Open Access Article
Open Access ArticleA highly selective protein adsorber via two-step surface-initiated molecular imprinting utilizing a multi-functional polymeric scaffold on a macroporous cellulose membrane†
Dejing Liu and
Mathias Ulbricht *
*
Lehrstuhl für Technische Chemie II, Universität Duisburg-Essen, 45141 Essen, Germany. E-mail: mathias.ulbricht@uni-essen.de
First published on 10th February 2017
Abstract
Although molecularly imprinted materials using small organic molecules as templates have been well established, development of such materials for protein separation is still rather challenging. We therefore describe herein a two-step surface imprinting method established with a hydrophilic macroporous cellulose membrane with relatively large specific surface area. In the first step, tailor-made multi-functional polymer chains were grafted on the cellulose membrane using photo-initiated graft copolymerization, enabled by a surface-immobilized photo-initiator. This scaffold allowed the preorganization of the template protein lysozyme (Lys) on the surface of the membrane pores. Notably, the scaffold-grafted membrane showed already a significant adsorption selectivity versus the very similar protein cytochrome C (CyC). In the second step, surface-initiated cross-linking copolymerization, enabled by a photo-initiator immobilized in the scaffold layer, resulted in a protein-imprinted cellulose membrane. Imprinting efficiency was further improved by optimization of monomer concentrations in the second step. Protein selectivity of the best imprinted cellulose membrane for Lys over CyC reached a very remarkable value of about 45, measured with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixtures of the two proteins. We envision that this property of a protein-imprinted cellulose membrane, which is based on the tailored binding selectivity achieved using the two-step functionalization method, could be largely beneficial for separation and purification of target proteins from complex mixtures.
1 mixtures of the two proteins. We envision that this property of a protein-imprinted cellulose membrane, which is based on the tailored binding selectivity achieved using the two-step functionalization method, could be largely beneficial for separation and purification of target proteins from complex mixtures.
1. Introduction
Molecularly imprinted materials have gained much interest in recent years.1–5 The introduction of template molecules during assembling and polymerization process results in the formation of binding sites in polymeric matrix. After template removal, such binding sites (often also called “cavities”) exhibit excellent ability for recognition and separation of target molecules. However, although the imprinting with small organic molecules, for example cholesterol, has been well established,6–8 development of imprinted materials for large biomolecules, especially proteins, is still rather challenging.4,9–11 Various types of molecularly imprinted polymers (MIP) have thus been developed to achieve selective and efficient protein separation.12–16 For example, the group of Sellergren had reported on the selective capture of human serum albumin (HSA) or immunoglobulin G (IgG) from blood serum using materials prepared by hierarchical surface imprinting method with wide-pore silica beads.10 The group of Shea had developed surface protein imprinted films using exposed protein epitopes as templates.17 Chou et al. explored protein stamping and micro-contact methods to prepare protein imprinted film between two glass slides.18 Notably, other promising strategies toward protein-imprinted materials such as scaffold imprinting and epitope imprinting using silica nanoparticles, polymeric hydrogel and gold supported substrate etc. have also been reported.19–26Despite the significant progress, there are still some problems, especially when proteins are applied as template/target.4 The major problem is the restricted mass transfer across the cross-linked polymer matrix due to the large sizes of proteins, which limits the ease of template removal as well as target rebinding. The complex and flexible structures of proteins also significantly reduced usable effects of non-covalent interactions between template and monomers in aqueous media.27
Accordingly, we previously reported a two-step method to obtain protein-imprinted macro-porous membrane as a highly promising strategy to overcome these obstacles.28,29 This method combined the advantages of surface grafting toward thin imprinted films with the scaffold approach where large templates can be pre-organized before the actual cross-linking step. The resulting membranes can also be used in flow-through mode, leading to very high overall separation efficiency because the binding sites are accessible by convective transport.30 Track-etched polyethyleneterephthalate (PET) membranes had been used because their well-defined pore structure enabled the detailed characterization of functionalization and protein binding.28,29,31 Through the thin surface-grafted hydrogel network layer within the large pores in PET membrane, the transfer of large template such as protein molecule was largely enhanced. Moreover, decoupling the optimization of several important parameters in surface imprinting by a two-step method provided an easy way to control protein binding capacity and selectivity relationship: the scaffold grafted on PET membrane through surface-initiated atom transfer radical polymerization (SI-ATRP) could control the amount of template adsorbed as well as its pre-organization; the thickness of hydrogel layer could easily be fine-tuned by the exposure time to UV irradiation in the second step. Additionally, the cross-linking density of hydrogel layer could also be varied through different monomer compositions in the second step. Success of the method had been demonstrated for lysozyme (Lys)28 and immunoglobulin G (IgG).29 Furthermore, the feasibility of the transfer of the SI-ATRP/UV grafting method to macroporous cellulose membranes had been demonstrated as well.32
However, the selectivity of the recognition cavity in these materials over other proteins was still far from perfect. The scaffold grafted from PET membrane only contained rather simple functionalities such as carboxylic acid groups. Although certain extent of template protein adsorption and pre-organization could be achieved through the binding between carboxylic acid and protein surface, such interactions are usually rather weak and unspecific. Considering the application of these types of materials in biologically relevant conditions in which various types of proteins co-exist in media, the specificity of protein adsorption needs to be improved. Also, the pore diameter of PET membrane was around 650 nm at a very low porosity of 10%, yielding an accessible specific surface area of only 1.2 m2 g−1; this limits its further application for protein capture due to the low capacity.
