Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
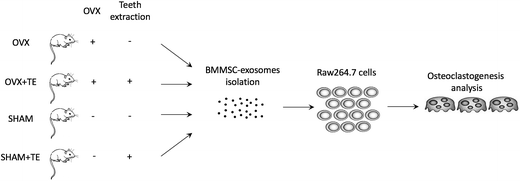
Bone marrow mesenchymal stem cell-derived exosomes enhance osteoclastogenesis during alveolar bone deterioration in rats†
Shuyu Xu and
Zuolin Wang
and
Zuolin Wang *
*
Shanghai Engineering Research Center of Tooth Restoration and Regeneration, Department of Oral Implant, School of Stomatology, Hospital of Stomatology, Tongji University, 399 Yanchang Road, Shanghai 200072, PR China. E-mail: zuolin@tongji.edu.cn; Fax: +86-21-66524025; Tel: +86-21-66313725
First published on 13th April 2017
Abstract
Healthy and functional alveolar bones to support implants or dentures are fundamental for dental restoration. Osteoporosis, which leads to alveolar bone resorption, is a common disease, especially in elderly patients with tooth loss. Enhanced osteoclastogenesis is the main pathogenesis of bone resorption, which is related to the paracrine regulation of BMMSCs (bone marrow mesenchymal stem cells). Furthermore, exosomes have been substantiated to be involved in the paracrine functions of BMMSCs. However, the underlying mechanisms of the regulation of exosomes in osteoclastogenesis remain unclear. The aim of this study is to investigate the role of BMMSC-derived exosomes in regulating osteoclastogenic differentiation of the osteoclast precursor cell line Raw264.7 to illustrate the role of exosomes in alveolar bone resorption. To establish the rat model of alveolar bone osteoporosis, thirty ten-week-old female SD rats were subjected to ovariectomy (OVX) or sham-operation. One month later, the right maxillary molars were extracted. Three months after tooth extraction (TE), the maxillae were collected. The BMMSCs of the alveolar bones from the SHAM + TE, OVX and OVX + TE groups were retrieved and cultured for the isolation of exosomes. The Raw264.7 cells were co-cultured with BMMSC-derived exosomes to investigate their osteoclastogenic differentiation. In vivo, OVX and tooth loss exerted a synergistic effect on the upregulation of osteoclastogenesis, which led to bone resorption. In vitro, the BMMSC-derived exosomes from the rats with bone deterioration (SHAM + TE, OVX and OVX + TE) increased the osteoclastogenesis of the Raw264.7 cells. These results suggest that BMMSC-derived exosomes could accelerate osteoclastogenesis in alveolar bone deterioration.
Introduction
Osteoporosis is a condition of low bone mass and microstructural changes in the bone tissue, leading to the increased likelihood of bone fragility and fracture.1–3 In most cases, osteoporosis occurs in the proximal femur and vertebral bodies. However, consensus is still lacking among published studies on the correlation between osteoporosis and alveolar bone loss. Alveolar bone loss has aroused considerable interest among dentists due to its possible impact on periodontal disease, residual alveolar ridge resorption, and implant success rate.4 Nevertheless, there is considerably less data and evidence with alveolar bone than with long bones. Oestrogen deficiency and mechanical unloading are believed to be the main causes of osteoporosis, which may also affect the facial bones.5–8 To the best of our knowledge, there is no consensus on whether these two factors exert synergistic or antagonistic effects on alveolar bone resorption. Clarification of such effects regarding the alveolar bone could provide guidance to dentists for the therapeutic decisions involving the treatment of postmenopausal women with missing teeth.The pathogenesis of bone loss mainly includes increased osteoclastogenesis and decreased osteogenesis. Oestrogen deficiency-induced osteoporosis involves elevated osteoclast formation as a result of increased production of molecules such as pro-inflammatory cytokines.9,10 The reduction of mechanical loading results in a dramatic loss of bone mass and deterioration of the bone microarchitecture, and it is attributed to the regulation of the receptor activator of nuclear factor-kappaB ligand (RANKL)/RANK/osteoprotegerin (OPG) system to increase osteoclastogenesis. Although it has been well reviewed that the mesenchymal stem cells (MSCs) have an important role in the regulation of induction factors and cytokines that could participate in osteoclastogenesis,11 the underlying mechanisms still need further investigation.
According to the traditional concept, the transfer of information between cells can be achieved by direct contact and cytokines. Recently, increasing attention has been given to exosomes, small vesicles of endocytic origin.12 Exosomes act in a paracrine or endocrine manner to facilitate various intracellular or intercellular signalling mechanisms. It has been reported that exosomes contain a variety of biological components, including membrane proteins, lipids, RNA and even DNA from donor cells.13 Once the exosomes are released, they can either target a neighbouring cell or reach the cells of distant organs after entering the bloodstream.14 There are several reports demonstrating that exosomes derived from MSCs (MSCs-exos) can be taken up by other cell types to influence their functions.15 However, to date, there is a lack of evidence on the effects of MSC-exos on osteoclast differentiation under osteoporotic conditions.
Therefore, the aim of this study was to investigate the potential effects of tooth loss on ovariectomy (OVX)-related alveolar bone deterioration, and the role of exosomes secreted by bone marrow derived stem cells (BMMSC-exos) in osteoclastic differentiation during this process. Our study may provide a new perspective of the pathogenic mechanism of osteoporosis in the alveolar bone and a potential guidance for therapeutic strategies in treating postmenopausal women with missing teeth.
