown to protect neurons via resisting 3-NP-induced striatal neurodegeneration.24 1 was also shown to suppress sidestream smoke-induced airway hyper responsiveness in mice.25 Based on its safety profile and hence high potential for clinical translation we proceeded to make a small library of this class of molecules following the synthetic strategy shown in Fig. 2A. A total of 15 analogs with subtle changes to the indolinone and benzylidene moieties were easily synthesized (Fig. 2B) and screened for DAC inhibition.
000 compound library containing pharmacologically active compounds in order to identify cell permeable inhibitors of DAC. From the screen we identified a benzylidene-indolinone derivative, compound 1 (Fig. 2) as an inhibitor of c-di-AMP synthesis (Fig. S1B†). The DAC inhibition was confirmed using 32P-ATP assays (Fig. S1C†). Compound 1 (also called GW5074), was originally developed as a selective c-Ras inhibitor. It is non-toxic to mammalian cells and has been used in a few mouse studies without showing any adverse effects.24,25 Compound 1 possesses neuroprotective properties and in an in vivo model of Huntington's disease, it was shown to protect neurons via resisting 3-NP-induced striatal neurodegeneration.24 1 was also shown to suppress sidestream smoke-induced airway hyper responsiveness in mice.25 Based on its safety profile and hence high potential for clinical translation we proceeded to make a small library of this class of molecules following the synthetic strategy shown in Fig. 2A. A total of 15 analogs with subtle changes to the indolinone and benzylidene moieties were easily synthesized (Fig. 2B) and screened for DAC inhibition.
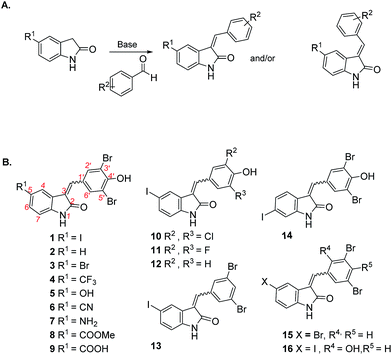 | ||
| Fig. 2 (A) Synthetic strategy for making hydroxybenzylidene-indolinones; (B) structures of hydroxybenzylidene-indolinones that were synthesized. | ||
The “hit” compound 1 is decorated with an iodo group at the 5-position of the indolinone. Compounds 2–9 were designed to identify which substituent at the 5-position of the indolinone core was optimal for DAC inhibition. Compound 2 did not have any substitution at position 5, whereas compounds 3 and 4 contained bromo and trifluoromethyl groups (both groups are similarly as hydrophobic as the iodo group). Compounds 3, 4, 5, 6 and 7 were 5-bromo, 5-trifluoromethyl, 5-hydroxy, 5-cyano and 5-amino substitutions respectively. Compounds 5–9 contained the polar groups OH (5), CN (6), NH2 (7), CO2Me (8) and CO2H (9). We expected the ester group to be converted into the acid moiety inside the cell, although the ester compound (a pro-drug) could have different permeation properties than the acid. Compound 14 contained an iodo moiety at the 6-position of the indolinone and is ideal for comparing 5- vs. 6-substituions of the indolinone. To investigate the importance of the bromo groups on the benzylidene portion of the molecule, we synthesized compounds 10–12 whereby the bromo groups were replaced with H, F and Cl. Finally, the importance of the hydroxyl group at the 4-position of the benzylidene was investigated by making compounds 13 and 15, which did not contain a phenol or compound 16, which had the OH group moved from the 4-position to the 2-position on the benzylidene. With these compounds in hand, we proceeded to investigate which compounds inhibited DisA, using the coralyne assay.23
DAC activity assay, in the presence of the synthesized compounds revealed that substitution of the indolinone moiety with hydrophilic groups (OH (5), CN (6), NH2 (7), CO2Me (8) and CO2H (9)) lead to compounds that were weak DAC inhibitors (Fig. 3A). On the contrary, substitution of indolinone moiety with iodo or CF3 group at the 5-position (compounds 1, 4 or 10) afforded potent inhibitors of DAC. At 20 μM, compounds 1 and 4 completely inhibited the synthesis of c-di-AMP by DisA (0.25 μM), see Fig. 3B. The substitution pattern of the benzylidene group was also critical for DAC inhibition. The 4-OH group on the benzylidene group was essential as compound 15, which lacked OH group or compound 16, bearing 2-OH (but not 4-OH) did not inhibit c-di-AMP synthesis. The nature of the halogen on the benzylidene moiety was also critical for DAC inhibition. Substitution of positions 3 and 5 of the benzylidene with Br or Cl afforded DAC inhibitors (compounds 1 and 10) whereas compounds bearing H or F substituents at the 3 and 5 positions of the benzylidene were not active (compounds 11 and 12) (Fig. S2†).