Recently, we demonstrated a heterogeneous photo-graft copolymerization method based on a covalently immobilized “type I” photo-initiator benzoin ether derivative.33 Various tailor-made acrylamide based functional monomers had been directly grafted on macro-porous cellulose membranes through radical polymerization. In fact, this direct photo-grafting method had also proven to be very efficient to obtain IgG-selective imprinted membranes by using a mixture of functional monomers, cross-linker monomer and an oligopeptide (epitope) derived from the chain end of IgG as template.34 Regenerated cellulose membrane is superior for the binding and recognition of large protein template. With a pore diameter of around 1 μm and a porosity of 70%, the accessible specific surface area thus can reach 6.3 m2 g−1; this is 5 fold larger than for previously used PET membranes. Therefore, we hypothesized that the combination of surface-selective photo-initiation, tailor-made functional monomers and macroporous cellulose membrane could provide an excellent molecular imprinted material for selectively protein separation.
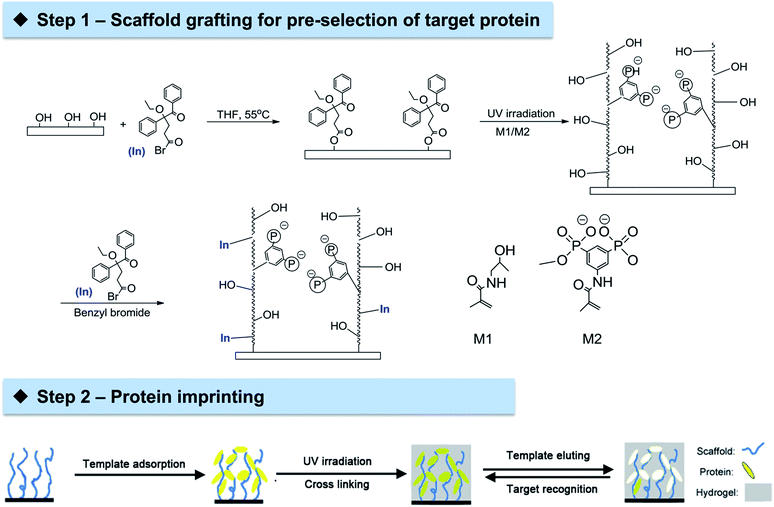
The general approach could be divided into two steps (Scheme 1). The first step is the grafting of cellulose membrane with a polymeric layer comprised of tailor-made functionalities through UV irradiation. Arginine-selective functional monomer bisphosphonato-m-xylylene methacrylamide (M2) was employed in copolymers with hydrophilic methacryloylamino-2-hydroxypropane (M1). This combination, grafted on PET membranes, was previously shown to be rather effective in the adsorption of the model protein lysozyme (Lys).35 The binding between the bisphosphonate groups in M2 and arginine residues on the protein surface may allow an ideal organization of template Lys which could facilitate subsequent imprinting. The second step is the imprinting of template Lys via an orthogonal initiator on cellulose membrane surface using conventional non-functional monomers acrylamide and methylene bisacrylamide. This novel two-step approach to prepare protein imprinted cellulose membrane exhibited excellent ability in achieving highly selective protein adsorption and separation.
