Open Access Article
Open Access ArticleMicrowave-assisted synthesis of fluorescent carbon quantum dots from an A2/B3 monomer set†
Ari Chaea,
Yujin Choia,
Seongho Joa,
Nur'aenia,
Peerasak Paoprasertd,
Sung Young Park *ac and
Insik In
*ac and
Insik In *ab
*ab
aDepartment of IT Convergence (Brain Korea PLUS 21), Korea National University of Transportation, Chungju 380-702, South Korea. E-mail: in1@ut.ac.kr
bDepartment of Polymer Science and Engineering, Korea National University of Transportation, Chungju 380-702, South Korea
cDepartment of Chemical and Biological Engineering, Korea National University of Transportation, Chungju 380-702, South Korea
dDepartment of Chemistry, Faculty of Science and Technology, Thammasat University, Pathumthani 12121, Thailand
First published on 22nd February 2017
Abstract
Strongly fluorescent carbon quantum dots (CQDs) were simply prepared by microwave-assisted synthesis using succinic acid and tris(2-aminoethyl)amine as an “A2 + B3” monomer set with a high mass yield of 17.3%. The prepared disc-like CQDs had average lateral dimensions ranging from 5 to 30 nm and a uniform thickness of 6.2 nm. The CQDs showed high solubility in protic polar solvents and demonstrated excitation-dependent emission with a high photoluminescence quantum yield of 49.9%. Because of the excellent cell viability of the CQDs, multicolor fluorescent imaging of cellular media (MDAMB and MDCK cells) was efficiently achieved.
Introduction
Carbon quantum dots (CQDs) show promising optoelectrical properties, and they have been intensively researched as next-generation carbon nanomaterials, in addition to zero-dimensional (OD) fullerene, one-dimensional (1D) carbon nanotubes, and two-dimensional (2D) graphene.1–3 In addition to their comparable fluorescence properties, CQDs are becoming comparable to those of inorganic quantum dots (QDs) through the exploitation of their biocompatibility, low toxicity, excellent photochemical stability, and facile surface functionalization.4–6 Until now, CQDs have been widely examined in various applications such as bioimaging,7–12 sensing,13,14 photo/electrocatalysis,15,16 light-harvesting,17–19 and drug-delivery systems.20–22Although both “top-down” and “bottom-up” approaches have been routinely used for the synthesis of CQDs depending on the type of precursor materials, “bottom-up” approaches utilizing various small molecules, polymers, and even biocompatible materials have advantages in terms of low cost and environmental friendliness because carbon nanotubes, nanodiamonds, and graphene oxides, which are frequently used as carbon-rich precursor materials in “top-down” approaches, are not necessary.23–28 Bottom-up CQD synthesis from small molecules through dehydration (or self-assembly) and carbonization has generally been carried out using harsh reaction processes, including hydrothermal processes, combustion, acid pyrolysis, etc.29–35
Quite recently, a very convenient method of synthesizing CQDs was introduced through the utilization of a typical household microwave oven. Within 5 min, highly photoluminescent CQDs showing good water solubility and biocompatibility were synthesized from citric acid and urea.36 However, the photoluminescence quantum yield (PLQY) of the prepared CQDs was less than 15%, which is not even close to the highest PLQY (∼80%) reported for other CQDs prepared by hydrothermal treatment of citric acid/ethylene diamine.31 Therefore, there has been a significant effort to produce CQDs with enhanced PLQYs and mass yields through simple microwave treatment of inexpensive materials. In terms of monomer-type precursor materials of polycondensation reactions, the abovementioned citric acid and urea (or ethylene diamine) can be regarded as A3 and B2 monomers (A: –COOH, B: –NH2), respectively. Upon microwave treatment, dehydration (polymerization) reactions between the A3 and B2 monomers initially occur, which is exactly the same as “A3 + B2” polyamidation.37 Although typical “A3 + B2” polycondensation reactions might result in crosslinked polymer networks or soluble hyperbranched polymers depending on the initial feeding ratio of the two monomers,38,39 a very complicated carbonization process would compete with the crosslinking/polymerization reactions during the microwave treatment of the “A3 + B2” monomer set because of the high local temperature generated by microwave irradiation.40
In our approach, succinic acid (SA) and tris(2-aminoethyl)amine (TAEA), which comprise a representative “A2 + B3” monomer set, were utilized for the synthesis of CQDs instead of the previously used “A3 + B2” monomer sets. Microwave treatment of the SA and TAEA monomer set produced CQDs with a PLQY of 49.9%, which is the highest PLQY value among CQDs prepared by microwave operation.36 Even in equimolar reactions, the number of B functional groups (–NH2) exceeds that of A functional groups (–COOH). Consequently, there is a much greater possibility of self-surface passivation by the amine moieties, as shown in previous studies, where various amine-containing small molecules or polymers were used as efficient surface passivation agents to increase the PLQY of CQDs through the amidation reaction with carboxylic acid groups on the surfaces of the CQDs.41–43 We believe that the “A2 + B3” monomer set is better than the “A3 + B2” monomer set, especially in terms of the mass production of CQDs, because a variety of natural or synthetic water-soluble dicarboxylic acids can be utilized as A2 monomers for the synthesis of CQDs. In comparison, water-soluble diamine monomers (B2 type) are not numerous, which limits the synthesis of CQDs with compositional variations from the “A3 + B2” monomer set.