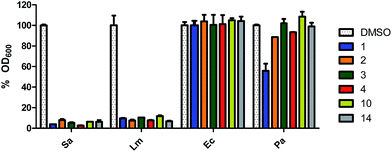
As previously stated, a myriad of physiological processes are regulated by c-di-AMP signalling, such as cell wall homeostasis9,10 and coupled with the essentiality of DACs of human pathogens,5,10,14,15 we hypothesized that cell permeable inhibitors of c-di-AMP synthesis, such as the ones identified above could also possess antibacterial properties. We therefore decided to investigate the effects of the hydroxybenzylidene-indolinones compounds on bacterial viability. Initially, we screened the compounds against S. aureus (ATCC 25923) and E. coli (ATCC 25923) (as representative Gram-positive and Gram-negative bacteria). The bacteria were cultured in the presence of 16 μg mL−1 of the hydroxybenzylidene-indolinones in Mueller Hinton broth (MHB) for 24 h at 37 °C with 250 rpm shaking. Post incubation, we measured the optical density at 600 nm (OD600) of the cultures. For each bacterial species, an equivalent amount of DMSO, not exceeding 0.1% was used as negative control. Compounds 1, 2, 3, 4, 10 and 14 (but not compounds 5–9 or 11–13 or 15–16 or 5-iodoindolin-2-one) significantly inhibited the growth of S. aureus. None of the compounds affected E. coli growth (Fig. 4 and S3–S5†).
To rule out the possibility that the lack of activity against E. coli was not due to permeation issue, we also investigated the activity of compound 1 (16 μg mL−1) in the presence of ¼ the MIC value of colistin (0.03125 μg mL−1). Treating E. coli with ¼ the MIC of colistin (a non-toxic concentration) would make the bacteria permeable to compounds. Even in the presence of colistin, compound 1 did not inhibit the growth of E. coli (Fig. S5†). This experiment suggests that the hydroxybenzylidene-indolinones work via a Gram-positive specific mechanism. For the active compounds, we expanded the panel of bacteria to include L. monocytogenes (ATCC 19115) and Pseudomonas aeruginosa (ATCC 27853) and also observed that they were active against the Gram-positive L. monocytogenes but not against the Gram-negative P. aeruginosa (Fig. 4).
Having established the susceptibility of bacteria to the compounds, we sought to determine their minimum inhibitory concentration (MIC). For this we also included antibiotic resistant strains MRSA ATCC 33592 and vancomycin-resistant E. faecalis (ATCC 51575). The MIC values obtained are as shown in Table 1.
| Test compounds | MIC (μg mL−1) | |||
|---|---|---|---|---|
| S. aureus | MRSA | L. monocytogenes | VRE faecalis | |
| a ND represents not determined. | ||||
| 1 | 8 | 8 | 8 | 16 |
| 2 | 8 | 16 | 16 | 128 |
| 3 | 8 | 8 | 16 | >128 |
| 4 | 8 | 4 | 2 | 32 |
| 10 | 8 | 4 | 2 | 64 |
| 14 | 4 | 4 | 4 | >128 |
| Vancomycin | 1 | 1 | 1 | >128 |
| Methicillin | 2 | >128 | ND | ND |
All compounds had good MIC values against S. aureus and MRSA, ranging from 4 μg mL−1 to 16 μg mL−1 but not against VRE faecalis. Compounds 4, 10 and 14 appeared to be particularly potent against L. monocytogenes, with MIC values ranging from 2 μg mL−1 to 4 μg mL−1 (Table 1).