2. Experimental section
Materials
Regenerated cellulose membranes with experimentally determined mean pore diameter of 1.2 μm and BET surface area of 6.3 m2 g−1 (cf. ref. 34) were purchased from Whatman. Photo-initiator derivative 4-ethoxy-5-oxo-4,5-diphenylpentanoyl bromide (In) which could be directly used for coupling to cellulose hydroxyl groups was synthesized following a published protocol (see ESI; including Fig. S1†).33 N-(2-Hydroxypropyl)-methacrylamide (M1) was from Polysciences Europe. 5-(Methacryloylamino)-m-xylylene bisphosphonic acid dimethylester dilithium salt (M2) was synthesized following a published protocol (see ESI; including Fig. S2†).35 Benzoyl bromide (97%), triethylamine (Et3N, 99%), tetrahydrofuran (THF, HPLC grade), hydrochloric acid (1 M), and N,N′-methylenebisacrylamide (MBAA; 99%), were obtained from Sigma Aldrich. Lysozyme (isoelectric point, pI ∼ 9.1, molecular weight, Mw ∼ 14 kDa) was from ICN Biomedicals; cytochrome C (pI ∼ 9.2, Mw ∼ 12 kDa) was from Sigma-Aldrich. Acrylamide (Am; 98.5%) was purchased from Acros. Sodium hydroxide was from VWR. The kit for BCA protein assay was from Thermo Scientific. Substrate for determination of Lys enzyme activity was Micrococcus lysodeikticus (ATCC no. 4698, Sigma-Aldrich). All chemicals were used as received. Water purified with a Milli-Q system (Millipore) was used for all experiments.Photo-initiator immobilization on cellulose membrane
Cellulose membrane (d = 47 mm) was immersed in a solution of 100 mg photo-initiator derivative (In) and 49.3 mg benzoyl bromide (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio) with 100 mg Et3N dissolved in 4 ml dry THF, and reacted in a gas-tight vessel for 4 h at 55 °C on a shaker with a speed of 300 rpm (cf. Scheme 1). Thereafter, the membrane was washed with 20 ml ethanol, 20 ml water and again 20 ml ethanol for at least 30 min each; then it was dried. Determination of the photo-initiator and benzoic acid density on the surface was done via acid ester cleavage from cellulose membranes. The membranes were reacted in 3 ml 0.1 M HCl for 2 h and the resulting solutions were analyzed with HPLC. The HPLC system comprised a P580 pump from Dionex and an auto-injector 235 from Gilson. A Cromasil C18 RP column, elution buffer with a mixture of methanol (70%), water (29.9%), and TFA (0.1%) and a flow rate of 1 ml min−1 were used. The detection was done with a UV detector (Shimadzu) at 250 nm.
1 molar ratio) with 100 mg Et3N dissolved in 4 ml dry THF, and reacted in a gas-tight vessel for 4 h at 55 °C on a shaker with a speed of 300 rpm (cf. Scheme 1). Thereafter, the membrane was washed with 20 ml ethanol, 20 ml water and again 20 ml ethanol for at least 30 min each; then it was dried. Determination of the photo-initiator and benzoic acid density on the surface was done via acid ester cleavage from cellulose membranes. The membranes were reacted in 3 ml 0.1 M HCl for 2 h and the resulting solutions were analyzed with HPLC. The HPLC system comprised a P580 pump from Dionex and an auto-injector 235 from Gilson. A Cromasil C18 RP column, elution buffer with a mixture of methanol (70%), water (29.9%), and TFA (0.1%) and a flow rate of 1 ml min−1 were used. The detection was done with a UV detector (Shimadzu) at 250 nm.
Photo-initiated graft copolymerization on cellulose membrane
The above prepared membrane was placed in a Petri dish, 1 ml degassed monomer solution (100 mg ml−1 in water) was added and the membrane was sealed with a second smaller Petri dish. UV irradiation was performed in an UV Cube 2000 from Hoenle for 25 min (intensity measured with the UVA meter of Hoenle). Different monomer ratios and UV intensities were evaluated. The membranes were washed two times. The degree of grafting (DG) was determined gravimetrically through the following equation, where W0 and W1 represent the sample weights before and after modification, respectively, and Sspec stands for specific surface area of the membrane (for the used membrane 6.3 m2 g−1; cf. above):| DG = (W1 − W0)/(W0Sspec). |
Thereafter, the photo-initiator was again attached on the grafted cellulose membrane as described above, now mainly reacting with hydroxyl groups of M1 segments of grafted chains (cf. Scheme 1).