Materials and methods
Animal experiments
Ten-week-old female SD rats from the Laboratory Animal Center of Tongji University were used in this study, following the International Guiding Principles for Animal Research (1985). The experimental protocol was approved by the Laboratory Animal Center of Tongji University (No. TJLAC-017-014).Thirty rats were randomly divided into two groups and anesthetized with an intraperitoneal injection of sodium pentobarbital. Fifteen rats were subjected to bilateral ovariectomy (OVX group), while the other fifteen rats were selected for sham operation (SHAM group), performed following the standard method.16 All animals were housed under the same conditions and fed a standard rodent diet with free access to water. One month later, all the rats were anesthetized with sodium pentobarbital and the right maxillary molars were extracted using dental tweezers (SHAM + TE and OVX + TE groups). The left maxillaries were taken as control (SHAM and OVX groups).
A total of 12 weeks after the surgery, the maxillaries and femur were dissected. Ten rats were used for the measurements of microstructure by micro-computerized tomography (micro-CT), ten rats for histological analysis, and ten rats for BMMSC culture.
Micro-CT and histological evaluation
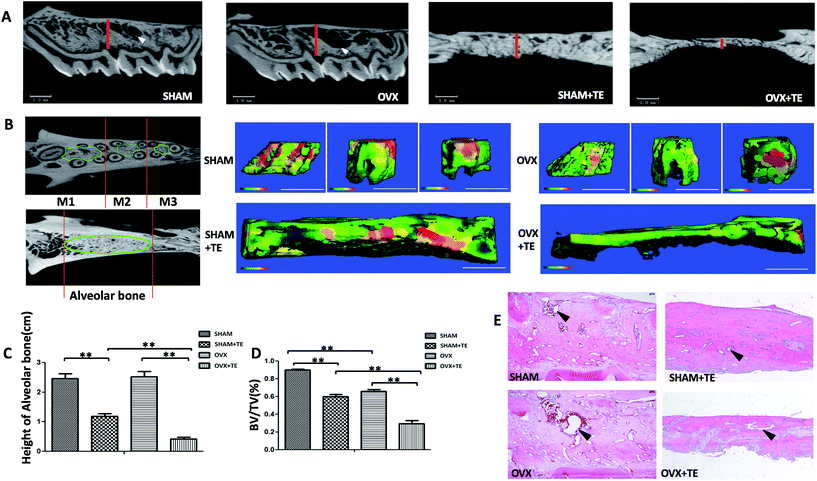
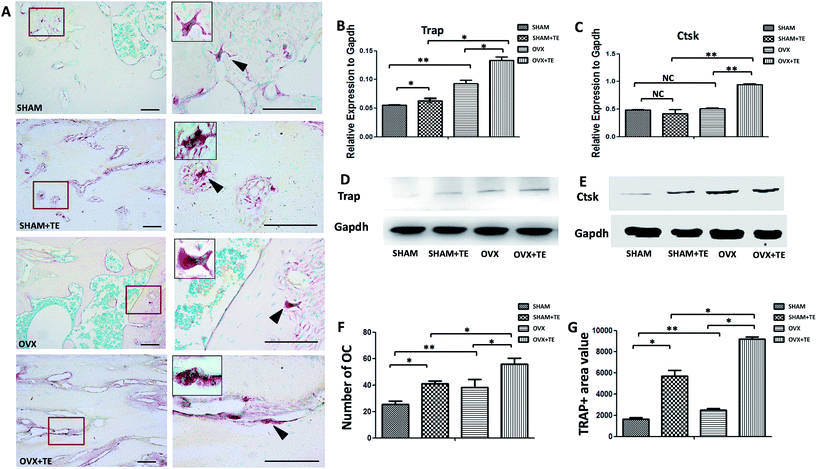
Four months after the surgery, all the rats were euthanized with an overdose of sodium pentobarbital and the maxillaries were extracted. Twenty samples (five SHAM, five SHAM + TE, five OVX, five OVX + TE) were scanned by a micro-CT scanner (micro-CT35, Scanco Medical AG, Bassersdorf, Switzerland) with a 10 μm voxel size using the following parameters: 114 mA, 70 kVp, and 300 ms exposure time. The region of interest (ROI) of the maxillary was selected according to a previously described method, which succeeded in showing the integral structure of the alveolar bone.17 The ROI of the alveolar bone was manually established. In the OVX and SHAM groups, we identified the coronal surface by passing the centres of the medial root of the first molar and the distal root of the third molar on two-dimensional images. In the SHAM + TE and OVX + TE groups, we identified the centre of the alveolar bone by passing the centres of the medial and distal sites (Fig. 1A and B). Then, we drew a contour from the middle portion of the alveolar bones on the horizontal plane of the maxillary, avoiding roots and other structures, on two-dimensional images. The green lines showed the ROI on the horizontal surfaces. Three-dimensional microarchitectures of the alveolar bones were analysed by the software accompanying the micro-CT machine. The alveolar bone histomorphometry was calculated directly from the ROI. We evaluated the bone volume/total volume (BV/TV, %) (Fig. 1D).The maxillary bone segments, containing the alveolar bone, were dissected for each group. Five-micron thick sections were cut from the maxillary alveolar bone. The sections were stained with haematoxylin and eosin (H&E). One section from each sample was stained with H&E and another section was stained with tartrate-resistant acid phosphatase (TRAP; Sigma-Aldrich, St. Louis, MO, USA). TRAP-positive multi-nucleated cells (TRAP + MNCs) were considered as osteoclasts.18,19
Preparation of BMMSCs
Three months after the tooth extraction (four months after OVX or Sham-op), the rats were euthanized. For the isolation of BMMSCs, the maxillary bones were dissected from the rats and cleaned with sterile phosphate buffered saline (PBS). After separating and discarding the teeth, the specimens were fully excised and incubated with 0.25% type I collagenase (Sigma-Aldrich, St. Louis, MO, USA) for 90 min at 37 °C with vigorous agitation. After neutralization of the collagenase, the cells released from the bone specimens were collected by centrifugation at 400g for 5 min. The cells were resuspended and seeded in tissue culture-treated flasks in Minimum Essential Medium alpha (αMEM, Life Technologies, California, USA) plus 10% foetal bovine serum and 1% penicillin–streptomycin.Identification of BMMSCs