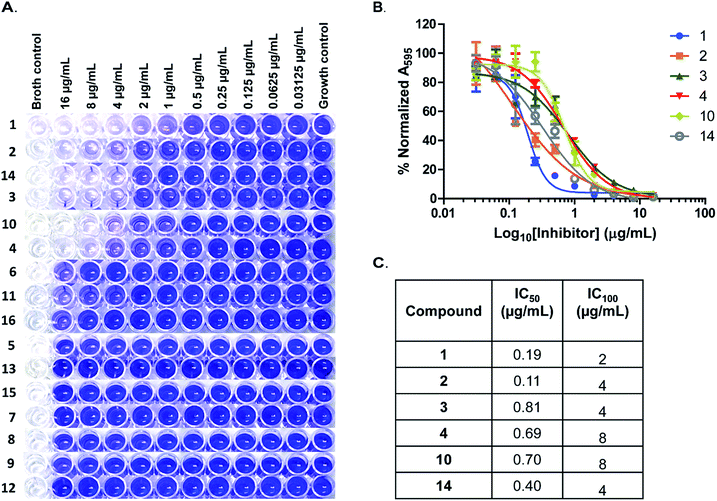
As earlier stated, biofilm-associated infections continue to be a major public health threat. Biofilms in general are difficult to treat, due in part to the reduced penetration of antibiotics into the biofilm.26 Very recently Peng et al. demonstrated that the deletion of pdeA, gene that encodes the S. pneumoniae PDE resulted in an increased S. pneumoniae biofilm formation.13 Also, Gründling and colleagues showed that in S. aureus, deletion of GdpP (PDE) resulted in increased biofilm formation relative to wildtype.4 These observations implicated c-di-AMP signaling in regulating biofilm formation.4,13 A report by the United States Centers for Disease Control and Prevention in 2013, characterized MRSA as being at the threat level of serious; implying that these require immediate attention.27 Staphylococcal infections are problematic in the healthcare setting primarily as a result of biofilm formation on host tissues, implants and medical devices.28 Others have pursued small molecules that inhibit MRSA biofilm formation.29,30 Motivated by these studies, we tested all 16 compounds for their effect on MRSA biofilm formation. The microtiter plate biofilm formation31 was employed using compounds at concentrations ranging from 16 μg mL−1 to 0.03125 μg mL−1. We observed that 6 compounds (1, 2, 3, 4, 10 and 14) potently inhibited biofilm formation (Fig. 5A). The IC50 values for biofilm inhibition for the compounds were 0.19 μg mL−1 for compound 1; 0.11 μg mL−1 for compound 2; 0.81 μg mL−1 for compound 3; 0.69 μg mL−1 for compound 4; 0.70 μg mL−1 for compound 10 and 0.40 μg mL−1 for compound 14 (Fig. 5B and C). The concentration of maximum biofilm inhibition, IC100, (Fig. 5C) were observed to be similar to the MIC values (Table 1), implying that the anti-biofilm activities were derived from growth inhibition.
‘Resurrecting’ antibiotics that have been rendered ineffective due to resistance by combining them with small molecules ”adjuvants” is now being pursued as a strategy to combat antibiotic resistance.32–34 Recently several groups have reported several small molecules that could re-sensitize MRSA or VRE to β-lactams or vancomycin respectively.35–38
Having observed that the compounds were active against MRSA and VRE faecalis, we investigated the ability of hydroxybenzylidene-indolinones to re-sensitize MRSA and VRE faecalis to methicillin and vancomycin respectively. MRSA is resistant to methicillin with an MIC of greater than 128 μg mL−1. In the presence of 2 μg mL−1 of compound 1 and 4, an MIC of 2 μg mL−1 was obtained for methicillin against MRSA; signifying a fold change in MIC of greater than 64-fold (Table 2). Compounds 3 and 10 also reduced the MIC of methicillin by greater than 32-fold from >128 μg mL−1 to 4 μg mL−1.