Preparation of Lys imprinted membrane
Afterwards, template adsorption on the grafted membranes was performed using 8 ml of a 1 g L−1 solution of Lys in PBS buffer (10 mM, pH 7.4) for 20 h. The membranes were then washed in 20 ml water for 1 h twice to remove protein weakly bound on membrane. Subsequently, the membranes were wetted with the imprinting monomer solution (Am and MBAA in varied ratios) and mounted into a filter holder (Swinnex, Millipore). The imprinting solution was gently pushed through the membrane with a 20 ml syringe (Omnifix, B. Braun) for 3 times, then the membranes were immersed in the solution for 1.5 h. UV initiation was done in a UV irradiation system UVA Cube 2000 (Hoenle). Afterwards, the membranes were taken out and washed 3 times in water to remove the unreacted monomers. Finally, template Lys was removed from the membranes using 10 mM phosphate buffer containing 1 M NaCl (pH = 7.4) via shaking with a rate of 250 rpm for 20 h. All membranes were stored in 10 mM phosphate buffer (pH = 7.4) until further use.Static binding capacity of proteins
Protein solutions were prepared by dissolving the corresponding protein in PBS buffer solution (10 mM, pH = 7.4). The static protein adsorption capacity was measured using batch experiments. Various membranes with the same uniform diameter of 47 mm were incubated overnight at room temperature in 8 ml protein solutions with 0.2 mg ml−1 initial concentration. The equilibrated membrane samples were taken out of the solution and washed 3 times using binding buffer (for 20 min each time). Thereafter, the protein adsorbed membranes were soaked in 5 ml elution solution (10 mM PBS buffer containing 1 M NaCl, pH = 7.4) to release bounded protein. The BCA protein assay was used for both Lys and CyC concentration determination in the eluted solution. The absorption spectrum was recorded using a UV-Vis spectrophotometer (JASCO V550), and quantification was done using respective calibration curves (see ESI Fig. S3 and S4†).In situ selective adsorption of proteins
The same procedures as above were followed to determine the separation property of G2-IPb membranes in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of lysozyme and cytochrome C with 0.2 mg ml−1 initial concentration, respectively. UV-Vis absorbance was used for quantification, making use of the fact that only CyC has an absorbance maximum at 410 nm while both proteins have another one at 280 nm. Hence, CyC concentration was determined through its UV-Vis calibration curve at 410 nm, and Lys concentration was also determined by an independent method using it enzyme activity with Micrococcus lysodeikticus as substrate (see ESI, Fig. S5 and S6†). For comparison, spectra of the lysozyme and cytochrome C solutions with the same concentration as the ones eluted from the membranes were also measured.
1 mixture of lysozyme and cytochrome C with 0.2 mg ml−1 initial concentration, respectively. UV-Vis absorbance was used for quantification, making use of the fact that only CyC has an absorbance maximum at 410 nm while both proteins have another one at 280 nm. Hence, CyC concentration was determined through its UV-Vis calibration curve at 410 nm, and Lys concentration was also determined by an independent method using it enzyme activity with Micrococcus lysodeikticus as substrate (see ESI, Fig. S5 and S6†). For comparison, spectra of the lysozyme and cytochrome C solutions with the same concentration as the ones eluted from the membranes were also measured.
Water permeability measurements
The water flux of membranes was measured using stirred cells with 10 ml volume and 4.15 cm2 effective membrane area (Amicon Model 8010, Millipore). 0.12 bar transmembrane pressure was used by adjusting the nitrogen pressure from a gas flask.3. Results and discussion
Grafting of monomer M1 and M2 on cellulose membrane and resulting protein binding properties
The same base membrane as in previous studies had been used here.33,34 The photo-initiator had been covalently coupled to the surface. The density of photo-initiator was found to be 15.8 pmol cm−2 (specific surface area) as determined by analytical HPLC through its hydrolysis from cellulose membrane (ESI; Fig. S7†). Regenerated cellulose membrane was then in the first photo-grafting step functionalized using tailor-made acrylamide based functional monomers as shown in Scheme 1, step 1. Different combinations of functional monomers M1 and M2 and conditions were used for grafting (Table 1). Owing to the different reactivity of M1 and M2, their overall concentration was kept constant while varying the ratio of each monomer to obtain efficient grafting. Fig. 1 shows that the degree of grafting (DG) was correlated with the relative ratio of M1 and M2. This result is not surprising as monomer M2 has a much lower reactivity in radical polymerization than M1. Increasing the amount of M1 would naturally lead to a higher degree of grafting. However, the increased DG did not guarantee a higher binding capacity of cellulose membrane. As can also be seen in Fig. 1, the membrane grafted with a lower DG (membrane G1) actually exhibited a higher lysozyme binding capacity compared to the one with higher DG (membrane G2). The selectivity of membrane G1 was lower than that of membrane G2. The binding capacity for cytochrome C (CyC), a protein structurally similar to lysozyme (almost identical size and isoelectric point) was almost three fold higher in case of membrane G1 than of G2. Consequently, the Lys vs. CyC selectivity was much lower for membrane G1. This indicates that a too high density of M2 segments on the membrane surface leads to an over-proportional increase of less selective binding; in this case caused by ion-exchange binding of the positively charged proteins.| Scaffold-grafted membrane | Monomer ratio | Monomer concentration | UV irradiation |
