Open Access Article
Open Access ArticleMicrobiome-based carboxylic acids production: from serum bottles to bioreactors†
Richard Hegner,
Christin Koch *,
Vanessa Riechert and
Falk Harnisch
*,
Vanessa Riechert and
Falk Harnisch *
*
Helmholtz Center for Environmental Research – UFZ, Department of Environmental Microbiology, Permoserstraße 15, 04318 Leipzig, Germany. E-mail: christin.koch@ufz.de; falk.harnisch@ufz.de
First published on 7th March 2017
Abstract
Microbiome-based anaerobic fermentations are promising platform technologies to convert low grade biomass into chemical building blocks. However, systematic investigations on their scalability are scarce. Here, microbiome-based production of medium-chain carboxylic acids from acetate and ethanol was systematically investigated across scales from serum bottles (110 mL) to standard bioreactors (2.2 L) in batch mode. Microbiome cultivation on serum bottle level for high throughput screening led to a considerable total medium-chain carboxylic acids concentration of 843.2 ± 20 C mM with 77.0 ± 2 mM n-butyrate and 89.2 ± 2 mM n-caproate. Introducing agitation significantly enhanced the maximum product formation rates of n-butyrate (rC4,max) and n-caproate (rC6,max) and was identified as a key parameter for further upscaling. The highest total medium-chain carboxylic acids concentration of 977.8 ± 22.8 C mM with 98.5 ± 2.1 mM n-butyrate and 97.3 ± 2.4 mM n-caproate was reached in the 2.2 L standard bioreactor and was only restricted by end-product inhibition. Further, a carbon recovery of up to 94% was independent of the reactor scale proving the transferability of the microbiome and its functions. Based on cloning and sequencing the most abundant microorganisms were closest related to the model organism for microbial medium-chain carboxylic acid formation, Clostridium kluyveri. The current study demonstrates that time and resource efficient screening of functional microbiomes for relevant cultivation conditions on a small scale can be combined with its subsequent upscaling without performance loss.
1 Introduction
A bio-based economy, i.e. bio-based fuels and chemicals, needs tailored resource management strategies. For management of the resource biomass among others anaerobic fermentation technologies based on microbiomes, i.e., open mixed microbial communities, can play a key role.1,2 Microbiome-based anaerobic fermentations, which allow upgrading low grade, i.e. non-food, non-feed, and chemically diverse biomass to chemical building blocks, are promising platform technologies. Chemical building blocks resulting as key intermediates and products from anaerobic fermentations include hydrogen (H2), carbon dioxide (CO2), methane (CH4) and medium-chain carboxylic acids (CA), i.e., n-carboxylic acids possessing carbon chain lengths of four (C4) to eight (C8) carbon atoms.2 Microbiomes themselves have already been proven to act as stable and cost-effective whole cell biocatalysts in anaerobic fermentations.3,4 Due to their metabolic diversity they are able to treat complex and variable organic waste streams highly efficiently and without the need of sterilization or antibiotic treatment.5,6 However, the reactor microbiome composition and activity depends on food-webs and species interactions, and these in turn strongly depend on the substrates and intermediates available as well as on other environmental factors.7Accordingly, for an efficient production of medium-chain CA the microbiome-based chain elongation of n-carboxylic acids is of particular interest.6,8 Several pathways for microbial chain elongation are described4 with the reverse β-oxidation performed by Clostridium kluyveri as model organism being most intensively studied.9–13 The reverse β-oxidation is a cyclic process comprising an oxidative and a reductive part. In the reductive part acetyl-CoA, derived from oxidation of ethanol by NAD+, is added to a CoA-activated carboxylic acid.4 This leads to a chain elongation of the acid by two carbon atoms per reaction cycle, e.g., acetic acid (C2) is elongated to n-butyric acid (C4) (eqn (1)) or n-butyric acid to n-caproic acid (C6) (eqn (2)). In the oxidative part ATP is generated via substrate level phosphorylation while in the reductive part ATP is generated via electron-transport phosphorylation. The ratio of the substrates ethanol and acetate theoretically determines to which extend ATP is produced (substrate level phosphorylation coupled to the oxidative part vs. electron-transport phosphorylation via Rnf complex/ATP synthase coupled to the reductive part).6 In this study, like in many others (Table 1), two model substrates are used, i.e. ethanol serves as electron donor as well as source of reducing equivalents and acetate as electron acceptor.6,14
| Biocatalysts | Experimental conditions | Max. production rate [g L−1 d−1] concentration [g L−1] (concentration [mM])a | Total carbon bound in medium-chain CA [C mM] | Reference | ||
|---|---|---|---|---|---|---|
| n-Butyrate | n-Caproate | n-Caprylate | ||||
| a Recalculation from g L−1 into mM with Mn-butyrate = 88.11 g mol−1, Mn-caproate = 116.16 g mol−1, and Mn-caprylate = 144.21 g mol−1.b Taken from ref. 6.c Average ± standard deviation of three replicates. | ||||||
| Microbiome | Batch, stirred bioreactor, no pH control, duration: 82 h | 6.32 ± 0.23c | 8.53 ± 0.50c | — | 977.8 ± 22.8 | This study |