| Resistant bacteria | Antibiotic-compound combinations | MIC (μg mL−1) | Fold change |
|---|---|---|---|
| a a = 2 μg mL−1, b = 4 μg mL−1 and c = 8 μg mL−1 of compound NA stands for not applicable. | |||
| MRSA | Methicillin (Meth) | >128 | NA |
| Meth + 1a | 2 | >64 | |
| Meth + 3a | 4 | >32 | |
| Meth + 4a | 2 | >64 | |
| Meth + 10a | 4 | >32 | |
| VRE faecalis | Vancomycin (Van) | >128 | NA |
| Van + 1b | 2 | >64 | |
| Van + 3c | 2 | >64 | |
| Van + 4c | 2 | >64 | |
| Van + 10c | 2 | >64 | |
| Van + 14c | 1 | >128 | |
Similarly, the MIC of vancomycin for VRE faecalis was determined to be greater than 128 μg mL−1. When combined with compound 1 at 4 μg mL−1 (1/4 MIC) we observed a greater than 64-fold improvement in MIC of vancomycin (Table 2). On their own, compounds 3, 4, 10 and 14 have weak activity against VRE faecalis (Table 1). Interestingly, at 8 μg mL−1 of 3, 4 and 10, the MIC of vancomycin improved by greater than 64-fold. At that same concentration we observed that compound 14 could reduce the MIC of vancomycin for VRE faecalis from >128 μg mL−1 to 1 μg mL−1 (Table 2). In both instances, we observed that compound 2 could not potentiate the activity of either methicillin or vancomycin. Due to the interesting biological activities displayed by these compounds, we attempted to generate mutant bacteria to these compounds in order to confirm the mechanism of action. Unfortunately we have been unable to generate mutants that are resistant to these compounds, despite numerous efforts, in order to identify mechanism of action.
Conclusions
In our continual efforts to identify compounds that inhibit cyclic dinucleotide signalings17,39 we uncovered hydroxybenzylidene-indolinones as new inhibitors of c-di-AMP synthesis in vitro. Interestingly these compounds could sensitize resistant bacteria to methicillin and vancomycin. Further works in our laboratory are focused on mode of action studies and also lead optimization to arrive at more potent analogs of the compounds reported in this manuscript.Methods
General synthesis of hydroxybenzylidene-indolinones
For the synthesis and characterization of hydroxybenzylidene-indolinones, see ESI.†Bacterial strains and growth conditions
Standard strains of S. aureus (ATCC 25923 and MRSA ATCC 33592), L. monocytogenes ATCC 19115, vancomycin-resistant E. faecalis ATCC 51575, E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were routinely cultured in Mueller–Hinton broth 37 °C unless otherwise stated.Protein expression and purification
The enzyme used in this study were expressed from E. coli BL21 (DE3) cells harboring specific plasmids.3 Briefly, protein expression was induced at OD600 of 0.6 by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and the culture temperature reduced to 16 °C. Expression was performed for 18 hours after which cultures were centrifuged at 5000 rpm for 15 min at 4 °C. The cell pellets were then resuspended in lysis buffer (50 mM sodium phosphate buffer, pH 8.0, 300 mM NaCl) and lysed by sonication. The supernatant of the lysate was collected by centrifugation and proteins were purified by nickel affinity chromatography. Aliquots of purified proteins were stored in 10% glycerol at −80 °C.Screening
The screening was performed using the coralyne assay as previously described.18 Briefly, the compounds were stored as 10 mM DMSO stock solutions. Aliquots of the compounds (20 μM) or DMSO were added to a reaction mixture containing 10 μM coralyne, 300 μM ATP and 3 mM KI in a 40 mM Tris–HCl pH 7.5, 100 mM NaCl and 10 mM MgCl2 reaction buffer. We then initiated the reactions by adding DisA at 0.5 μM and incubated at 30 °C. The change in coralyne fluorescence at 475 nm when excited at 420 nm was monitored on a BioTek Cytation 5 Cell Imaging Multi-Mode Reader for 30 min. Compounds that reduced the fluorescence of coralyne were selected and further analysed by HPLC. The reaction components (as above but without coralyne and KI) were mixed up 30 °C. After 2 hours, the reaction was heated at 95 °C for 5 min and the precipitated proteins were filtered off. Components of the filtrate were then analyzed on a COSMOSIL C18-MS-II Packed column (5 μm) using 0.1 M TEAA in water (Buffer A) and acetonitrile (Buffer B). The samples will be eluted with 99% → 87% Buffer A at 0 to 16 min, 87% → 10% Buffer A at 16 to 22 min and kept at 10% Buffer A till 25 min, detecting signals at room temperature with a 260 nm UV detector.For radioactive TLC assay, compound 1 (20 μM) was incubated with 100 μM ATP, 11.1 nM 32P-ATP and DisA (0.25 μM) in the same reaction buffer as above for 2 h. An equal volume of DMSO was used as control. Aliquots of the reaction were spotted on TLC plates and separated using a saturated (NH4)2SO4 and 1.5 M KH2PO4 buffer. The spots were imaged on a Typhoon FLA 9500 scanner.