Experimental
Materials and characterization
Both SA (ACS reagent, ≥99.0%) and TAEA (96%) were purchased from Sigma-Aldrich Corp. A household microwave oven (700 W) (Daewoo Electronics, South Korea) was used throughout the experiments. Dialysis was carried out by using a molecular weight cut-off (MWCO) membrane (3500 Da) of Spectrum Laboratories, Inc. MDAMB-231, A549, and MDCK cells were obtained from Korean Cell Line Bank (Seoul, Korea). Penicillin–streptomycin, phosphate-buffered saline (PBS), 0.05% (w/v) trypsin EDTA (1×) solution, and RPMI-1640 medium were purchased from Gibco BRL (Carlsbad, CA, USA).Ultraviolet-Visible (UV-Vis) spectra were measured by an Optizen Alpha UV-Vis Smart spectrometer of (Mecasys, South Korea). The photoluminescence (PL) spectra of the CQD solutions were examined by using a FS-2 Fluorescence Spectrometer (Scinco Corp., South Korea) with a xenon lamp excitation source (150 W). High-resolution transmission electron microscopy (HRTEM) images were taken on a TECNAI F20 system (Philips) at an operating voltage of 200 kV. Atomic Force Microscopy (AFM) images were acquired by a Multimode-N3-AM nanoscope three-dimensional (3D) scanning probe microscopy (SPM) system (Bruker Corp.). Fourier transform infrared (FT-IR) spectra were acquired on a Nicolet iS10 FT-IR spectrometer (Thermo Scientific). Raman spectroscopy measurements were performed on an ARAMIS Raman spectrometer (Horiba Jobin Yvon, France) by using 514.5 or 785 nm laser radiation. X-ray diffraction (XRD) measurements of the powder samples were recorded on a D8-Advance X-ray powder diffractometer (Bruker) using Cu Kα radiation (λ = 1.5406 Å). The X-ray photoelectron spectroscopy (XPS) spectra were recorded with a Scientific Sigma Probe spectrometer (Thermo VG). Fluorescence microscopy images of CQD-labelled cells were obtained using LSM 510 confocal laser scanning microscope (ZEISS).
Microwave-assisted synthesis of CQDs
For the microwave-assisted synthesis of CQDs from SA and TAEA, 3 g (1 eq.) of SA and 5.572 g (1.5 eq.) of TAEA were completely dissolved in 10 mL of deionized water in a 250 mL flat-bottomed flask. After loosely wrapping the flask with polyethylene wrapping film, the mixture solution was treated inside a microwave oven for 5 min (CAUTION! Carry out the whole experiment inside a fume hood because of the emission of organic volatiles during the microwave treatment). After the microwave treatment and complete cooling, dark-brown solids were obtained, indicating the formation of CQDs. Extraction of the CQDs was carried out by adding 100 mL of deionized water to the solids and subsequent ultrasonication in a bath sonicator (500 W) for 30 min. Centrifuging at 4000 rpm for 30 min was carried out to remove any insoluble precipitates or agglomerates from the CQD solution. Finally, 1.48 g of brown CQD2 powders was obtained after freeze-drying of the CQD solution obtained after dialysis using 3500 Da MWCO membranes for 3 days.Cell viability test and bioimaging
The cell viability of the CQDs was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)44 or dehydrogenase (LDH) assay45 method. In MTT assay method, 200 μL of MDA-MB-231 or MDCK cells at a density of 1 × 105 cells per mL was placed in each well of a 96-well plate. Afterwards, the cells were incubated for 24 h at 37 °C in a humidified 5% CO2 atmosphere. To determine the cellular viability, a stock solution of CQD2 was dissolved in RPMI medium at a concentration of 1 mg mL−1 and the stock solution was diluted up to 0.001 mg mL−1. The media was removed and the cells were treated with different concentrations of the CQD2 solution. Then, the cells were incubated for another 24 h. The media containing CQD2 were replaced with 180 mL of fresh medium and 20 μL of a stock solution containing 15 mg of MTT in 3 mL of PBS and incubated for another 4 h. Finally, the medium was removed and a 200 μL solution of an MTT solubilizing agent was added to the cells and accurate shaking was performed for 15 min. The optical absorbance was measured at a wavelength of 570 nm using a microplate reader (Varioskan® Flash, Thermo Electron Corporation). The relative cell viability was measured by comparing the samples with the 96-well control plate containing only cells. After cell viability measurements, confocal images were obtained to confirm the stained cells by using a laser scan microscope (CLSM) at 10× magnification with excitation wavelengths of 405, 488, and 543 nm. In LDH assay method, 200 μL of MDA-MB-231 or MDCK cells at a density of 1 × 105 cells per mL was incubated with different concentrations of CQD2 at 37 °C for 24 h in each well of a 96-well plate. Then, 20 μL of LDH standard solution (DoGen, South Korea) was transferred into each well for 1 h and the cells were incubated for 1 h at 37 °C in a humidified 5% CO2 atmosphere. After that, 100 μL of cell supernatant was aspirated and transferred into another plate according to the instruction. Finally, the optical absorbance of supernatant was monitored at 490 nm to analyze relative cell viability.Results and discussion