| 8.68 ± 0.18c | 11.30 ± 0.28c | |||||
| (98.5 ± 2.1)c | (97.3 ± 2.4)c | |||||
| Fed-batch, stirred reactor, pH 7, duration: 120 d | — | 0.5b | 0.1b | 540.4 | 5 | |
| 2.2c | 8.2b | 0.3b | ||||
| (25.0) | (70.6) | (2.1) | ||||
| Continuous, upflow filter, pH 6.5–7.0, duration: 80 d | — | 16.6b | 0.9b | 702.4 | 22 | |
| 2.1 | 11.1b | 0.6b | ||||
| (23.8) | (95.6) | (4.2) | ||||
| Continuous, upflow filter, pH 6.5–7.2, duration: 60 d to 69 d | — | 57.4b | 1.8b | 728.6 | 41 | |
| 1.3 | 12.0 | 0.9 | ||||
| (14.8) | (103.3) | (6.2) | ||||
| C. kluyveri | Batch, Hungate tubes, no pH control, duration: 3 d | — | — | — | 675.2 | 13 |
| 2.6 | 12.8 | — | ||||
| (30.5) | (110.2) | |||||
Stoichiometry:4
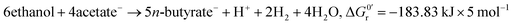 | (1) |
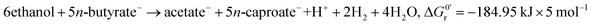 | (2) |
However, the stoichiometry of the reverse β-oxidation is not fixed as suggested by eqn (1) and (2) as the absolute concentrations and ratio of ethanol and acetate and the concentration of hydrogen in the medium have a strong impact on the overall reactions6 and, thus, the yield and ratio of the produced n-butyrate and n-caproate.9,12
A couple of studies have been devoted to the identification and characterization of microbiomes performing medium-chain CA formation from ethanol and acetate (Table 1). However, among other factors hindering a comparative assessment these studies were exclusively performed using individual reactor setups without parallelization and only at one scale. Therefore, it remains unclear whether the performance differences in microbial chain elongation are based on differences in composition and/or activity of the exploited microbiomes or the reactor configurations and cultivation conditions used.
Small scale cultivation in serum bottles can serve as a time and resource efficient way for identifying functional microbiomes and relevant cultivation conditions with high degree of parallelization. However, the question can be raised, if the performance during serum bottle cultivation can successfully be transferred to bioreactor scale which is compatible with further upscaling to industrial scale. Therefore, we systematically investigated the microbiome-based chain elongation across scales. We demonstrate that screening of microbiome cultivation conditions in serum bottles (110 mL) is a time and resource efficient way for identifying a functional microbiome. Subsequently we show that upscaling from serum bottles (110 mL) via 440 mL reactors to 2.2 L standard bioreactors without performance loss is feasible, demonstrating the transferability of the microbiome and its functions and identifying further relevant process parameters for respective upscaling.
2 Material and methods
2.1 Inoculum, chemicals and media
All chemicals were of at least analytical grade and were supplied from Carl Roth GmbH (Karlsruhe, Germany) and Merck KGaA (Darmstadt, Germany). Microbial media were prepared in de-ionized water (Millipore, Darmstadt, Germany). All experiments and sub-cultivations were carried out at 37 °C. Carrier gases and calibration gases for gas chromatography were purchased from Air Products GmbH (Berlin, Germany).2.2 Reactor setup and operation
Three different scales of reaction vessels allowing different degree of process monitoring were investigated (Table 2). Serum bottles (250 mL) with a final working volume of 110 mL were the smallest scale, without agitation but the bottles were shaken once a day. The medium was poured into the serum bottles in an anaerobic glove box. Ethanol and sodium acetate were added from sterile anoxic stock solutions followed by gas purging of the medium with a N2/CO2 mixture as stated above. The same procedure was applied to the tailor-made lab flasks (600 mL) with a final working volume of 440 mL. Here stirring was performed with a magnetic stir bar (150 rpm). For the third scale the liquid medium in standard bioreactors (3 L, UniVessel® double-walled glass culture vessel, Sartorius, Goettingen, Germany) was made anoxic by first purging with N2 till no dissolved oxygen was measured (VisiFerm DO 225, Hamilton, Switzerland) followed by switching to N2/CO2 mixture. In this context “standard bioreactor” refers to the standard setup of a conventional commercially available bioreactor with defined and reproducible operation parameters (e.g. cylindrical reactor geometry, agitators, gas sparger, gas outlet, sampling port, removable reactor lid etc.). The final working volume was 2.2 L and stirring was applied by a disk-type six-blade impeller (from reactor top, 150 rpm). On all scales the growth of the microbiome was monitored by measuring the OD600 nm (1.5 mL semi-micro cuvettes, UviLine 9400 spectrophotometer, Mainz, Germany). Furthermore, liquid samples (1 mL) for HPLC and GC-MS analysis were taken regularly and centrifuged (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 15 min, 4 °C). For the serum bottles and the lab flasks the pH was determined offline with a pH meter (H138 miniLab™ Elite (HACH-Lange, Germany)) which was calibrated on a daily basis. The pH of the fermentation broth in the standard bioreactors was measured online in situ with a pH probe (EasyFerm VP Plus 225, Hamilton, Switzerland). All experiments were performed without pH control.