Effect of hydroxybenzylidene-indolinones on bacterial viability
The effect on the growth of S. aureus, L. monocytogenes, P. aeruginosa and E. coli strains was determined by assessing culture turbidity after 24 h. Briefly, overnight cultures of the bacteria were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in MHB and cultured for 2–3 h (early exponential) at 37 °C. Aliquots were then dispensed into sterile glass tubes containing stock solutions of compounds in DMSO to yield a final concentration of 16 μg mL−1. For the E. coli with colistin experiment, 0.03125 μg mL−1 colistin was added to the cultures before adding either compound 1 or an equal volume of DMSO. The cultures were incubated at 37 °C for 24 h and the OD600 of each culture was measured using a BioTek Cytation 5 Cell Imaging Multi-Mode Reader.
000 in MHB and cultured for 2–3 h (early exponential) at 37 °C. Aliquots were then dispensed into sterile glass tubes containing stock solutions of compounds in DMSO to yield a final concentration of 16 μg mL−1. For the E. coli with colistin experiment, 0.03125 μg mL−1 colistin was added to the cultures before adding either compound 1 or an equal volume of DMSO. The cultures were incubated at 37 °C for 24 h and the OD600 of each culture was measured using a BioTek Cytation 5 Cell Imaging Multi-Mode Reader.
Minimum inhibitory concentration determination
The MIC of active compounds were determined according to the guideline of the CLSI. Cation-adjusted Mueller–Hinton broth was routinely used. Aliquots of 0.5 McFarland standardized inoculum were dispensed into wells of 96 well plate to 5 × 105 CFU mL−1 with test compounds at a final concentrations from 128 μg mL−1 to 0.5 μg mL−1. Vancomycin and methicillin were routinely used as the antibiotic controls and were tested within the same range of concentrations. The cultures were incubated at 35 °C for 20 h after which wells were visually inspected for turbidity. The MIC was defined as the lowest concentration of compound or antibiotic to result in no visible growth.Biofilm studies
MRSA biofilm inhibition was performed in tissue culture treated 96 well plates (CellTreat Sci. Pdt, MA, USA). Overnight cultures of MRSA ATCC 33592 were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in fresh tryptic soy broth (TSB) supplemented with 1% glucose. The diluted culture was inoculated into wells with compound (at 16 μg mL−1 to 0.03125 μg mL−1). The plates were incubated at 37 °C for 24 h after which the medium was carefully removed and the unattached cells washed away. The biofilms were stained with 0.1% crystal violet for 30 min. The crystal violet was removed and wells washed until no crystal violet was present in the wash. The dye was solubilized with 100% ethanol for 1 h and the biofilm mass was quantified by measuring absorbance at 595 nm on a BioTek Cytation 5 Cell Imaging Multi-Mode Reader. The A595 value for any absorbance reading, A was normalized to the no compound (AT) and broth (Ao) controls using the equation