The synthesis of CQDs from SA and TAEA was carried out by microwave treatment of both monomers for 5 min with different molar feeding ratios of SA and TAEA: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5, 1
1.5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5, 1
2.5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, and 1
3, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.5. For the monomer feeding ratios of 1
3.5. For the monomer feeding ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5, 1
2.5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, and 1
3, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.5, negligible amounts of CQDs were obtained. CQDs obtained from SA and TAEA feeding ratios of 1
3.5, negligible amounts of CQDs were obtained. CQDs obtained from SA and TAEA feeding ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5, and 1
1.5, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
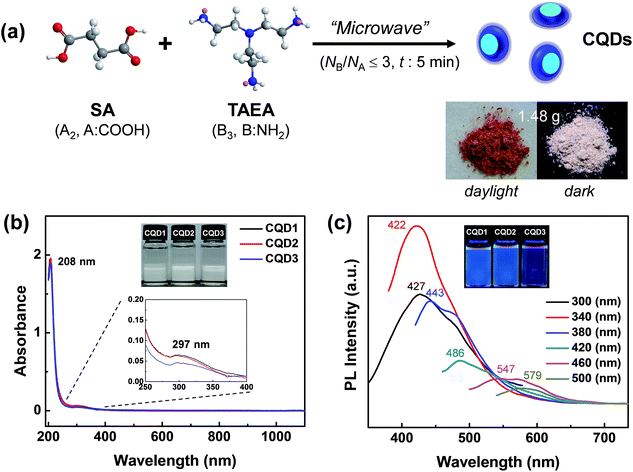
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 were denoted as CQD1, CQD2, and CQD3. CQD2 showed the highest mass yield of 17.3% (1.48 g of CQD2 was obtained from 8.5572 g of monomers) (Fig. 1a). The prepared CQDs were highly soluble in aqueous media. The CQD powders were highly hygroscopic, and long-term storage in ambient atmosphere resulted in CQD powders that stuck to the inner walls of the vials.
2 were denoted as CQD1, CQD2, and CQD3. CQD2 showed the highest mass yield of 17.3% (1.48 g of CQD2 was obtained from 8.5572 g of monomers) (Fig. 1a). The prepared CQDs were highly soluble in aqueous media. The CQD powders were highly hygroscopic, and long-term storage in ambient atmosphere resulted in CQD powders that stuck to the inner walls of the vials.
The ultraviolet-visible (UV-vis) spectra of an aqueous solution of CQDs showed no optical absorption beyond 400 nm (Fig. 1b). Therefore, the CQD solutions were colorless in daylight. The strong absorption at 208 nm and weak absorption at 297 nm originated from π–π* transition of the π-conjugated carbon framework and the n–π* transition of the C–O groups, respectively. The photoluminescence (PL) spectra of the CQDs showed excitation-dependent emission behavior, which is frequently observed in most CQDs prepared through a bottom-up approach (Fig. 1c).31,36 The different particle sizes2 and different levels of oxidation46 even in same size could contribute to the above excitation-dependent emission behavior. The highest emission was observed at 422 nm (blue) upon excitation by the 340 nm UV light. The lowest emission was observed at 579 nm (yellow) upon excitation of 500 nm. The highest PLQY of 49.9% (422 nm emission) was accomplished in the case of CQD2 with the use of quinine sulfate as a reference.31 The PLQYs of CQD1 and CQD3 were 24.0 and 43.5%, respectively. These PLQYs of CQDs prepared from the SA (A2) and TAEA (B3) monomer set in this study are the highest reported among CQDs prepared from the bottom-approach through microwave treatment (Table S1, ESI†).36,47–50 Therefore, the utilization of the “A2 + B3” monomer set in this study is seen to be very effective for the synthesis of highly fluorescent CQDs through microwave-assisted polyamidation/carbonization. From the highest mass yield and PLQY in the case of CQD2 (NA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NB = 1
NB = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.25, where NX means the number of X functional groups), is estimated that NB should be larger than NA to accomplish high PLQY through surface passivation by terminal B (–NH2) groups. However, too large NA
2.25, where NX means the number of X functional groups), is estimated that NB should be larger than NA to accomplish high PLQY through surface passivation by terminal B (–NH2) groups. However, too large NA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NB of 1
NB of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 in the case of CQD3 could produce very low mass yield of 3.13 wt% probably due to the too large mismatch between NA and NB. Because the formation of CQDs from either “A3 + B2” monomer set31 or “A2 + B3” monomer set in this study is regarded to proceed through concerted condensation/polymerization/carbonization reaction, mismatch in the feeding ratio of A to B functional groups might produce initial condensates with low molecular weight as reported in A2 and B3 polymerization study where different NA and NB ratio was adopted.39 Increasing of NA/NB resulted in increased mass yield more than 20% in the case of CQD4 and CQD5 where the molar feeding ratios of SA and TAEA are 3