|---|---|---|---|
| G1 | M1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M2 = 1 M2 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
100 mg ml−1 | 17.5 mW cm−2; t = 25 min |
| G2 | M1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M2 = 3 M2 = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
100 mg ml−1 | 55 mW cm−2; t = 25 min |
| Protein-imprinted membrane | Monomer (Am) concentration | Cross-linker monomer fraction | UV irradiation |
|---|---|---|---|
| G2-IPa | 0.5 M | 15% | 17.5 mW cm−2; t = 25 min |
| G2-IPb | 0.3 M | 15% | 55 mW cm−2; t = 25 min |
| 0.5 M | 15% | 55 mW cm−2; t = 25 min | |
| 0.56 M | 15% | 55 mW cm−2; t = 25 min | |
| G2-IPc | 0.56 M | 7.5% | 55 mW cm−2; t = 25 min |
| 0.56 M | 15% | 55 mW cm−2; t = 25 min | |
| 0.56 M | 20% | 55 mW cm−2; t = 25 min | |
| G2-IPd | 0.56 M | 15% | 55 mW cm−2; t = 8 min |
| 0.56 M | 15% | 55 mW cm−2; t = 15 min | |
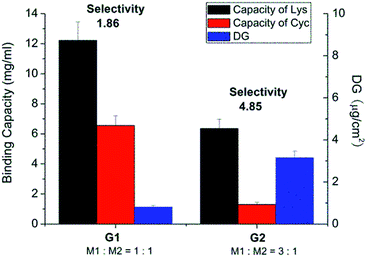 | ||
| Fig. 1 Results after optimization of photo-grafting conditions and the resulting protein binding capacities and Lys vs. CyC selectivity of different grafted membranes in pH 7.4 PBS buffer solution. | ||
Imprinting of lysozyme on cellulose membrane
Acrylamide (Am), a monomer with amide functionality, was used for the imprinting step because of its excellent hydrogen bonding ability and reactivity in radical polymerization. Scaffold-grafted membrane G2 was applied to prepare imprinted membranes G2-IPa. The binding capacity and selectivity towards lysozyme were then tested. However, surprisingly, membrane G2-IPa did not exhibit imprinting effect, i.e. the selectivity was not increased compared to scaffold-grafted membranes G2 (see exemplary data in Fig. 2). This is probably because the conditions applied in imprinting step had not been optimized. Cross-linking polymerization of monomer Am apparently did not lead to the formation of binding cavity on the surface of grafted cellulose membrane.
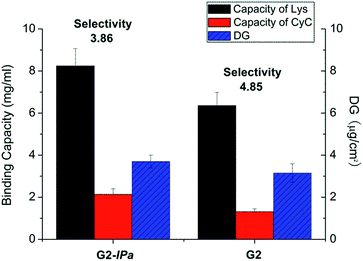 | ||
| Fig. 2 Protein binding capacity and Lys vs. CyC selectivity for imprinted membrane G2-IPa and its scaffold-grafted precursor membrane G2 (cf. Table 1). | ||
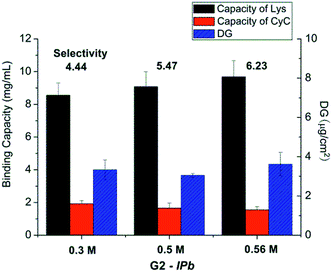 | ||
| Fig. 3 Results for optimization of monomer concentration in the imprinting step (scaffold-grafted membrane G2-IPb; cf. Table 1). | ||
The influences of cross-linking degree, via fraction of cross-linker monomer in the reaction mixture, and UV grafting time on imprinting, were also evaluated (cf. Table 1). However, there seemed no obvious relationship between binding selectivity and cross-linking degree (data not shown). Neither did the UV exposure time show any significant effect on the binding selectivity of resulting imprinted membranes (see Fig. 4). After all these variations, the DG had also not been influenced by the chosen conditions. This indicates that the reaction conditions at sufficiently high UV intensity, the highest Am concentration and sufficiently high cross-linker concentration and UV reaction time lead to similar structures in the cross-linked polymer layer. This can be related to the confinement of photo-initiator within the polymer layer and the cross-linking character of the polymerization within a grafted polymer layer, leading to a complex (semi) interpenetrating polymer network structure.
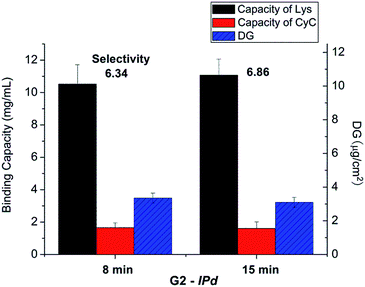 | ||
| Fig. 4 Results for optimization of UV irradiation time in the imprinting step for scaffold-grafted membrane G2; cf. Table 1. | ||
Protein binding for non-imprinted membranes prepared under optimized conditions had also been studied, and the results had been compared with those for the respective scaffold-grafted as well as the in parallel prepared imprinted membranes (ESI, Fig. S8†). Bound amounts for both Lys and CyC and, consequently, binding selectivity were within the range of error identical for scaffold-grafted and non-imprinted membrane. Lys binding capacity and hence Lys/CyC selectivity were significantly higher for the imprinted membranes. That protein binding capacities for non-imprinted membranes were similar to the ones for scaffold-grafted membranes indicates that the second step of the two-step molecular imprinting procedure mainly causes local cross-linking in the grafted layer. In presence of the template protein this leads to selective sites in a globally not much changed hydrogel layer. This interpretation is supported by the fact that the increase of degree of grafting for non-imprinted and imprinted membranes in the second step was very small (in some cases within the range of error; cf. ESI, Fig. S8†).