100 in fresh tryptic soy broth (TSB) supplemented with 1% glucose. The diluted culture was inoculated into wells with compound (at 16 μg mL−1 to 0.03125 μg mL−1). The plates were incubated at 37 °C for 24 h after which the medium was carefully removed and the unattached cells washed away. The biofilms were stained with 0.1% crystal violet for 30 min. The crystal violet was removed and wells washed until no crystal violet was present in the wash. The dye was solubilized with 100% ethanol for 1 h and the biofilm mass was quantified by measuring absorbance at 595 nm on a BioTek Cytation 5 Cell Imaging Multi-Mode Reader. The A595 value for any absorbance reading, A was normalized to the no compound (AT) and broth (Ao) controls using the equationAntibiotic-hydroxybenzylidene-indolinones combination analysis
We used the standard checkerboard assay to determine the ability of the hydroxybenzylidene-indolinones to potentiate the activity of antibiotics (methicillin and vancomycin).40 Briefly, the antibiotics were serially diluted along the abscissa of a 96 well microtiter plate to achieve a starting concentration of 128 μg mL−1 whilst the hydroxybenzylidene-indolinones were diluted along the ordinate. An inoculum equal to a 0.5 McFarland turbidity standard was prepared for both MRSA ATCC 33592 and VRE faecalis ATCC 51575 in sterile saline. Aliquots of the standardized inoculum were dispensed into the microtiter plates to give 5 × 105 CFU mL−1. The plates were then incubated at 35 °C for 20 h. Guided by the MIC values of the compounds and antibiotics, we calculated the fold change for wells with no visible turbidity.Acknowledgements
This work was funded by the National Science Foundation (CHE1307218 and CHE1636752), Purdue University. We thank Dr Karl-Peter Hopfner and Dr Angelika Gründling for plasmids encoding DisA.Notes and references
- M. E. de Kraker, A. J. Stewardson and S. Harbarth, PLoS Med., 2016, 13, e1002184 Search PubMed.
- G. D. Wright, ACS Infect. Dis., 2015, 1, 80–84 CrossRef CAS PubMed.
- G. Witte, S. Hartung, K. Buettner and K.-P. Hopfner, Mol. Cell, 2008, 30, 167–178 CrossRef CAS PubMed.
- R. M. Corrigan, J. C. Abbott, H. Burhenne, V. Kaever and A. Gründling, PLoS Pathog., 2011, 7, e1002217 CAS.
- J. J. Woodward, A. T. Iavarone and D. A. Portnoy, Science, 2010, 328, 1703–1705 CrossRef CAS PubMed.
- T. Kamegaya, K. Kuroda and Y. Hayakawa, Nagoya J. Med. Sci., 2011, 73, 49–57 CAS.
- Y. Bai, J. Yang, X. Zhou, X. Ding, L. E. Eisele and G. Bai, PLoS One, 2012, 7, e35206 CAS.
- J. R. Barker, B. J. Koestler, V. K. Carpenter, D. L. Burdette, C. M. Waters, R. E. Vance and R. H. Valdivia, mBio, 2013, 4, e00018-13 CrossRef PubMed.
- Y. Luo and J. D. Helmann, Mol. Microbiol., 2012, 83, 623–639 CrossRef CAS PubMed.
- C. E. Witte, A. T. Whiteley, T. P. Burke, J.-D. Sauer, D. A. Portnoy and J. J. Woodward, mBio, 2013, 4, e00282-13 CrossRef PubMed.
- L. Zhang, W. Li and Z.-G. He, J. Biol. Chem., 2013, 288, 3085–3096 CrossRef CAS PubMed.
- Y. Bai, J. Yang, T. M. Zarrella, Y. Zhang, D. W. Metzger and G. Bai, J. Bacteriol., 2014, 196, 614–623 CrossRef PubMed.
- X. Peng, Y. Zhang, G. Bai, X. Zhou and H. Wu, Mol. Microbiol., 2016, 99, 945–959 CrossRef CAS PubMed.
- J. H. Song, K. S. Ko, J. Y. Lee, J. Y. Baek, W. S. Oh, H. S. Yoon, J. Y. Jeong and J. Chun, Mol. Cells, 2005, 19, 365–374 CAS.
- R. R. Chaudhuri, A. G. Allen, P. J. Owen, G. Shalom, K. Stone, M. Harrison, T. A. Burgis, M. Lockyer, J. Garcia-Lara, S. J. Foster, S. J. Pleasance, S. E. Peters, D. J. Maskell and I. G. Charles, BMC Genomics, 2009, 10, 291 CrossRef PubMed.
- C. T. Walsh and T. A. Wencewicz, J. Antibiot., 2014, 67, 7–22 CrossRef CAS PubMed.
- C. Opoku-Temeng, J. Zhou, Y. Zheng, J. M. Su and H. O. Sintim, Chem. Commun., 2016, 52, 9327–9342 RSC.