3 in the case of CQD3 could produce very low mass yield of 3.13 wt% probably due to the too large mismatch between NA and NB. Because the formation of CQDs from either “A3 + B2” monomer set31 or “A2 + B3” monomer set in this study is regarded to proceed through concerted condensation/polymerization/carbonization reaction, mismatch in the feeding ratio of A to B functional groups might produce initial condensates with low molecular weight as reported in A2 and B3 polymerization study where different NA and NB ratio was adopted.39 Increasing of NA/NB resulted in increased mass yield more than 20% in the case of CQD4 and CQD5 where the molar feeding ratios of SA and TAEA are 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (NA
2 (NA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NB of 1
NB of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and 3
1) and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (NA
1 (NA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NB of 2
NB of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), respectively (Table S1, ESI†). However, both CQD4 and CQD5 presented very low PLQY values probably due to inefficient surface passivation originating from rich terminal carboxylic acid groups. An average PL relaxation time (τavg) of 2.73 ns was obtained for the CQDs, regardless of the monomer feeding ratio, showing that the microenvironments near the emitting sites in CQDs were similar to each other (Fig. S1, ESI†). Additionally, all CQDs showed similar multi-exponential decay, with slower decay in the tailing, which might originate from the significant intra/interelectronic interaction among emitting sites in the CQD(s).3
1), respectively (Table S1, ESI†). However, both CQD4 and CQD5 presented very low PLQY values probably due to inefficient surface passivation originating from rich terminal carboxylic acid groups. An average PL relaxation time (τavg) of 2.73 ns was obtained for the CQDs, regardless of the monomer feeding ratio, showing that the microenvironments near the emitting sites in CQDs were similar to each other (Fig. S1, ESI†). Additionally, all CQDs showed similar multi-exponential decay, with slower decay in the tailing, which might originate from the significant intra/interelectronic interaction among emitting sites in the CQD(s).3
Then, the compositional features of the CQDs were examined by Fourier-transform infrared (FT-IR) spectroscopy, nuclear magnetic resonance (NMR) spectroscopy, and X-ray photoelectron spectroscopy (XPS). The FT-IR spectrum of CQD2 showed a broad N–H/O–H stretching peak at 3424 cm−1 (Fig. 2a). Also, together with the C–H stretching peaks at 3086 and 2944 cm−1, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O vibrational peaks were observed at 1648 and 1552 cm−1 (amide I and II, respectively).31 Therefore, the build-up of a significant number of amide linkages during the microwave treatment of SA and TAEA is clearly indicated. The carboxylic acid groups of SA are regarded to have been completely consumed during microwave treatment. The 13C-NMR spectrum of the CQDs showed the complete disappearance of carbonyl peaks of SA at 177 ppm (Fig. S2†). Instead, a newly formed amide C
O vibrational peaks were observed at 1648 and 1552 cm−1 (amide I and II, respectively).31 Therefore, the build-up of a significant number of amide linkages during the microwave treatment of SA and TAEA is clearly indicated. The carboxylic acid groups of SA are regarded to have been completely consumed during microwave treatment. The 13C-NMR spectrum of the CQDs showed the complete disappearance of carbonyl peaks of SA at 177 ppm (Fig. S2†). Instead, a newly formed amide C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peak was observed at 175 ppm. In addition, both the 13C- and 1H-NMR spectra of the CQDs showed the complete disappearance of monomer peaks (Fig. S3†). The formation of a graphene-like framework during CQD formation was traced by finding aromatic protons and carbons in the NMR spectra of the CQDs. However, no aromatic protons (at 9–6 ppm) or carbon peaks (150–100 ppm) were observed in any of the cases, revealing that the construction of a graphitized carbon framework might not have been required for obtaining strongly photoluminescent CQDs in this study. XPS survey scans of the CQDs showed that they were mainly composed of rich carbon (83 at%), together with nitrogen (∼7 at%) and oxygen (∼10 at%) (Fig. 2b). The high resolution C1s binding peak revealed the presence of two different carbon bonds in the CQDs (Fig. 2c). The deconvolution of the raw C1s binding peak showed 87% C–C/C