The binding of bovine serum albumin (BSA) at pH 7.4 (i.e., same conditions as for all other binding analyses) to non-imprinted and imprinted membranes had also been analyzed. The data were below detection limit for both membranes. This can be related to the net negative charge of the membrane surface due the fraction of monomer M2 in the grafted functional layer (cf. Scheme 1) and the negative charge of BSA (isoelectric point 5.8).
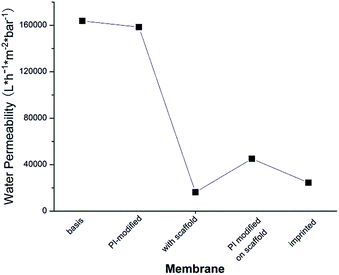
As can be seen in Fig. 5, water permeability of cellulose membrane was only slightly reduced after PI immobilization. This can be related to the more hydrophobic surface after this step. Water permeability largely decreased after polymer scaffold grafting. This is because liquid flow through the pores of cellulose membrane was partially hindered by the relatively long polymer chains which were well solvated by water. This is analogous to previous work where a similar grafted copolymer had been prepared by a different photo-initiation method.35 Grafted chains have a broad molecular weight distribution, and few long chains can cause a relatively large drop in permeability. In contrast to this outcome of surface-initiated free radical polymerization (in this and the above mentioned previous work), SI-ATRP what had been used in the original work to establish the two-step surface imprinting method is more controlled; but the efforts to control the reaction conditions are higher (cf. ref. 28 and 29). After the second coupling of photo-initiator on the grafted chains, the thus introduced hydrophobic groups of the PI caused a significant de-swelling of the polymer chains. The de-swelling resulted in the enlargement of the membrane pores which led to the increase of water permeability. After imprinting, a cross-linked polymer gel layer had been grafted on the pore walls. Because the mass per surface area slightly increased in this step, the water permeability decreased again, but due to the cross-linked character of the grafted layer the values were still larger than for the scaffold-grafted layer containing linear grafted chains.
In situ separation of protein mixture (Lys/CyC) by Lys-imprinted membrane
The best performing imprinted cellulose membrane G2-IPb (cf. Table 1) with a lysozyme binding capacity of 9.2 mg ml−1 and a selectivity of Lys vs. CyC of 6.2, all measured with solutions of single proteins (0.2 g L−1, 10 mM PBS buffer, pH = 7.4) was used to directly separate Lys from its mixture with CyC. Membrane G2-IPb was immersed into Lys/CyC mixture (0.2 g L−1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
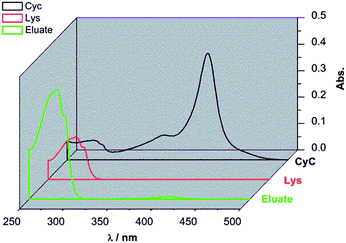
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) overnight to ensure full saturation of all binding sites. The membrane was thoroughly washed with binding buffer to remove all the weakly bound proteins. Afterwards, the membrane was eluted with elution buffer with high salt concentration (pH = 7.4, 10 mM PBS buffer containing 1 M NaCl). As shown in Fig. 6, the eluate (green curve) had strong absorption around 280 nm corresponding to Lys. Meanwhile, the absorption peak around 410 nm for CyC was very weak. According to additional quantification versus the calibration curves for Lys and CyC (cf. ESI; Fig. S5 and S6†), the binding capacities of G2-IPb membrane were 8.0 mg ml−1 for Lys and 0.18 mg ml−1 for CyC. This result clearly showed that imprinted G2-IPb cellulose membrane can highly efficiently separate Lys from the protein mixture. Because quantification of Lys was based on its enzyme activity, data also confirm that the target protein can be recovered in its native structure.
1 ratio) overnight to ensure full saturation of all binding sites. The membrane was thoroughly washed with binding buffer to remove all the weakly bound proteins. Afterwards, the membrane was eluted with elution buffer with high salt concentration (pH = 7.4, 10 mM PBS buffer containing 1 M NaCl). As shown in Fig. 6, the eluate (green curve) had strong absorption around 280 nm corresponding to Lys. Meanwhile, the absorption peak around 410 nm for CyC was very weak. According to additional quantification versus the calibration curves for Lys and CyC (cf. ESI; Fig. S5 and S6†), the binding capacities of G2-IPb membrane were 8.0 mg ml−1 for Lys and 0.18 mg ml−1 for CyC. This result clearly showed that imprinted G2-IPb cellulose membrane can highly efficiently separate Lys from the protein mixture. Because quantification of Lys was based on its enzyme activity, data also confirm that the target protein can be recovered in its native structure.