000 × g, 15 min, 4 °C). For the serum bottles and the lab flasks the pH was determined offline with a pH meter (H138 miniLab™ Elite (HACH-Lange, Germany)) which was calibrated on a daily basis. The pH of the fermentation broth in the standard bioreactors was measured online in situ with a pH probe (EasyFerm VP Plus 225, Hamilton, Switzerland). All experiments were performed without pH control.
| a Includes the volume of the gas bag which was constantly connected to the reactor vessels.b Calculation based on three independent biological replicates (n = 3) as only for three replicates on each scale the sampling frequency was dense enough to accurately calculate the respective number. | |||
|---|---|---|---|
 |
 |
 |
|
| Reactor type | Serum bottle | Tailor-made lab flask | Standard bioreactor |
| Number of replicates | 4 | 4 | 3 |
| Reaction volume | 110 mL | 440 mL | 2.2 L |
| Reactor volume | 250 mL | 600 mL | 3.0 L |
| Run time [h] | 85 | 85 | 82 |
| Agitation | No (shaking once a day) | Yes (magnetic stir bar, 150 rpm) | Yes (disk-type six-blade impeller, 150 rpm) |
| Reynolds number | — | 3.8 × 103 | 9.4 × 103 |
| Headspace volume | 140 mL | 410 mLa | 1.6 La |
| Working volume/headspace ratio | 2.3 | 1.1 | 1.4 |
| Ethanol/acetate ratio | 1.2 | 1.3 | 1.1 |
| Substrate [mM] | Ethanol: 305 ± 8 | Ethanol: 318 ± 11 | Ethanol: 322 ± 3 |
| Acetate: 252 ± 8 | Acetate: 254 ± 4 | Acetate: 300 ± 2 | |
| Carbon provided [C mM] | 1104 ± 57 | 1152 ± 26 | 1245 ± 5 |
| Products [mM] | C4: 77.0 ± 2.0 | C4: 78.0 ± 3.0 | C4: 98.5 ± 2.1 |
| C6: 89.2 ± 2.0 | C6: 90.0 ± 1.0 | C6: 97.3 ± 2.4 | |
| Carbon recovered in C4 and C6 [C mM] | 811 ± 21 | 781 ± 71 | 921 ± 33 |
| Substrate carbon remaining [C mM] | 384 ± 29 | 383 ± 19 | 353 ± 28 |
| Carbon consumed [C mM] | 727 ± 70 | 769 ± 29 | 892 ± 26 |
| μmaxb [h−1] | 0.041 ± 0.004 | 0.051 ± 0.002 | 0.059 ± 0.000 |
| C recovery [%] | 94.06 ± 7.74 | 90.04 ± 5.67 | 92.87 ± 5.66 |
| Max. medium-chain CA formation rate rx,maxb [mmol L−1 h−1] | C4: 1.94 ± 0.05 | C4: 2.24 ± 0.27 | C4: 2.99 ± 0.11 |
| C6: 2.27 ± 0.06 | C6: 3.10 ± 0.21 | C6: 3.06 ± 0.08 | |
2.3 Analytical methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, inlet temperature 220 °C, helium as carrier gas with a constant flow of 1.2 mL min−1) into a ZB-WAX plus capillary column (30 m; 0.25 mm; 0.25 μm, Phenomenex, USA). The column temperature program was set 2 min at 50 °C followed by a heating rate of 15 °C min−1 to 250 °C. The temperature of the transfer line to the MS was 240 °C and the electron impact ionization source was set to 230 °C, the MS quadrupole to 150 °C. For stabilization retention time locking (RTL) was used. Mass scanning was performed in the range of m/z 18–300. Each peak was identified by library (NIST 2014 Mass Spectral Library, NIST, USA) and standard substance based comparison. The internal standard used was propionic acid. Calibrated substances were ethanol, acetate, n-butyrate, n-valerate, n-caproate, n-heptanoate and n-caprylate with four point calibration (R2 ≥ 0.99; calibration limits: ethanol: 2.0 mM; acetate: 1.5 mM; n-butyrate: 0.4 mM; n-valerate: 0.4 mM; n-caproate: 0.3 mM; n-heptanoate: 0.015 mM; n-caprylate: 0.014 mM).
20, inlet temperature 220 °C, helium as carrier gas with a constant flow of 1.2 mL min−1) into a ZB-WAX plus capillary column (30 m; 0.25 mm; 0.25 μm, Phenomenex, USA). The column temperature program was set 2 min at 50 °C followed by a heating rate of 15 °C min−1 to 250 °C. The temperature of the transfer line to the MS was 240 °C and the electron impact ionization source was set to 230 °C, the MS quadrupole to 150 °C. For stabilization retention time locking (RTL) was used. Mass scanning was performed in the range of m/z 18–300. Each peak was identified by library (NIST 2014 Mass Spectral Library, NIST, USA) and standard substance based comparison. The internal standard used was propionic acid. Calibrated substances were ethanol, acetate, n-butyrate, n-valerate, n-caproate, n-heptanoate and n-caprylate with four point calibration (R2 ≥ 0.99; calibration limits: ethanol: 2.0 mM; acetate: 1.5 mM; n-butyrate: 0.4 mM; n-valerate: 0.4 mM; n-caproate: 0.3 mM; n-heptanoate: 0.015 mM; n-caprylate: 0.014 mM).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 4 °C). Subsequently, the supernatant was removed and the cells were washed in 5 mL 7.8 mM phosphate buffer. This was followed by a second centrifugation and washing step. After the final centrifugation step the supernatant was discarded and the cell pellets were dried at 65 °C for at least 48 h until constant weight was reached.