O peak was observed at 175 ppm. In addition, both the 13C- and 1H-NMR spectra of the CQDs showed the complete disappearance of monomer peaks (Fig. S3†). The formation of a graphene-like framework during CQD formation was traced by finding aromatic protons and carbons in the NMR spectra of the CQDs. However, no aromatic protons (at 9–6 ppm) or carbon peaks (150–100 ppm) were observed in any of the cases, revealing that the construction of a graphitized carbon framework might not have been required for obtaining strongly photoluminescent CQDs in this study. XPS survey scans of the CQDs showed that they were mainly composed of rich carbon (83 at%), together with nitrogen (∼7 at%) and oxygen (∼10 at%) (Fig. 2b). The high resolution C1s binding peak revealed the presence of two different carbon bonds in the CQDs (Fig. 2c). The deconvolution of the raw C1s binding peak showed 87% C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds. The other 13% were estimated to be C–O/–N/C
C bonds. The other 13% were estimated to be C–O/–N/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds. However, the relative amount of each bond was difficult to determine because of the severe overlapping. The high-resolution O1s and N1s binding peaks showed that both oxygen and nitrogen elements were uniformly distributed in the CQDs (Fig. S4, ESI†). The major functional groups at the surfaces of the CQDs are regarded not as hydroxyl groups but as amine groups. Zeta-potential measurements showed positive potential values of +8.7 and +26.2 mV for CQD1 and CQD2, respectively (Fig. S5, ESI†). The increased zeta-potential value of CQD2 as compared to that of CQD1 might originate from the higher TAEA (B3) feeding ratio in the case of CQD2. The presence of rich amine moieties at the surfaces of the CQDs contribute to the high solubility of CQDs in other protic solvents such as methanol and ethanol. Similar to the aqueous solution of the CQDs, alcoholic solutions of CQDs revealed intensive blue emission upon the irradiation of UV light (Fig. S6, ESI†).
O bonds. However, the relative amount of each bond was difficult to determine because of the severe overlapping. The high-resolution O1s and N1s binding peaks showed that both oxygen and nitrogen elements were uniformly distributed in the CQDs (Fig. S4, ESI†). The major functional groups at the surfaces of the CQDs are regarded not as hydroxyl groups but as amine groups. Zeta-potential measurements showed positive potential values of +8.7 and +26.2 mV for CQD1 and CQD2, respectively (Fig. S5, ESI†). The increased zeta-potential value of CQD2 as compared to that of CQD1 might originate from the higher TAEA (B3) feeding ratio in the case of CQD2. The presence of rich amine moieties at the surfaces of the CQDs contribute to the high solubility of CQDs in other protic solvents such as methanol and ethanol. Similar to the aqueous solution of the CQDs, alcoholic solutions of CQDs revealed intensive blue emission upon the irradiation of UV light (Fig. S6, ESI†).
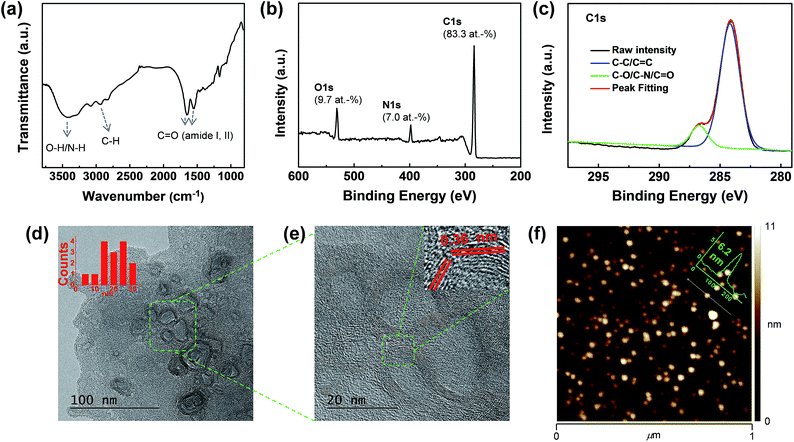 | ||
| Fig. 2 (a) FT-IR spectrum (KBr pellet), (b) XPS survey scan, (c) high-resolution C1s binding peak, (d) HRTEM image, (e) magnified HRTEM image of selected area (f) AFM image of CQD2. | ||
The structural and morphological features of the CQDs were examined by high-resolution transmission electron microscopy (HRTEM), atomic force microscopy (AFM), X-ray diffraction (XRD), and Raman spectroscopy. The HRTEM images of the CQDs showed the presence of semi-spherical particles with variable sizes and shapes, with lateral dimensions ranging from 5 to 30 nm (Fig. 2d). The internal areas of the CQDs seemed to be different, with the outside wall-like area showing a graphene-like lattice structure. The wall thickness of each particle was very uniform (4.1 ± 0.4 nm). The Fourier transform of the HRTEM image revealed the presence of a main lattice in the walls of the CQDs whose d-spacing was mainly 0.36 nm in addition to weak d-spacing of 0.21 nm (Fig. 2e and S7, ESI†). AFM images of the CQDs showed their dot-like morphology, with a uniform thickness of 6.2 nm (Fig. 2f). Therefore, it seems that the prepared CQDs had a disk-like morphology with different morphologies either inside or outside the particles. XRD analysis of the CQDs demonstrated the presence of broad scattering peaks at 11.8° (0.75 nm) and 20.3° (0.44 nm) (Fig. S8, ESI†). The Raman spectra of the prepared CQDs did not reveal the presence of either G or D band peaks upon irradiation with visible (514.5 nm) or near-IR laser (785 nm) (Fig. S9, ESI†). All these results indicate that the CQDs prepared from SA and TAEA through microwave treatment were composed of a carbon framework whose lattice structure was not graphitic but rather amorphous-like throughout this study.