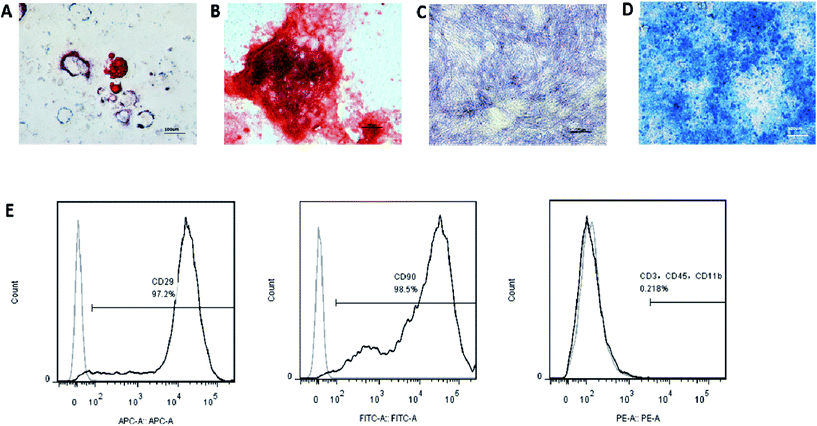
For fluorescence activated cell sorting (FACS) analyses, the cells were stained with antibodies CD29-APC, CD90-FITC, CD3-PE, CD11b-PE, and CD45-PE; isotype-matched normal IgG (BD®, San Diego, CA, USA) was used as controls. Quantitative fluorescence analysis was performed using the FACSCalibur platform (Becton Dickinson, Franklin Lakes, N.J., USA) and the CellQuest software. To test the multipotency of the cells, the BMMSCs were cultured with either osteogenic, chondrogenic or adipogenic media (Cyagen Biosciences Inc., Guangzhou, China) for 21 d. Standard Alizarin Red, Alcian Blue and Oil Red O staining were used to identify osteoblasts, cartilage and adipocyte-like cells, respectively. Alkaline phosphatase (ALP) staining was used on the seventh day of culture by osteogenic media to analyse the osteogenic activity.Preparation of exosomes from the BMMSCs
Exosome preparation was performed using previously described methods with some modifications.20,21 Briefly, the BMMSCs were resuspended in αMEM with 10% FBS to 1 × 106 cells per mL. The medium was changed to serum-free αMEM 24 h later, and the conditioned medium (CM) of BMMSCs was collected after the cells were cultured for another 5 d. The CM was centrifuged at 2000g for 10 min at 4 °C then 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min at 4 °C, and the supernatant was filtered through a 0.2 μm filter unit (Kurabo, Osaka, Japan) to remove cellular debris, larger vesicles, such as microvesicles/microparticles, and apoptotic bodies. The CM was then transferred for exosome isolation using the Total Exosome Isolation kit (Life Technologies, California, USA) according to the manufacturer's protocol. The pelleted exosomes were resuspended in a convenient volume of serum-free αMEM or 1X PBS. The protein concentration of the exosome fraction was measured using the Micro BCA Protein Assay Kit (Life Technologies) according to the manufacturer's instructions.
000g for 30 min at 4 °C, and the supernatant was filtered through a 0.2 μm filter unit (Kurabo, Osaka, Japan) to remove cellular debris, larger vesicles, such as microvesicles/microparticles, and apoptotic bodies. The CM was then transferred for exosome isolation using the Total Exosome Isolation kit (Life Technologies, California, USA) according to the manufacturer's protocol. The pelleted exosomes were resuspended in a convenient volume of serum-free αMEM or 1X PBS. The protein concentration of the exosome fraction was measured using the Micro BCA Protein Assay Kit (Life Technologies) according to the manufacturer's instructions.
Morphology and size of exosomes
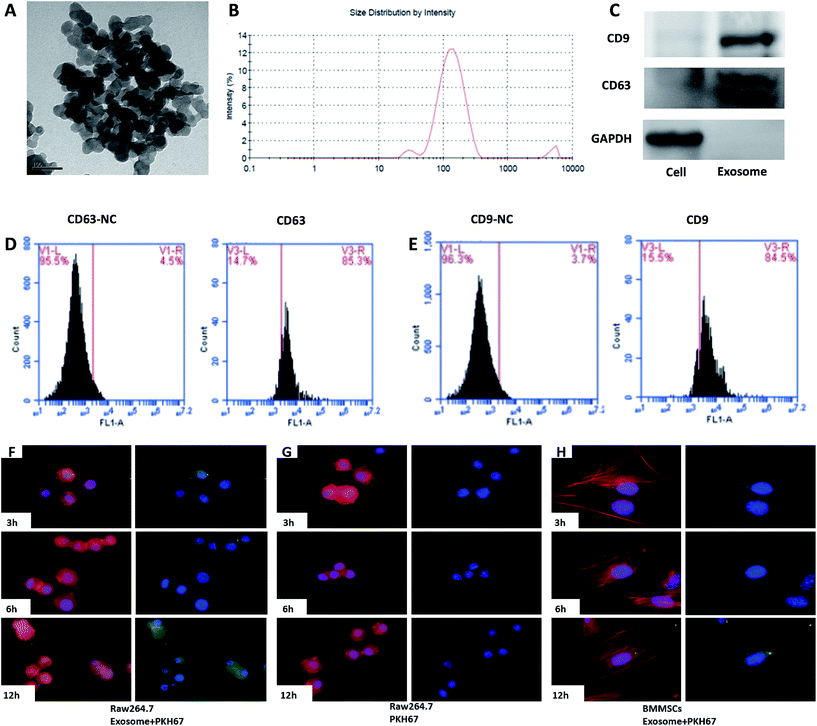
Morphology of the exosomes was observed by transmission electron microscopy (TEM) (Hitachi H-7100; Hitachi High-Technologies Corporation, Tokyo, Japan). The samples were prepared by dropping 5 μl of exosomes resuspended in PBS onto an elastic carbon-support film grid with 100 mm pitch and visualized with 2% ammonium molybdate. The exosome particle size was measured by dynamic light scattering (DLS). DLS was carried out with a Zetasizer Nano ZS instrument (Malvern Instruments Ltd, Malvern, Worcestershire, UK) equipped with a temperature control system.For FACS analysis, the exosomes were coated onto 4 μm-diameter aldehyde/sulphate latex beads, using a previously described method.22 Briefly, 5 μg exosomes was incubated with 10 μl 4 μm-diameter aldehyde/sulphate latex beads for 15 min at room temperature in PBS. The mixture was then transferred to 1 mL PBS with gentle shaking for 2 h. After centrifugation, the pellet was blocked by incubation with 110 μl of 1 M glycine (100 mM final) for 30 min. The exosome-coated beads were washed two times in PBS and resuspended in 100 μl PBS. Afterwards, the beads were incubated with PE-labelled CD9 antibody for 1 h at room temperature in the dark. The beads were analysed by flow cytometry using an FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA, USA) and the FlowJo software (Tree Star, Ashland, OR, USA).