000 × g, 4 °C). Subsequently, the supernatant was removed and the cells were washed in 5 mL 7.8 mM phosphate buffer. This was followed by a second centrifugation and washing step. After the final centrifugation step the supernatant was discarded and the cell pellets were dried at 65 °C for at least 48 h until constant weight was reached.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H
H![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) O
O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N) of the yeast extract (Merck KGaA (Darmstadt, Germany)) that was used in the experiments was determined using CHNS analysis according to DIN EN ISO 16948:2015 with a VarioMacroCube macro analyzer (Elementar, Langenselbold, Germany). Oxygen was determined indirectly by mass difference calculation.
N) of the yeast extract (Merck KGaA (Darmstadt, Germany)) that was used in the experiments was determined using CHNS analysis according to DIN EN ISO 16948:2015 with a VarioMacroCube macro analyzer (Elementar, Langenselbold, Germany). Oxygen was determined indirectly by mass difference calculation.2.4 Calculations
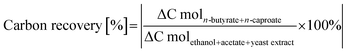 | (3) |
The potential contribution of yeast extract to the carbon recovery as indirect substrate was determined in an additional experiment. Microbiomes (n = 3) cultivated in DSM 52 medium without acetate and ethanol in serum bottles for 85 h were analyzed for the formation of substrates which could contribute to the reverse β-oxidation pathway (eqn (1) and (2)). Formate and acetate formation were observed with a maximum of 31.3 ± 0.7 C mM in total and therefore included as substrate carbon.
The carbon bound in cellular biomass was calculated based on a theoretical molecular composition of cellular biomass proposed by Roels (1980) of CH1.8O0.5N0.2 (MW = 24.6 g mol−1)20 and on the determined cell dry weight (for details please see Material and method section 2.3.5).
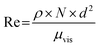 | (4) |
| μvis = ν × ρ | (5) |
3 Results and discussion
First, a systematic screening of the microbiome for optimal n-butyrate and n-caproate production was conducted in serum bottles. Afterwards, upscaling from serum bottles (110 mL) via lab flasks (440 mL) to standard bioreactors (2.2 L) was performed to assess the transferability of the performance of the microbiome into biotechnologically more relevant reaction systems with further upscale potential.3.1 Screening experiments
Screening experiments for suitable media and substrate concentrations were performed. In a first step DSM 52 medium and medium according to Zhang16 were compared for microbial medium-chain CA formation. Cultivation in the medium according to Zhang led to a very long lag-phase of six weeks whereas medium-chain CA formation in the DSM 52 medium started within ten days. The long lag-phase in the medium according to Zhang is most likely caused by the lack of yeast extract. Yeast extract is a complex mixture of amino acids, salts and growth factors like vitamins and it was already found that higher yeast extract concentration increased medium-chain CA formation by improving protein biosynthesis.22,23 Accordingly, the DSM 52 medium containing yeast extract was used as base medium for all subsequent experiments.A systematic screening of the microbiome for optimal n-butyrate and n-caproate production was conducted in serum bottles by using four different ethanol/acetate ratios (molar ratios of 1.32 to 4.13) (see Table S2†). The ethanol/acetate ratio was shifted by keeping a constant ethanol concentration of approximately 320 mM and varying the concentration of acetate.
As Fig. 1 and Table S2† show using an ethanol/acetate ratio of 1.3, i.e. starting with 311 mM ethanol and 235 mM acetate, the highest final carbon recovery of 61.69% was achieved, representing also the highest n-butyrate and n-caproate concentrations of 68.3 ± 0.2 mM and 65.6 ± 1.0 mM, respectively. By increasing the ethanol/acetate ratio up to 3.25 the carbon recovery decreased to 51.20% with a more selective production of n-caproate (59.2 ± 0.6 mM) than n-butyrate (23.4 ± 0.5 mM). The observed behavior of the microbiome regarding product formation at the certain ethanol/acetate ratios is in agreement with the results of earlier studies for pure cultures of C. kluyveri10,12 and a recently published theoretical approach.6 The application of too high ethanol concentrations of 463 mM and 712 mM strongly reduced medium-chain CA formation probably due to inhibition of microbial activity.24 As the aim of this study was investigating the transferability of the production performance of the microbiome, an ethanol/acetate ratio of 1.2 ± 0.1 was used to maximize the total medium-chain CA production for the upscaling experiments.