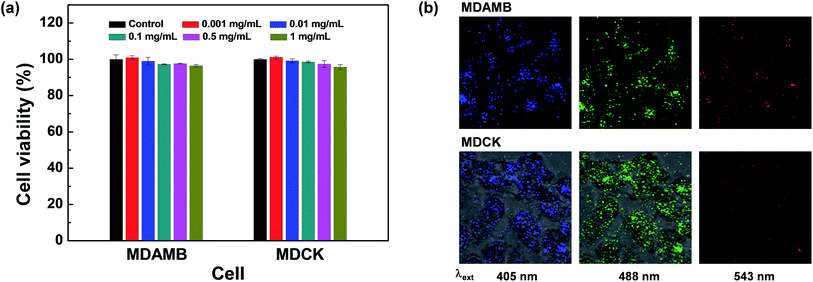
The strong fluorescent and high dispersion stability of the prepared CQDs prompted us to investigate the bioapplication of CQDs as non-toxic fluorescence imaging agents. Actually, various forms of fluorescent carbon nanodots (CDs) are abundantly present in daily foods including coffee and bread,51,52 showing the promising biocompatibility of CQDs that is not easily attainable without proper surface modification in the case of inorganic QDs such as PbS QDS.53 Both MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide)- and LDH (lactate dehydrogenase)-mediated cell viability assay were performed with MDA-MB-231 (breast cancer cells) and Madin Darby canine kidney (MDCK) epithelial cells. MTT assay demonstrated that the prepared CQDs show high cell viabilities of 96.3% and 95.9% for MDAMB and MDCK cells, respectively, even in high concentrations of 1 mg mL−1 (Fig. 3a). Surprisingly, slightly decreased cell viability was observed for CQDs in the case of LDH assay (Fig. S10, ESI†). Cell viability slightly decreases along the increase of CQD concentration. 87.6–92.5% of cell viability was observed in 1.0 mg mL−1 solution of all CQDs. Cross checking of cell toxicity of carbon based nanomaterials could be helpful for the evaluation of reliable cell viability.45 This low cytotoxicity of the prepared CQDs is in line with the high biosafety of other CQDs or carbon dots (CDs) treated with surface passion agents.2 After incubation of MDAMB and MDCK cells with the prepared CQDs for 24 h, most cells were wholly labeled with blue-, green-, or red-emitting CQDs depending on the excitation wavelength, showing the successful uptake of CQDs prepared from the SA and TAEA monomer set through endocytosis (Fig. 3b). However, red-colored fluorescence images were not vivid compared with blue- or green-colored fluorescence images due to the lower fluorescence intensity of CQDs with the excitation of longer wavelength, which is comparable to other blue emitting CQDs presenting excitation-dependent emission behavior.6,54 All these results demonstrate that CQDs are strong candidates for use in cellular imaging for diagnosis purposes.
Conclusion
In summary, strongly fluorescent CQDs were prepared in gram scale through the simple microwave-assisted synthesis from SA and TAEA as an ‘A2 + B3’ monomer set. The prepared CQDs were disk-like in their morphology and crystalline lattice structures were observed in the outside area of CQDs. Among CQDs prepared through microwave-assisted synthesis, CQDs prepared in this study revealed the highest PLQY of 49.9%. The excitation-dependent fluorescence behavior and high cell viability of prepared CQDs enable multi-color fluorescence imaging of cellular media. The success of ‘A2 + B3’ monomer set for the microwave-assisted synthesis of CQDs showing high PLQY could be widely and easily utilized for the mass production of other CQDs because naturally or synthetically abundant A2-type dicarboxylic acids can be integrated into the construction of CQDs. For a comparison, less abundant B2-type diamines (mainly synthetic) could be used in the previous bottom-up CQD synthesis from ‘A3 + B2’ monomer set.Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1D1A3A01020192), a Grant (No. 10049064) by Industrial Technology Innovation Program and funded by Ministry of Trade, Industry & Energy (MI, Korea), and International joint technology development project (N0002123) through the Ministry of Trade, Industry & Energy (MI, Korea) and Korea Institute for Advancement of Technology (KIAT).Notes and references
- S. Zhu, Y. Song, X. Zhao, J. Shao, J. Zhang and B. Yang, Nano Res., 2014, 8, 335 Search PubMed.