Incorporation of exosomes
For the uptake studies, the purified exosomes were labelled with a PKH67 (green) kit (Sigma-Aldrich, St. Louis, MO, USA) according to previously reported protocols.23 In parallel, the PKH67 dye was added to 0.5 mL Diluent C and incubated without the exosome solution taken as a negative control. To bind excess dye, 2 mL 0.5% bovine serum albumin/PBS was added. The labelled exosomes were washed and the exosome pellet was resuspended with PBS and used for the uptake experiments. The PKH67-labelled exosomes were co-cultured with Raw264.7 cells for the indicated time, BMMSCs were used as control. Then, the cells were fixed and stained with phalloidin and DAPI (Life Technologies, California, USA). Images were obtained by an immunofluorescence microscope.Cell counting kit 8 (CCK8) assay
RAW264.7 cells were seeded at 4 × 103 per well into 96-well microplates and incubated with DMEM containing BMMSC-exos (SHAM, SHAM + TE, OVX and OVX + TE groups) for 3 d and 7 d. CCK8 (Dojindo, Japan) solution (10 μl) was added to each well and the plate was incubated for an additional 1 h. Finally, the absorbance of each well was measured with a Multiskan Spectrum microplate reader (Thermo Scientific, Waltham, MA, USA) at 450 nm wavelength.Osteoclast formation assay
Raw264.7 cells were seeded at a density of 1000 per well in 24-well plates and cultured with CM or DMEM containing BMMSC-exos (SHAM, SHAM + TE, OVX and OVX + TE groups) derived from same volume of CM, and 15 ng mL−1 sRankl was added to restore the original CM-volume. DMEM containing 15 ng mL−1 sRankl was used as the negative control. The medium was replaced every 2 d for 10 d. The cells were stained for TRAP according to the manufacturer's protocol. TRAP-positive cells containing no less than three nuclei were counted as osteoclast-like cells (TRAP + MNCs). mRNA was collected from the cells on the third and seventh days to evaluate the expression of the osteoclastogenesis-related genes, including Nfatc1 and cathepsin K (Ctsk). Protein was isolated from the cells on the fifth and ninth days to investigate the expression of the markers.Real-time qPCR
Total RNA was extracted using TRIzol reagent (Life Technologies, California, USA) from homogenized tissues or cells. Two micrograms of total RNA was reverse-transcribed into cDNA using the superscript first-strand synthesis system (Roche, Basel, Switzerland) according to the manufacturer's protocol. Reactions were conducted in a 20 μl reaction mixture using a Real-Time PCR System (LightCycler®96, Roche, Switzerland). The expression of the various genes was presented as 2 (−ΔCT). The primers used are listed in Table 1 for rat and Table 2 for mouse. The target genes were normalized against glyceraldehydes-3-phosphatedehydrogenase (Gadph) expression.| Gene (rat) | Direction | Sequence (5′–3′) |
|---|---|---|
| Ctsk | Forward | GACCCGTCTCTGTGTCCATC |
| Reverse | ACGGTCGCAGTTTTCGTCAT | |
| Trap | Forward | GCTTCCACCCTGAGATTCGT |
| Reverse | ATGATGAAGTCAGCGCCCAT | |
| Gapdh | Forward | GGCACAGTCAAGGCTGAGAATG |
| Reverse | ATGGTGGTGAAGACGCCAGTA |
| Gene (mouse) | Direction | Sequence (5′–3′) |
|---|---|---|
| Ctsk | Forward | CAGCAGAGGTGTGTACTATG |
| Reverse | GCGTTGTTCTTATTCCGAGC | |
| Nfatc1 | Forward | TGAGGCTGGTCTTCCGAGTT |
| Reverse | CGCTGGGAACACTCGATAGG | |
| Trap | Forward | TGCCTACCTGTGTGGACATGA |
| Reverse | CACATAGCCCACACCGTTCTC | |
| Gapdh | Forward | GACGGCCGCATCTTCTTGTGC |
| Reverse | TGCAAATGGCAGCCCTGGTGA |
Western blot analysis
The isolated exosome pellets and cultured Raw264.7 cells were harvested and lysed in RIPA buffer supplemented with complete protease inhibitor cocktail solution (Sigma-Aldrich, St. Louis, MO, USA). Cell debris was removed by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400g for 20 min. Then, the lysates of the exosomes or cells were separated using SDSPAGE gels, transferred to PVDF membranes (Bio-Rad, California, USA), and incubated with relevant antibodies as indicated. Antibodies against CD63 and Nfatc1 were purchased from Santa Cruz Biotechnology (California, USA) and those against Ctsk, Trap, and Gapdh were purchased from Abcam (Cambridge, UK).