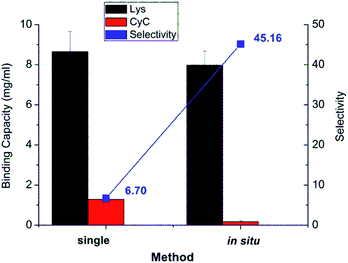
The results from in situ separation experiment with protein mixture were also compared with the binding profile when using pure protein in solution, as shown in Fig. 7. The recognition property was obviously much more enhanced in in situ experiment compared to two individual protein experiments. The selectivity of in situ experiment was 45.2, much higher than that from the two single protein experiments of 6.7. The reason for that is that in the binding of protein mixture, target protein Lys bound with imprinted membrane much faster and stronger than CyC. The bound Lys thus occupied the imprinted cavity which prevented the unspecific interaction between the cavity and CyC; this further confirmed the effect of imprinting. This was much higher than that of only simple cation-exchange polymer scaffold, obtained via SI-ATRP, and subsequent photo-grafting on the same cellulose base membrane (≈4).32
Overall, cellulose-based membranes prepared via two-step imprinting method under conditions used for G2-IPb exhibit a very high potential for application for selective separation of target proteins in its mixture.
4. Conclusions
In conclusion, a two-step surface grafting had been established to hydrophilic cellulose membrane with a relatively large specific surface area. The performance of the imprinted cellulose membranes was improved by optimization of monomer concentration in the second step. It had further been shown that the UV irradiation time and cross-linking degree in the second step did not significantly influence final imprinting effect. Through the combination of multifunctional polymeric scaffold grafting and UV-grafting/crosslinking copolymerization for lysozyme imprinting, both with an immobilized photo-initiator, protein selectivity of imprinted cellulose membrane G2-IPb for lysozyme over cytochrome C reached to around 6.5. Moreover, in situ separation experiment showed amazing separation ability. The selectivity in the recognition of Lys from its mixture with CyC was as high as 45. These results are significantly better than what had been achieved on the same base membrane with a different version of the two-step method; the here reported new version is superior due to the multi-functional scaffold (compared to simple carboxyl-containing polymer28,29,32) and the relatively easier implementation of free radical compared to controlled radical polymerization. The grafted scaffold has a significant contribution to binding specificity; therefore further variations can be used to adapt the protein-imprinting to specific targets. Overall, we envision that this property of imprinted cellulose membrane could largely facilitate future separation and purification of proteins from complex mixtures.Acknowledgements
DL acknowledges the financial support by a postdoctoral scholarship provided by the Chinese Scholarship Council (CSC).Notes and references
- L. X. Chen, X. Y. Wang, W. H. Lu, X. Q. Wu and J. H. Li, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- S. J. Li, S. S. Cao, M. J. Whitcombe and S. A. Piletsky, Prog. Polym. Sci., 2014, 39, 145–163 CrossRef CAS.
- Y. Q. Lv, T. W. Tan and F. Svec, Biotechnol. Adv., 2013, 31, 1172–1186 CrossRef CAS PubMed.
- E. Verheyen, J. P. Schillemans, M. van Wijk, M. A. Demeniex, W. E. Hennink and C. F. van Nostrum, Biomaterials, 2011, 32, 3008–3020 CrossRef CAS PubMed.
- L. X. Chen, S. F. Xu and J. H. Li, Chem. Soc. Rev., 2011, 40, 2922–2942 RSC.
- C. W. Hsu and M. C. Yang, J. Non-Cryst. Solids, 2008, 354, 4037–4042 CrossRef CAS.
- S. F. Wang, J. W. Xu, Y. J. Tong, L. Wang and C. B. He, Polym. Int., 2005, 54, 1268–1274 CrossRef CAS.
- N. Perez, M. J. Whitcombe and E. N. Vulfson, Macromolecules, 2001, 34, 830–836 CrossRef CAS.
- R. X. Gao, X. Kong, X. Wang, X. W. He, L. X. Chen and Y. K. Zhang, J. Mater. Chem., 2011, 21, 17863–17871 RSC.
- A. Nematollahzadeh, W. Sun, C. S. A. Aureliano, D. Lutkemeyer, J. Stute, M. J. Abdekhodaie, A. Shojaei and B. Sellergren, Angew. Chem., Int. Ed., 2011, 50, 495–498 CrossRef CAS PubMed.