- S. Y. Lim, W. Shen and Z. Gao, Chem. Soc. Rev., 2015, 44, 362 RSC.
- C. J. Reckmeier, J. Schneider, A. S. Susha and A. L. Rogach, Opt. Express, 2016, 24, A312 CrossRef CAS PubMed.
- J. Zhang and S. H. Yu, Mater. Today, 2016, 19, 382 CrossRef CAS.
- Z. Wang, H. Zeng and L. Sun, J. Mater. Chem. C, 2015, 3, 1157 RSC.
- O. S. Wolfbeis, Chem. Soc. Rev., 2015, 44, 4743 RSC.
- L. Cao, S. T. Yang, X. Wang, P. G. Luo, J. H. Liu, S. Sahu, Y. Liu and Y. P. Sun, Theranostics, 2012, 2, 295 CrossRef CAS PubMed.
- Y. Xu, M. Wu, Y. Liu, X. Z. Feng, X. B. Yin, X. W. He and Y. K. Zhang, Chem.–Eur. J., 2013, 19, 2276 CrossRef CAS PubMed.
- S. K. Bhunia, A. Saha, A. R. Maity, S. C. Ray and N. R. Jana, Sci. Rep., 2013, 3, 1473 Search PubMed.
- K. Jiang, S. Sun, D. L. Zhang, Y. Lu, A. Wu, C. Cai and H. Lin, Angew. Chem., Int. Ed., 2015, 54, 5360 CrossRef CAS PubMed.
- C. J. Jeong, A. K. Roy, S. H. Kim, J. E. Lee, J. H. Jeong, I. In and S. Y. Park, Nanoscale, 2014, 6, 15196 RSC.
- A. K. Roy, S. M. Kim, P. Paoprasert, S. Y. Park and I. In, RSC Adv., 2015, 5, 31677 RSC.
- J. L. Chen and X. P. Yan, Chem. Commun., 2011, 47, 3135 RSC.
- X. Jia, J. Li and E. Wang, Nanoscale, 2012, 4, 5572 RSC.
- H. Li, X. He, Z. Kang, H. Huang, Y. Liu, J. Liu, S. Lian, C. H. A. Tsang, X. Yang and S. T. Lee, Angew. Chem., Int. Ed., 2010, 49, 4430 CrossRef CAS PubMed.
- J. Shen, Y. Li, Y. Su, Y. Zhu, H. Jiang and X. Yanga, Nanoscale, 2015, 7, 2003 RSC.
- Y. Li, Y. Hu, Y. Zhao, G. Shi, L. Deng, Y. Hou and L. Qu, Adv. Mater., 2011, 23, 776 CrossRef CAS PubMed.
- P. Mirtchev, E. J. Henderson, N. Soheilnia, C. M. Yip and G. A. Ozin, J. Mater. Chem. A, 2012, 22, 1265 RSC.
- V. Gupta, N. Chaudhary, R. Srivastava, G. D. Sharma, R. Bhardwaj and S. Chand, J. Am. Chem. Soc., 2011, 133, 9960 CrossRef CAS PubMed.
- H. Tao, K. Yang, Z. Ma, J. Wan, Y. Zhang, Z. Kang and Z. Liu, Small, 2012, 8, 281 CrossRef CAS PubMed.
- H. U. Lee, S. Y. Park, E. S. Park, B. Son, S. C. Lee, J. W. Lee, Y. C. Lee, K. S. Kang, M. I. Kim, H. G. Park, S. Choi, Y. S. Huh, S. Y. Lee, K. B. Lee, Y. K. Oh and J. Lee, Sci. Rep., 2014, 4, 4665 Search PubMed.
- H. Ding, F. Du, P. Liu, Z. Chen and J. Shen, ACS Appl. Mater. Interfaces, 2015, 7, 6889 CAS.
- H. Peng and J. T. Sejdic, Chem. Mater., 2009, 21, 5563 CrossRef CAS.
- J. Zhou, Z. Sheng, H. Han, M. Zou and C. Li, Mater. Lett., 2012, 66, 222 CrossRef CAS.
- S. Sahu, B. Behera, T. K. Maiti and S. Mohapatra, Chem. Commun., 2012, 48, 8835 RSC.
- M. P. Sk, A. Jaiswal, A. Paul, S. S. Ghosh and A. Chattopadhyay, Sci. Rep., 2013, 3, 1473 Search PubMed.
- S. K. Bhunia, A. Saha, A. R. Maity, S. C. Ray and N. R. Jana, Sci. Rep., 2012, 2, 383 Search PubMed.
- S. Mitra, S. Chandra, S. H. Pathan, N. Sikdar, P. Pramanik and A. Goswami, RSC Adv., 2013, 3, 3189 RSC.