400g for 20 min. Then, the lysates of the exosomes or cells were separated using SDSPAGE gels, transferred to PVDF membranes (Bio-Rad, California, USA), and incubated with relevant antibodies as indicated. Antibodies against CD63 and Nfatc1 were purchased from Santa Cruz Biotechnology (California, USA) and those against Ctsk, Trap, and Gapdh were purchased from Abcam (Cambridge, UK).
Statistical analysis
Data are expressed as the mean ± standard deviation (SD). Statistical analyses were performed by one-way ANOVA followed by Student's t-test and are shown as the mean ± SD. *, #, +p < 0.05; **, ##, ++p < 0.01; n.s., non-significant (p > 0.05).Results
Establishment of the osteoporotic model
The micro-CT and histological analyses were used to confirm the establishment of the osteoporotic model. Compared to the SHAM group, the 3D images of the femurs in the OVX group showed a significant decrease of the subchondral trabecular bone volume, thickness, and density at 1 month and a more severe decrease at 4 months (ESI Fig. S1†). H&E staining of the distal femurs in the three groups also showed a significant decrease in subchondral trabecular bone volume in the OVX group compared to the sham group at both 1 month and 4 months. Micro-CT analysis of the femurs in the OVX group demonstrated a significant decrease in bone volume over total volume (BV/TV%) compared to that in the SHAM group, consistent with the histological results. However, the alveolar bone loss was not significant at 1 month (data not show) but was significant at 4 months (ESI Fig. S1†).The synergistic effects of tooth loss and OVX on bone loss
In this study, the three maxillary molars on the right side were extracted after 1 month of OVX/SHAM operation, and the left side without tooth extraction was taken as control. Three months after the tooth extraction, two-dimensional images generated by micro-CT showed that the alveolar bone height in the OVX + TE and SHAM + TE groups decreased significantly compared to the groups without tooth extraction (OVX and SHAM groups), whereas the alveolar bone height did not change notably between the SHAM and OVX groups. The alveolar bone height in the OVX + TE group decreased most significantly. In addition, the marrow spaces, especially the inter-radicular alveolar bone encompassing the three maxillary molars, in the OVX group increased compared to that in SHAM group (Fig. 1A and C). The bone morphometry by micro-CT of the SHAM + TE, OVX, OVX + TE groups demonstrated a significant decrease in the thickness and BV/TV% compared to the SHAM group (Fig. 1B and D). Moreover, H&E staining showed an enlarged marrow space in the OVX group compared to that of the SHAM group. Additionally, in the H&E staining, the alveolar bone height of the SHAM + TE and OVX + TE groups decreased notably, especially in the OVX + TE group, which is consistent with the results of the micro-CT (Fig. 1E).Upregulation of osteoclastogenesis in the bone loss of maxillaries
Osteoclastogenesis in the maxillary bones was analysed by TRAP staining (Fig. 2). In comparison with the SHAM group, the SHAM + TE, OVX, OVX + TE groups demonstrated an increase in the marrow medullary areas of alveolar bones and a pronounced osteoclastogenesis, as indicated by the increased osteoclast counts (TRAP + MNCs), value of TRAP + area (Fig. 2F and G) and osteoclastogenesis-related gene and protein expressions (Trap, Ctsk) (p < 0.05; Fig. 2B–E). The OVX + TE group showed the strongest osteoclastogenesis, as the highest amount TRAP + MNCs were found in this group (Fig. 2F and G), accompanied with the highest level of Trap, Ctsk mRNA and protein expression and value of the TRAP + area (Fig. 2B–E and G).Existence of exosomes in MSCs derived from rat maxillary bones
The surface markers of the BMMSCS derived from the rat maxillary bones were identified by flow cytometry. The cells were positive for MSC markers, such as CD29 and CD90, and negative for CD11b (monocyte and macrophage marker), CD3 (lymphocyte marker), and CD45 (haematopoietic progenitor and endothelial cell marker) (Fig. 3G). The cells were positive for Alizarin Red S and ALP staining, Oil Red O staining, or Alcian Blue staining, when the cells were induced with osteogenic, adipogenic, or chondrogenic media, respectively (Fig. 3A–F). Taken together, these results indicated that these cells had phenotypic and functional characteristics of BMMSCs.After the exosomes were isolated, their ultrastructure was observed using transmission electron microscopy, revealing a typical diameter smaller than 200 nm24 (Fig. 4A). To further investigate the size distribution profile of the BMMSC-exos, we performed size detection using the DLS, revealing a size peak of 150 nm. Based on the morphology and size, the vesicles were exosomes (Fig. 4B). Then, the expression of exosomal markers CD9 and CD63 was confirmed by Western blotting. Since CD63 is highly glycosylated, the bands appeared over a wide range of weights between 30 and 60 kDa under non-reducing conditions in Western blot analysis, as previously reported25 (Fig. 4C). To test the purity of exosomes, we also tested the expression of CD9 and CD63 by FACS, which indicated an extremely high proportion (84.5% and 85.3%, respectively) of exosome-positive cells in the MSCS derived from rat maxillary bones (Fig. 4D and E).26 Collectively, these data indicated that the BMMSCS derived from rat maxillary bones contained and secreted exosomes into the culture medium.