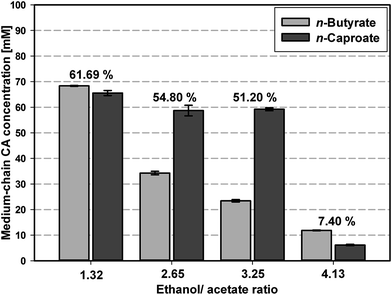 | ||
| Fig. 1 Screening of microbiome-based n-butyrate and n-caproate formation depending on initial ethanol/acetate ratio in DSM 52 medium performed in serum bottles. The numbers above each pair of bars represent the calculated carbon recovery of substrate carbon into the medium-chain CA. Reported values are averages of four replicates and the error bars represent the standard deviation. For further details regarding ethanol and acetate concentrations see Table S2.† | ||
3.2 Functional enrichment of the microbiome
The microbiome originating from a leach bed reactor was cultivated on serum bottle level in DSM 52 medium with a regular sub-cultivation every two weeks. This led to a functional enrichment of the microbiome over time which is reflected in an increased carbon recovery from the first transfer to the 26th transfer (which was used for upscaling experiments, Fig. 2C and Table 2) as well as a change in the community composition (Fig. S3†). Further, methanogenic archaea, which could be major substrate competitors for acetate under the applied cultivation conditions,22 were outcompeted during this sub-cultivation procedure. While other studies required the addition of cost-intensive agents (e.g. 2-bromoethanesulfonate (BES)5,25) to inhibit methanogenesis, we did not detect any methane (calibration limit for CH4: 0.1%) on serum bottle level after the third transfer. This means that the applied procedure is adequate to eliminate methanogenic archaea from the microbiome to optimize the process efficiency.3.3 Upscaling experiments
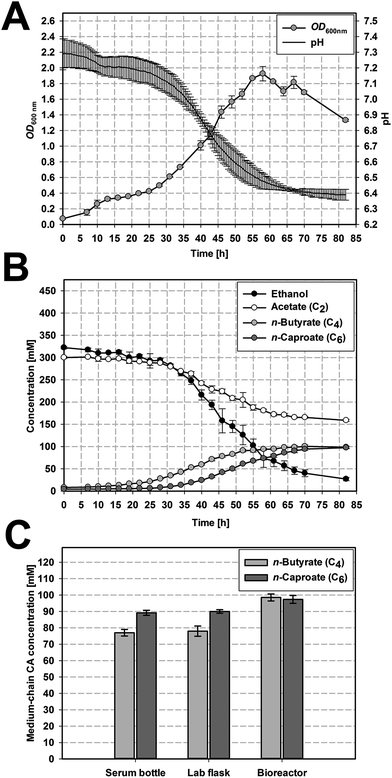
A high formation of n-butyrate and n-caproate was observed on all three scales (cf. Fig. 2 and S1†). It started 15 h to 20 h after inoculation for n-butyrate and after 31 to 36 h for n-caproate (cf. Fig. 2B, S2B and S2C†). A significantly higher maximum n-butyrate production rate rC4,max was found for the bioreactors (2.99 ± 0.11 mmol L−1 h−1, Fig. 2B) compared to serum bottles (1.94 ± 0.05 mmol L−1 h−1, p = 0.006) and lab flask (2.24 ± 0.27 mmol L−1 h−1, p = 0.045) which did not differ significantly (p = 0.801). The maximum n-caproate production rate rC6,max was highest in the continuously agitated reactors (3.10 ± 0.21 mmol L−1 h−1 in the lab flask, 3.06 ± 0.08 mmol L−1 h−1 in the bioreactor) and differed significantly from the serum bottles (2.27 ± 0.06 mmol L−1 h−1, p = 0.031).Serum bottles and lab flasks reached similar final concentrations of n-butyrate and n-caproate (Table 2) resulting in similar total acid production values (843.2 ± 20 C mM serum bottles, 852.0 ± 18 C mM lab flasks).
The highest total medium-chain CA concentration was reached in the bioreactors with 977.8 ± 22.8 C mM based on significantly higher concentrations of n-butyrate (98.5 ± 2.1 mM) and n-caproate (97.3 ± 2.4 mM) compared to the two other scales (p = 0.012 vs. serum bottle and p = 0.021 vs. lab flask). This represents an increase by factor 1.26 for n-butyrate and 1.12 for n-caproate. Furthermore, on all three scales the OD600 nm increased with medium-chain CA formation. This growth dependent formation of n-butyrate and n-caproate is in accordance with the assumption that microbial chain elongation of ethanol and acetate to n-butyrate and n-caproate is performed by the reverse β-oxidation pathway.