- J. Wang, C. F. Wang and S. Chen, Angew. Chem., Int. Ed., 2012, 51, 9297 CrossRef CAS PubMed.
- P. C. Hsu, Z. Y. Shih, C. H. Lee and H. T. Chang, Green Chem., 2012, 14, 917 RSC.
- S. Zhu, Q. Meng, L. Wang, J. Zhang, Y. Song, H. Jin, K. Zhang, H. Sun, H. Wang and B. Yang, Angew. Chem., Int. Ed., 2013, 52, 3953 CrossRef CAS PubMed.
- Y. Xu, M. Wu, X. Z. Feng, X. B. Yin, X. W. He and Y. K. Zhang, Chem.–Eur. J., 2013, 19, 6282 CrossRef CAS PubMed.
- L. Zhu, Y. Yin, C. F. Wang and S. Chen, J. Mater. Chem. C, 2013, 1, 4925 RSC.
- C. J. Jeong, A. K. Roy, S. H. Kim, J. E. Lee, J. H. Jeong, I. In and S. Y. Park, Nanoscale, 2014, 6, 15196 RSC.
- A. M. Alam, B. Y. Park, Z. K. Ghouri, M. Park and H. Y. Kim, Green Chem., 2015, 17, 3791 RSC.
- S. Qu, X. Wang, Q. Lu, X. Liu and L. Wang, Angew. Chem., Int. Ed., 2012, 51, 12215 CrossRef CAS PubMed.
- C. Gao and D. Yan, Prog. Polym. Sci., 2004, 29, 183 CrossRef CAS.
- H. Chen and J. Kong, Polym. Chem., 2016, 7, 3643 RSC.
- I. In and S. Y. Kim, PMSE Prepr., 2004, 91, 969 Search PubMed.
- S. Horikoshi, A. Osawa, M. Abe and N. Serpone, J. Phys. Chem. C, 2011, 115, 23030 CAS.
- Y. P. Sun, B. Zhou, Y. Lin, W. Wang, K. A. S. Fernando, P. Pathak, M. J. Meziani, B. A. Harruff, X. Wang, H. Wang, P. G. Luo, H. Yang, M. E. Kose, B. Chen, L. M. Veca and S. Y. Xie, J. Am. Chem. Soc., 2006, 128, 7756 CrossRef CAS PubMed.
- X. Wang, L. Cao, S. T. Yang, F. Lu, M. J. Meziani, L. Tian, K. W. Sun, M. A. Bloodgood and Y. P. Sun, Angew. Chem., Int. Ed., 2010, 49, 5310 CrossRef CAS PubMed.
- H. Ding, L. W. Cheng, Y. Y. Ma, J. L. Kong and H. M. Xiong, New J. Chem., 2013, 37, 2515 RSC.
- S. H. Kim, S. M. Sharker, H. Lee, I. In, K. D. Lee and S. Y. Park, RSC Adv., 2016, 6, 61482 RSC.
- J. M. Worle-Knirsch, K. Pulskamp and H. F. Krug, Nano lett., 2006, 6, 1261 CrossRef CAS PubMed.
- H. Ding, S. B. Yu, J. S. Wei and H. M. Xiong, ACS Nano, 2016, 10, 484 CrossRef CAS PubMed.
- Y. Choi, N. Thongsai, A. Chae, S. Jo, E. B. Kang, P. Paoprasert, S. Y. Park and I. In, J. Ind. Eng. Chem., 2017, 47, 329 CrossRef CAS.
- L. Tang, R. Ji, X. Cao, J. Lin, H. Jiang, X. Li, K. S. Teng, C. M. Luk, S. Zeng, J. Hao and S. P. Lau, ACS Nano, 2012, 6, 5102 CrossRef CAS PubMed.
- L. Zhao, F. Di, D. Wang, L. H. Guo, Y. Yang, B. Wan and H. Zhang, Nanoscale, 2013, 5, 2655 RSC.
- C. M. Luk, L. B. Tang, W. F. Zhang, S. F. Yu, K. S. Teng and S. P. Lau, J. Mater. Chem., 2011, 21, 2445 RSC.
- M. P. Sk, A. Jaiswal, A. Paul, S. S. Ghosh and A. Chattopadhyay, Sci. Rep., 2012, 2, 383 Search PubMed.
- C. Jiang, H. Wu, X. Song, X. Ma, J. Wang and M. Tan, Talanta, 2014, 127, 68 CrossRef CAS PubMed.
- F. M. Winnik and D. Maysinger, Acc. Chem. Res., 2013, 46, 672 CrossRef CAS PubMed.
- E. B. Kang, S. M. Sharker, I. In and S. Y. Park, J. Ind. Eng. Chem., 2016, 43, 150 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Detail experimental procedure, fluorescence decay profiles, XRD spectrum, zeta potential profiles, Raman spectra, and PL spectra of CQDs. See DOI: 10.1039/c6ra28176a |
| This journal is © The Royal Society of Chemistry 2017 |