- Y. Zheng, J. Zhou, D. A. Sayre and H. O. Sintim, Chem. Commun., 2014, 50, 11234–11237 RSC.
- C. Opoku-Temeng and H. O. Sintim, Chem. Commun., 2016, 52, 3754–3757 RSC.
- C. Opoku-Temeng and H. O. Sintim, Sci. Rep., 2016, 6, 25445 CrossRef CAS PubMed.
- J. D. Lambert and C. S. Yang, J. Nutr., 2003, 133, 3262S–3267S CAS.
- M. Müller, T. Deimling, K. P. Hopfner and G. Witte, Biochem. J., 2015, 469, 367–374 CrossRef PubMed.
- J. Zhou, D. A. Sayre, Y. Zheng, H. Szmacinski and H. O. Sintim, Anal. Chem., 2014, 86, 2412–2420 CrossRef CAS PubMed.
- P. C. Chin, L. Liu, B. E. Morrison, A. Siddiq, R. R. Ratan, T. Bottiglieri and S. R. D'Mello, J. Neurochem., 2004, 90, 595–608 CrossRef CAS PubMed.
- Y. Lei, Y. X. Cao, C. B. Xu and Y. Zhang, Respir. Res., 2008, 9, 71 CrossRef PubMed.
- D. Davies, Nat. Rev. Drug Discovery, 2003, 2, 114–122 CrossRef CAS PubMed.
- CDC, Antibiotic resistance threats in the United States, Report CS239559-B, 2013, http://www.cdc.gov/drugresistance/threats-report-2013.
- J. L. Lister and A. R. Horswill, Front. Cell. Infect. Microbiol., 2014, 4, 178 Search PubMed.
- R. J. Worthington, J. J. Richards and C. Melander, Org. Biomol. Chem., 2012, 10, 7457–7474 CAS.
- A. A. Yeagley, Z. Su, K. D. McCullough, R. J. Worthington and C. Melander, Org. Biomol. Chem., 2013, 11, 130–137 CAS.
- G. A. O'Toole, J. Visualized Exp., 2011, 47, e2437 Search PubMed.
- L. Kalan and G. D. Wright, Expert Rev. Mol. Med., 2011, 13, e5 CrossRef PubMed.
- R. Laxminarayan, A. Duse, C. Wattal, A. K. Zaidi, H. F. Wertheim, N. Sumpradit, E. Vlieghe, G. L. Hara, I. M. Gould, H. Goossens, C. Greko, A. D. So, M. Bigdeli, G. Tomson, W. Woodhouse, E. Ombaka, A. Q. Peralta, F. N. Qamar, F. Mir, S. Kariuki, Z. A. Bhutta, A. Coates, R. Bergstrom, G. D. Wright, E. D. Brown and O. Cars, Lancet Infect. Dis., 2013, 13, 1057–1098 CrossRef PubMed.
- G. D. Wright, Trends Microbiol., 2016, 24, 862–871 CrossRef CAS PubMed.
- T. L. Harris, R. J. Worthington and C. Melander, Angew. Chem., Int. Ed. Engl., 2012, 51, 11254–11257 CrossRef CAS PubMed.
- H. Yamazaki, K. Nonaka, R. Masuma, S. Omura and H. Tomoda, J. Antibiot., 2009, 62, 431–434 CrossRef CAS PubMed.
- C. M. Brackett, R. J. Melander, I. H. An, A. Krishnamurthy, R. J. Thompson, J. Cavanagh and C. Melander, J. Med. Chem., 2014, 57, 7450–7458 CrossRef CAS PubMed.
- G. Chiosis and I. G. Boneca, Science, 2001, 293, 1484–1487 CrossRef CAS PubMed.
- Y. Zheng, G. Tsuji, C. Opoku-Temeng and H. O. Sintim, Chem. Sci., 2016, 7, 6238–6244 RSC.
- G. Orhan, A. Bayram, Y. Zer and I. Balci, J. Clin. Microbiol., 2005, 43, 140–143 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary figures, methods for synthesis of compounds and characterization of compounds. See DOI: 10.1039/c6ra28443d |
| This journal is © The Royal Society of Chemistry 2017 |