Incorporation of exosomes into Raw264.7 cells
We investigated whether the BMMSC-exos were incorporated into Raw264.7 cells. The BMMSC-exos were labelled with the PKH67 green fluorescent membrane linker-dye and added to the Raw264.7 culture medium. After 3 h, the cells began to incorporate the BMMSC-exos (Fig. 4F left), and both the number of exosomes and the cells with exosomes increased in a time-dependent manner. After 12 h, the fluorescence intensity increased and almost all the cells had incorporated the labelled exosomes (Fig. 4F left). On the other hand, in the BMMSCs group, extremely low fluorescence intensity was obtained even after 12 h (Fig. 4F right). In the control group, none of the cells exhibited a fluorescent signal (Fig. 4F middle). These data suggested that the BMMSC-exos were incorporated into Raw264.7 cells in a time-dependent manner.Exosomes from the BMMSCs of alveolar bone induced osteoclastic differentiation of Raw264.7 cells
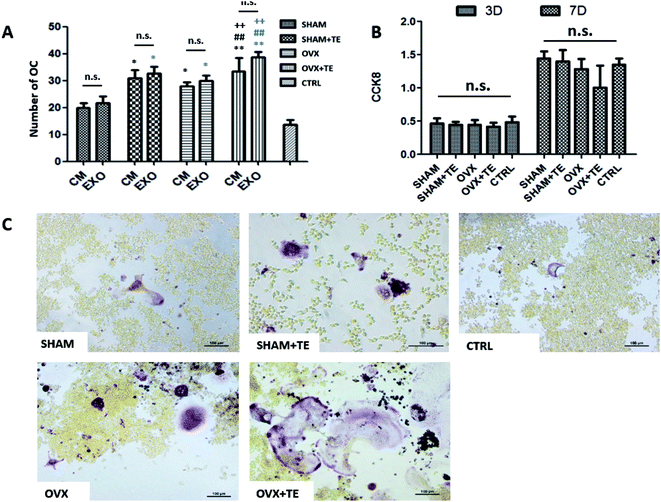
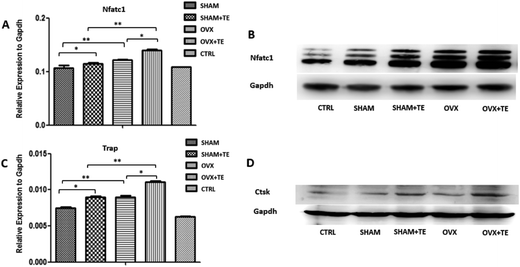
We further investigated the effect of CM or BMMSC-exos on the Raw264.7 cells (Fig. 5). The Raw264.7 cells were cultured with CM or DMEM containing different groups (SHAM, SHAM + TE, OVX, OVX + TE) of BMMSC-exos and 15 ng mL−1 sRANKL, and DMEM containing 15 ng mL−1 sRANKL was taken as the negative control (NC).After TRAP staining, TRAP + MNCs appeared in the Raw264.7 cells cultured in all types of CM or BMMSC-exos media without obvious morphological differences (Fig. 6C). As the exos/CM-free medium (NC group) induced the lowest number of TRAP + MNCs among all the groups, the CM and DMEM with BMMSC-exos appeared to increase the osteoclastogenic potential. The SHAM + TE, OVX, OVX + TE groups showed stronger osteoclastogenic potential than the SHAM group, as exhibited by the generation of significantly more TRAP + MNCs. The OVX + TE group induced the highest number of TRAP + MNCs among all groups (Fig. 6E). The levels of Nfatc1 and Ctsk mRNA and protein expression in the Raw264.7 cells were consistent with the TRAP + MNC counts (Fig. 7). The number of TRAP + MNCs was not significantly different in the Raw264.7 cells whether they were cultured with CM and exosomes from the same group (Fig. 6A).
Consistent with these observations, the CCK8 assay showed that the BMMSC-exos did not alter cell proliferation rates on 3 d or 7 d (Fig. 6B). These results indicate that the BMMSC-exos promote osteoclastogenesis by enhancing differentiation without affecting precursor proliferation.
Discussion
Osteoporosis is attracting more and more attention in dentistry. Oestrogen deficiency and tooth loss have been suggested as the main reasons for alveolar bone loss, and the number of elderly patients with missing teeth is increasing. Oestrogen deficiency, injury and mechanical unloading are expected to increase bone resorption and impair bone formation, which result in bone loss.27 Previous studies have shown the presence of osteoporosis in treatments such as orthodontic movement, implant restoration, periodontal treatment.28 The aim of this study was to investigate the potential role of BMMSC-exos in tooth loss and ovariectomy (OVX)-related alveolar bone deterioration. The results of this study demonstrated that BMMSC-exos could regulate the osteoclastic differentiation of Raw264.7 cells, and thus exosomal regulation might be a novel perspective to explain the mechanism of alveolar bone osteoporosis.An OVX and tooth loss rat model was established in this study to experimentally induce osteoporosis in the alveolar bone. In this model, 1 month after OVX/SHAM experiment, the molars were extracted from the maxillae to create mechanical unloading of the alveolar bone. Thus, bone loss was induced by both unloading and oestrogen deficiency. The trabecular bone of the distal femur and the alveolar bone significantly decreased in size 1 month after OVX as indicated by reduced BV/TV%. However, the bone mass of the alveolar bones did not decrease significantly in the OVX group compared to the SHAM group (ESI Fig. S1†). The difference could have been caused by the shape, mechanical loading, and embryological difference. One month after the OVX/SHAM experiment, the teeth were extracted from the right side of the maxillae. Hence, the mechanical loading of the alveolar bone during mastication may have disappeared, and thus bone loss was induced by both unloading and oestrogen deficiency. Tooth loss (SHAM + TE, OVX + TE groups) caused a decrease in bone height and bone mass, and oestrogen deficiency led to bone loss by enlarging marrow spaces and reducing bone thickness (Fig. 1). In addition, tooth loss and OVX both exerted effects on bone resorption (SHAM + TE, OVX, OVX + TE groups) by increasing the osteoclast number and promoting osteoclastogenesis-related gene and protein expressions (Fig. 2). Therefore, tooth loss could accelerate bone deterioration caused by oestrogen deficiency, i.e., tooth loss and oestrogen deficiency exert a synergistic effect on bone loss by improving osteoclastogenesis.