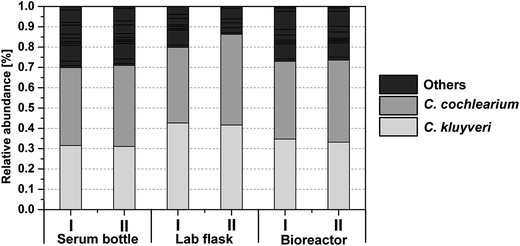 | ||
| Fig. 3 Community composition based on the DNA fingerprinting technique T-RFLP. The bars represent the major terminal restriction fragments (tRFs) after restriction digestion with HaeIII in two out of three (standard bioreactor, see Table 2) or four (serum bottle and lab flask, see Table 2) biological replicates of each scale at the end of the fermentation (85 h for serum bottle and lab flask and 82 h for the standard bioreactor, see Table 2). The grey scale in the legend represents the different tRFs with the corresponding tRFs of C. kluyveri and C. cochlearium being identified by T-RFLP of the corresponding single clones after sequencing. | ||
3.4 Implications for application
The microbiome composition was independent from the reactor scale. Consequently, a high and reproducible carbon recovery as well as a similar, but not identical, time course of cultivation (Fig. 2B and S1†) was observed. While the carbon recovery is not influenced by the reactor scale and the agitation regime (carbon recovery from 90.40 ± 5.67% to 94.06 ± 7.74% across the scales), the final medium-chain CA concentration seems to be influenced. Introducing agitation increases rC4,max and rC6,max in the lab flask and the bioreactor (Table 2) compared to the serum bottles without agitation. In accordance with these results, μmax increased in the agitated vessels compared to the serum bottles. The difference in performance (rC4,max, rC6,max and μmax) between shaking once a day in the serum bottle and constantly mixing in the lab flask is not statistically significant although a flow regime in the lab flask, as expressed by the Reynolds number of 3.8 × 103, close to transitional flow (102 < Re < 103)32 was implemented. However, the most turbulent flow regime, i.e. close to full turbulent flow in stirred tank reactors (Re > 104),32 applied in the bioreactor (Re = 9.4 × 103) led to significantly higher μmax, rC4,max and rC6,max and finally to a significantly higher product titer. In this way, agitation was identified as a potential key parameter for further upscaling as an increase of the mass transfer coefficient by increasing the power input for mixing improved the performance of the overall process. A further increase of the agitation will be a matter of subject in future studies. Nevertheless a higher power input has to be considered for the economic feasibility of the overall process.33In this study, n-butyrate and n-caproate formation stagnate at concentrations of 98.5 ± 2.1 mM and 97.3 ± 2.4 mM in the bioreactor probably due to end-product inhibition. For a biotechnological production platform a continuous medium-chain CA formation should be aimed at and would be the next step for process optimization. With the current cultivation conditions the pH drops with medium-chain CA accumulation. The closer the pH is to the pKa value of the acids, the higher is the concentration of the undissociated form of the medium-chain CA. These uncharged molecules then can easily pass the cell membrane14 leading to acidification of the cytoplasm due to dissociation, as the cytoplasmic pH is above the pKa,34 and thereby may have cytotoxic effects on energy metabolism and cell growth.35 Also the anion itself can be cytotoxic as its hydrophilic interaction potentially affects essential pathways.36–38 When assuming ideal conditions and using the Henderson–Hasselbalch equation the final concentrations of undissociated n-butyrate (n-butyric acid) and n-caproate (n-caproic acid) in the bioreactor can be estimated to be 3.3 mM n-butyric acid and 3.0 mM n-caproic acid. End-product inhibition was reported to start at higher concentrations of undissociated n-butyrate (10.8 mM,39 48 mM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) or n-caproate (7.5 mM
40) or n-caproate (7.5 mM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) in other microbiome studies. A reason why the highest agitation led to higher final product titers could also be that improved mixing reduced the medium-chain CA concentration in close vicinity to the cells compared to the bulk medium. However, the microbiome investigated in this study might become less susceptible to medium-chain CA inhibition after continuous cultivation or fed-batch cultivation, with increased exposure time to medium-chain CA. Alternative limitations like availability of trace elements, vitamins, yeast extract, substrates (ethanol or acetate individually and as mixture) were tested by addition of these components to the medium after stagnation of substrate consumption and medium-chain CA formation, but did not result in further growth or product formation (data not shown). These findings support the hypothesis that end-product inhibition is the major process limitation.
6) in other microbiome studies. A reason why the highest agitation led to higher final product titers could also be that improved mixing reduced the medium-chain CA concentration in close vicinity to the cells compared to the bulk medium. However, the microbiome investigated in this study might become less susceptible to medium-chain CA inhibition after continuous cultivation or fed-batch cultivation, with increased exposure time to medium-chain CA. Alternative limitations like availability of trace elements, vitamins, yeast extract, substrates (ethanol or acetate individually and as mixture) were tested by addition of these components to the medium after stagnation of substrate consumption and medium-chain CA formation, but did not result in further growth or product formation (data not shown). These findings support the hypothesis that end-product inhibition is the major process limitation.
When comparing the results of this microbiome-based medium-chain CA formation to other studies using acetate and ethanol (Table 1) its high potential as a biotechnological production platform becomes evident. In comparison with all studies from Table 1 the highest total medium-chain CA concentration of 977.8 ± 22.8 C mM was reached in our experiments on 2.2 L-standard bioreactor scale – after previous screening for the optimal cultivation conditions on serum bottle level.