- J. Orozco, A. Cortes, G. Z. Cheng, S. Sattayasamitsathit, W. Gao, X. M. Feng, Y. F. Shen and J. Wang, J. Am. Chem. Soc., 2013, 135, 5336–5339 CrossRef CAS PubMed.
- M. J. Whitcombe, I. Chianella, L. Larcombe, S. A. Piletsky, J. Noble, R. Porter and A. Horgan, Chem. Soc. Rev., 2011, 40, 1547–1571 RSC.
- K. G. Yang, L. H. Zhang, Z. Liang and Y. K. Zhang, Anal. Bioanal. Chem., 2012, 403, 2173–2183 CrossRef CAS PubMed.
- M. S. Zhang, J. R. Huang, P. Yu and X. Chen, Talanta, 2010, 81, 162–166 CrossRef CAS PubMed.
- T. Chen, M. W. Shao, H. Y. Xu, S. J. Zhuo, S. S. Liu and S. T. Lee, J. Mater. Chem., 2012, 22, 3990–3996 RSC.
- X. T. Shen, T. C. Zhou and L. Ye, Chem. Commun., 2012, 48, 8198–8200 RSC.
- H. Nishino, C. S. Huang and K. J. Shea, Angew. Chem., Int. Ed., 2006, 45, 2392–2396 CrossRef CAS PubMed.
- P. C. Chou, J. Rick and T. C. Chou, Anal. Chim. Acta, 2005, 542, 20–25 CrossRef CAS.
- W. Li, K. Dong, J. S. Ren and X. G. Qu, Angew. Chem., Int. Ed., 2016, 55, 8049–8053 CrossRef CAS PubMed.
- Y. Zhang, C. Y. Deng, S. Liu, J. Wu, Z. B. Chen, C. Li and W. Y. Lu, Angew. Chem., Int. Ed., 2015, 54, 5157–5160 CrossRef CAS PubMed.
- S. W. Li, K. G. Yang, B. F. Zhao, X. Li, L. K. Liu, Y. B. Chen, L. H. Zhang and Y. K. Zhang, J. Mater. Chem. B, 2016, 4, 1960–1967 RSC.
- Y. Kamon, N. Inoue, E. Mihara, Y. Kitayama, T. Ooya and T. Takeuchi, Appl. Surf. Sci., 2016, 378, 467–472 CrossRef CAS.
- S. W. Li, K. G. Yang, N. Deng, Y. Min, L. K. Liu, L. H. Zhang and Y. K. Zhang, ACS Appl. Mater. Interfaces, 2016, 8, 5747–5751 CAS.
- Y. H. He, Y. Y. Huang, Y. L. Jin, X. J. Liu, G. Q. Liu and R. Zhao, ACS Appl. Mater. Interfaces, 2014, 6, 9634–9642 CAS.
- D. J. Liu, Q. Yang, S. S. Jin, Y. Y. Song, J. F. Gao, Y. Wang and H. F. Mi, Acta Biomater., 2014, 10, 769–775 CrossRef CAS PubMed.
- J. Wackerlig and R. Schirhagl, Anal. Chem., 2016, 88, 250–261 CrossRef CAS PubMed.
- N. Adrus and M. Ulbricht, Polymer, 2012, 53, 4359–4366 CrossRef CAS.
- D. X. Yin and M. Ulbricht, J. Mater. Chem. B, 2013, 1, 3209–3219 RSC.
- D. X. Yin and M. Ulbricht, Biomacromolecules, 2013, 14, 4489–4496 CrossRef CAS PubMed.
- M. Ulbricht, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2004, 804, 113–125 CrossRef CAS PubMed.
- M. Ulbricht, Polymer, 2006, 47, 2217–2262 CrossRef CAS.
- D. X. Yin, Protein-selective adsorbers by molecular imprinting via a novel two-step surface grafting method, Ph.D. thesis, Duisburg-Essen, 2013.
- S. Schwark and M. Ulbricht, Eur. Polym. J., 2012, 48, 1914–1922 CrossRef CAS.
- S. Schwark, W. Sun, J. Stute, D. Lutkemeyer, M. Ulbricht and B. Sellergren, RSC Adv., 2016, 6, 53162–53169 RSC.
- D. M. He, W. Sun, T. Schrader and M. Ulbricht, J. Mater. Chem., 2009, 19, 253–260 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis and analytical characterization of photo-initiator and functional monomer; additional data such as chromatograms for determination of surface-immobilized photo-initiator, degree of grafting for experiments to verify efficiency of second photo-initiator immobilization, and calibration curves for protein analyses. See DOI: 10.1039/c6ra28403e |
| This journal is © The Royal Society of Chemistry 2017 |