Osteoclasts are derived from haematopoietic progenitors within the monocyte/macrophage lineage that differentiate into mature osteoclasts in response to signals provided directly by osteoblast/marrow stromal cells through the influence of systemic hormones and factors produced within the bone marrow microenvironment.29–32 It has been demonstrated that osteoblast/marrow stromal cells and osteocytes are the major sources of signals and cytokines that are essential not only for the maintenance and formation of bone matrix but also for the simultaneous orchestration of processes involved in osteoclast differentiation and bone resorption.33 The proximity of marrow stromal cells and haematopoietic cells in the marrow suggests that the contact between mesenchymal and haematopoietic cells may signal important intracellular events that, in turn, can regulate the biological activities of these two cell lineages. In support of this idea, several studies have demonstrated that cell-to-cell interactions between osteoclast progenitors and stromal cells are mediated through surface molecules including adhesion molecules and cytokines, such as cadherin-6/2, macrophage-colony stimulating factor (MCSF), and RANK/RANKL/OPG.34,35 Furthermore, BMMSC-derived proteins can influence BMM-derived osteoclast differentiation. The culture medium of BMMSCs contains various cytokines that act as trophic mediators to regulate neighbouring cells.36,37
Recent studies have demonstrated that exosomes are important in the paracrine activity of stem cells.38 Exosomes have received increased attention for their role in the intercellular transfer of certain molecules. Recent reports have also unveiled that MSC-derived exosomes could epigenetically change the target cells.39 Exosomes can horizontally transfer functional proteins, mRNAs and miRNAs to neighbouring cells and serve as mediators of intercellular communication.40 In this study, we found that BMMSCs possessed and secreted exosomes into the culture medium (Fig. 4A–D) and thus identified a novel mode of communication between BMMSC and osteoclasts. Exosomes secreted by BMMSCs may encapsulate miRNAs and proteins and subsequently get transferred and fused to pre-osteoclasts to regulate their function. To investigate whether this fusion entails specific binding or is a result of random combination, we co-cultured BMMSCs with BMMSC-exos (Fig. 4E). The results showed that the BMMSC-exos were more likely to be taken by Raw264.7 cells than BMMSCs. These data indicated that exosomes from BMMSCs were specifically taken by the target cells such as Raw264.7. On the other hand, we barely observed BMMSC-exos uptake by the BMMSCs themselves (Fig. 4E), suggesting that the exosomes were more likely to be taken up by other cell types rather than by homologous cells. In addition, the exosomes were not only incorporated into pre-osteoclast cell line Raw264.7 cells but also increased the osteoclastogenesis of pre-osteoclast cells and enhanced their osteoclastic differentiation. The number of TRAP + MNCs in exos-free medium was the lowest among all the groups, indicating that the contents of the BMMSC-exos may enhance osteoclastic differentiation. Moreover, the number of osteoclasts after incubation of Raw264.7 cells in medium containing exosomes from the SHAM + TE, OVX, OVX + TE groups was higher than that after incubation with the SHAM group (Fig. 6). Meanwhile, the osteoclastogenesis-related gene expression was consistent with TRAP + MNCs count (Fig. 6). These data indicated that BMMSC-exos may containing protein/mRNA/miRNAs that can improve osteoclastogenesis of pre-osteoclasts, and thus osteoporosis caused by oestrogen deficiency and tooth loss may partly be caused by exosomes delivery. So BMMSC-exos may be used in a drug delivery system like other nanoparticles.41–43 This may be an excellent theranostic platform for BONJ (bisphos-phonate related osteonecrosis of the jaw), orthodontic treatment and other cases need improved osteoclastogenesis.
Proteins such as inflammatory factors and cytokines may be transported in exosomes to regulate the target cells.23,44 Recently, it has been reported that exosomes bind to cells through receptor–ligand interactions.45 Sun et al.46 have identified a pair of key proteins ephrinA2/EphA2 that are involved in the binding of osteoclast-derived exosomes to osteoblasts, and this interaction suppresses osteoblast functions by releasing miR-214. However, the receptor–ligand interactions between BMMSC-exos and osteoclasts remain unclear and need more research.
Conclusions
This study demonstrated that tooth loss could accelerate OVX-related alveolar bone loss in rats, and BMMSC-derived exosomes were involved in accelerated osteoclastogenesis during alveolar bone deterioration partly because of released exosomes, which could activate the osteoclastic differentiation of Raw264.7 cells via the upregulation of Nfatc1, Trap and Ctsk expression in vitro. Our findings extend our current knowledge of how BMMSCs regulate the differentiation of osteoclasts and provide a potential target for preventing alveolar bone loss in osteoporosis patients in the clinic.Acknowledgements
This study was financially supported by grants from the National Natural Science Foundation of China (No. 81271110), Natural Science and Technology Support Program (No. 2014BAI04B07) and Fundamental Research Funds for the Central Universities of China (No. 20152957).References
- J. A. Kanis, E. V. McCloskey, H. Johansson, C. Cooper, R. Rizzoli, J. Y. Reginster, C. Scientific Advisory Board of the European Society for, O. Economic Aspects of, Osteoarthritis and F. the Committee of Scientific Advisors of the International Osteoporosis, Osteoporosis Int., 2013, 24, 23–57 CrossRef CAS PubMed.