In contrast to other microbiome-based studies,5,22,41,42 however, caprylate (C8) formation was not observed (Fig. S5†). Reasons for this could be differences in the cultivation conditions (e.g. continuous cultivation and pH control5) or a general limitation in the metabolic potential of the microbiome. Steinbusch et al. (2011)5 and Zhang et al. (2013)16 suggested H2 as additional alternative electron donor for caprylate formation in microbiome-based medium-chain CA formation. A continuous increase of H2 in the headspace gas was found in our experiments but was obviously not utilized. Therefore, a general metabolic restriction of the C. kluyveri strain in our experiments is most likely26 and was also confirmed in long-term incubations up to 18 months (data not shown). Our result is in accordance with other studies,43,44 while other microbiomes showing C8 formation might have contained other strains5,22,41 or no C. kluyveri at all.42
4 Conclusion
We have investigated the predictability and transferability of microbiome-based medium-chain CA formation across scales from the simple serum bottle cultivation up to cultivation in standard bioreactors. The stable microbiome composition established by serum bottle cultivation was the basis for the transferability of the microbiome performance and the upscaling of the whole process to standard bioreactors (2.2 L). Hence, small scale cultivation on serum bottle level was demonstrated to serve as a time and resource efficient approach for targeted screening of suitable cultivation conditions and microbiome adaptation. The described experimental approach can be transferred to microbiomes exploiting complex feedstocks and producing platform chemicals by anaerobic fermentation. Introducing agitation enhanced the product formation rates rC4,max and rC6,max and should be considered as a key parameter for further upscaling. It yielded maximum total medium-chain CA concentration of 977.8 ± 22.8 C mM with competitively high n-butyrate (98.5 ± 2.1 mM) and n-caproate (97.3 ± 2.4 mM) concentrations from acetate and ethanol at a carbon recovery close to 95% and was only restricted by end-product inhibition. This can be circumvented in future, e.g. by (1) in-line extraction in a single-phase fermentation system at low pH,45 or by (2) two-phase fermentation systems at a neutral pH to achieve a continuous process.46 After extraction, the direct use of the CA is limited. For instance n-caproate can be used as animal feed and green antimicrobial substance.45 The utilization of CA as biofuels is not possible, due to their high oxygen-to-carbon ratio and their low energy density. For this reason, they have to be further converted to liquid biofuels like energy-dense alkanes and esters. This can already be realized by electroorganic synthesis, e.g. Kolbe and non-Kolbe electrolysis,47,48 thus expanding the application potential for medium-chain CA production as valuable platform for chemical and biofuel production.Acknowledgements
We thank Anne Kuchenbuch and Desiree Schmidt for very valuable technical assistance in molecular biology. Furthermore we thank Dr Claus Härtig and Angelika Wichmann for GC-MS measurements. Further we thank Dr Jana Mühlenberg for CHNS analysis. F. H. acknowledges support by the BMBF (Research Award “Next generation biotechnological processes – Biotechnology 2020+”) and the Helmholtz-Association (Young Investigators Group). This work was supported by the Helmholtz-Association within the Research Programme Renewable Energies. There is no conflict of interests.References
- R. Kleerebezem and M. C. M. van Loosdrecht, Curr. Opin. Biotechnol., 2007, 18(3), 207 CrossRef CAS PubMed.
- M. T. Agler, B. A. Wrenn, S. H. Zinder and L. T. Angenent, Trends Biotechnol., 2011, 29(2), 70 CrossRef CAS PubMed.
- P. Weiland, Appl. Microbiol. Biotechnol., 2010, 85(4), 849 CrossRef CAS PubMed.
- C. M. Spirito, H. Richter, K. Rabaey, A. J. M. Stams and L. T. Angenent, Curr. Opin. Biotechnol., 2014, 27, 115 CrossRef CAS PubMed.
- K. J. J. Steinbusch, H. V. M. Hamelers, C. M. Plugge and C. J. N. Buisman, Energy Environ. Sci., 2011, 4(1), 216 CAS.
- L. T. Angenent, H. Richter, W. Buckel, C. M. Spirito, K. J. J. Steinbusch, C. M. Plugge, D. P. B. T. B. Strik, T. I. M. Grootscholten, C. J. N. Buisman and H. V. M. Hamelers, Environ. Sci. Technol., 2016, 50(6), 2796 CrossRef CAS PubMed.
- C. Koch, S. Müller, H. Harms and F. Harnisch, Curr. Opin. Biotechnol., 2014, 27, 65 CrossRef CAS PubMed.
- C. W. Marshall, E. V. LaBelle and H. D. May, Curr. Opin. Biotechnol., 2013, 24(3), 391 CrossRef CAS PubMed.
- H. A. Barker and S. M. Taha, J. Bacteriol., 1942, 43(3), 347 CAS.
- B. T. Bornstein and H. A. Barker, J. Biol. Chem., 1948, 172, 659 CAS.
- W. R. Kenealy, Y. Cao and P. J. Weimer, Appl. Microbiol. Biotechnol., 1995, 44(3–4), 507 CrossRef CAS PubMed.
- G. M. Smith, B. W. Kim, A. A. Franke and J. D. Roberts, J. Biol. Chem., 1985, 260(25), 13509 CAS.
- P. J. Weimer and D. M. Stevenson, Appl. Microbiol. Biotechnol., 2012, 94(2), 461 CrossRef CAS PubMed.
- K. Choi, B. S. Jeon, B. C. Kim, M. K. Oh, Y. Um and B. I. Sang, Appl. Biochem. Biotechnol., 2013, 171(5), 1094 CrossRef CAS PubMed.
- H. Sträuber, R. Lucas and S. Kleinsteuber, Appl. Microbiol. Biotechnol., 2016, 100(1), 479 CrossRef PubMed.
- F. Zhang, J. Ding, Y. Zhang, M. Chen, Z. W. Ding, M. C. M. van Loosdrecht and R. J. Zeng, Water Res., 2013, 47(16), 6122 CrossRef CAS PubMed.
- C. Koch, D. Popiel and F. Harnisch, ChemElectroChem, 2014, 1(11), 1923 CrossRef CAS.
- Q. Wang, G. M. Garrity, J. M. Tiedje and J. R. Cole, Appl. Environ. Microbiol., 2007, 73(16), 5261 CrossRef CAS PubMed.
- S. McGinnis and T. L. Madden, Nucleic Acids Res., 2004, 32, W20–W25 CrossRef CAS PubMed.
- J. A. Roels, Biotechnol. Bioeng., 1980, 22(12), 2457 CrossRef CAS.
- R. K. Sinnott, J. M. Coulson and J. F. Richardson, Chemical engineering design, Elsevier Butterworth-Heinemann Amsterdam, Amsterdam, 2006 Search PubMed.
- T. I. M. Grootscholten, K. J. J. Steinbusch, H. V. M. Hamelers and C. J. N. Buisman, Bioresour. Technol., 2013, 135, 440 CrossRef CAS PubMed.
- M. E. Martin, H. Richter, S. Saha and L. T. Angenent, Biotechnol. Bioeng., 2016, 113(3), 531 CrossRef CAS PubMed.
- S. Lonkar, Z. Fu and M. Holtzapple, Biotechnol. Bioeng., 2016, 113, 2597 CrossRef CAS PubMed.
- M. T. Agler, C. M. Spirito, J. G. Usack, J. J. Werner and L. T. Angenent, Water Sci. Technol., 2014, 69(1), 62 CrossRef CAS PubMed.
- W. B. Whitman, Bergey's Manual® of Systematic Bacteriology: Volume Three The Firmicutes, Springer-Verlag New York, New York, NY, 2009 Search PubMed.
- G. C. Mead, J. Gen. Microbiol., 1971, 67(1), 47 CrossRef CAS PubMed.
- H. J. Nanninga, W. J. Drent and J. C. Gottschal, FEMS Microbiol. Lett., 1986, 38(6), 321 CrossRef CAS.
- N. Tomlinson and H. A. Barker, J. Biol. Chem., 1954, 209(2), 585 CAS.
- B. T. Bornstein and H. A. Barker, J. Bacteriol., 1948, 55(2), 223 CAS.
- H. A. Barker, Antonie van Leeuwenhoek, 1947, 12(1–4), 167 CrossRef CAS PubMed.
- J. M. Smith, Chemical engineering kinetics, McGraw-Hill Inc., New York, N.Y., 1981 Search PubMed.
- M. P. C. Marques, J. M. S. Cabral and P. Fernandes, J. Chem. Technol. Biotechnol., 2010, 85(9), 1184 CrossRef CAS.
- J. Zigová and E. Šturdík, J. Ind. Microbiol. Biotechnol., 2000, 24(3), 153 CrossRef.
- P. J. Henderson, Annu. Rev. Microbiol., 1971, 25, 393 CrossRef CAS PubMed.
- J. B. Russell and F. Diez-Gonzalez, Adv. Microb. Physiol., 1998, 39, 205 CrossRef CAS PubMed.
- S. Stolte, J. Arning, U. Bottin-Weber, M. Matzke, F. Stock, K. Thiele, M. Uerdingen, U. Welz-Biermann, B. Jastorff and J. Ranke, Green Chem., 2006, 8(7), 621 RSC.
- A. J. Roe, C. O'Byrne, D. McLaggan and I. R. Booth, Microbiology, 2002, 148(7), 2215 CrossRef CAS PubMed.
- H. Richter, N. Qureshi, S. Heger, B. Dien, M. A. Cotta and L. T. Angenent, Biotechnol. Bioeng., 2012, 109(4), 913 CrossRef CAS PubMed.
- J. C. van den Heuvel, P. G. Verschuren, H. H. Beeftink and D. de Beer, Biotechnol. Tech., 1992, 6(1), 33 CrossRef CAS.
- T. I. M. Grootscholten, K. J. J. Steinbusch, H. V. M. Hamelers and C. J. N. Buisman, Bioresour. Technol., 2013, 136, 735 CrossRef CAS PubMed.
- L. A. Kucek, C. M. Spirito and L. T. Angenent, Energy Environ. Sci., 2016, 9(11), 3482 CAS.
- P. J. Weimer, M. Nerdahl and D. J. Brandl, Bioresour. Technol., 2014, 175, 97 CrossRef PubMed.
- M. Coma, R. Vilchez-Vargas, H. Roume, R. Jauregui, D. H. Pieper and K. Rabaey, Environ. Sci. Technol., 2016, 50(12), 6467 CrossRef CAS PubMed.
- M. T. Agler, C. M. Spirito, J. G. Usack, J. J. Werner and L. T. Angenent, Energy Environ. Sci., 2012, 5(8), 8189 CAS.
- T. I. M. Grootscholten, D. P. B. T. B. Strik, K. J. J. Steinbusch, C. J. N. Buisman and H. V. M. Hamelers, Appl. Energy, 2014, 116, 223 CrossRef CAS.
- C. Stang and F. Harnisch, ChemSusChem, 2016, 9(1), 50 CrossRef CAS PubMed.
- P. F. Levy, J. E. Sanderson, R. G. Kispert and D. L. Wise, Enzyme Microb. Technol., 1981, 3(3), 207 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra28259h |
| This journal is © The Royal Society of Chemistry 2017 |