- C. F. Streckfus, R. B. Johnson, T. Nick, A. Tsao and M. Tucci, J. Gerontol., Ser. A, 1997, 52, M343–M351 CrossRef CAS.
- Y. Takaishi, S. Arita, M. Honda, T. Sugishita, A. Kamada, T. Ikeo, T. Miki and T. Fujita, Adv. Ther., 2013, 30, 487–502 CrossRef PubMed.
- C. F. Hildebolt, Dentomaxillofac. Radiol. Suppl., 1997, 26, 3–15 CrossRef CAS PubMed.
- A. Yoshihara, Y. Seida, N. Hanada and H. Miyazaki, J. Clin. Periodontol., 2004, 31, 680–684 CrossRef PubMed.
- M. Tezal, J. Wactawski-Wende, S. G. Grossi, J. Dmochowski and R. J. Genco, J. Periodontol., 2005, 76, 1123–1128 CrossRef PubMed.
- U. H. Lerner, J. Dent. Res., 2006, 85, 596–607 CrossRef CAS PubMed.
- K. Nicopoulou-Karayianni, P. Tzoutzoukos, A. Mitsea, A. Karayiannis, K. Tsiklakis, R. Jacobs, C. Lindh, P. van der Stelt, P. Allen, J. Graham, K. Horner, H. Devlin, S. Pavitt and J. Yuan, J. Clin. Periodontol., 2009, 36, 190–197 CrossRef PubMed.
- S. H. Tella and J. C. Gallagher, J. Steroid Biochem. Mol. Biol., 2014, 142, 155–170 CrossRef CAS PubMed.
- L. Xia, Z. Yin, L. Mao, X. Wang, J. Liu, X. Jiang, Z. Zhang, K. Lin, J. Chang and B. Fang, Sci. Rep., 2016, 6, 22005 CrossRef CAS PubMed.
- J. R. Lavoie and M. Rosu-Myles, Biochimie, 2013, 95, 2212–2221 CrossRef CAS PubMed.
- H. Xin, Y. Li, B. Buller, M. Katakowski, Y. Zhang, X. Wang, X. Shang, Z. G. Zhang and M. Chopp, Stem Cells, 2012, 30, 1556–1564 CrossRef CAS PubMed.
- M. Yanez-Mo, P. R. Siljander, Z. Andreu, A. B. Zavec, F. E. Borras, E. I. Buzas, K. Buzas, E. Casal, F. Cappello, J. Carvalho, E. Colas, A. Cordeiro-da Silva, S. Fais, J. M. Falcon-Perez, I. M. Ghobrial, B. Giebel, M. Gimona, M. Graner, I. Gursel, M. Gursel, N. H. Heegaard, A. Hendrix, P. Kierulf, K. Kokubun, M. Kosanovic, V. Kralj-Iglic, E. M. Kramer-Albers, S. Laitinen, C. Lasser, T. Lener, E. Ligeti, A. Line, G. Lipps, A. Llorente, J. Lotvall, M. Mancek-Keber, A. Marcilla, M. Mittelbrunn, I. Nazarenko, E. N. Nolte-'t Hoen, T. A. Nyman, L. O'Driscoll, M. Olivan, C. Oliveira, E. Pallinger, H. A. Del Portillo, J. Reventos, M. Rigau, E. Rohde, M. Sammar, F. Sanchez-Madrid, N. Santarem, K. Schallmoser, M. S. Ostenfeld, W. Stoorvogel, R. Stukelj, S. G. Van der Grein, M. H. Vasconcelos, M. H. Wauben and O. De Wever, J. Extracell. Vesicles, 2015, 4, 27066 CrossRef PubMed.
- M. Mittelbrunn and F. Sanchez-Madrid, Nat. Rev. Mol. Cell Biol., 2012, 13, 328–335 CAS.
- R. C. Lai, F. Arslan, M. M. Lee, N. S. Sze, A. Choo, T. S. Chen, M. Salto-Tellez, L. Timmers, C. N. Lee, R. M. El Oakley, G. Pasterkamp, D. P. de Kleijn and S. K. Lim, Stem Cell Res., 2010, 4, 214–222 CrossRef CAS PubMed.
- J. Wactawski-Wende, S. G. Grossi, M. Trevisan, R. J. Genco, M. Tezal, R. G. Dunford, A. W. Ho, E. Hausmann and M. M. Hreshchyshyn, J. Periodontol., 1996, 67, 1076–1084 CrossRef CAS PubMed.
- Q. G. Dai, P. Zhang, Y. Q. Wu, X. H. Ma, J. Pang, L. Y. Jiang and B. Fang, Oral Dis., 2014, 20, 514–520 CrossRef PubMed.
- H. C. Wang, W. P. Jiang, Z. H. Sima and T. J. Li, Oral Dis., 2015, 21, 170–177 CrossRef PubMed.
- H. C. Wang and T. J. Li, Oral Dis., 2013, 19, 162–168 CrossRef PubMed.
- Y. Kawamichi, C. H. Cui, M. Toyoda, H. Makino, A. Horie, Y. Takahashi, K. Matsumoto, H. Saito, H. Ohta, K. Saito and A. Umezawa, J. Cell. Physiol., 2010, 223, 695–702 CAS.
- Y. Huang, Z. Miao, Y. Hu, Y. Yuan, Y. Zhou, L. Wei, K. Zhao, Q. Guo and N. Lu, Oncotarget, 2017, 8, 883 Search PubMed.
- S. Dai, T. Wan, B. Wang, X. Zhou, F. Xiu, T. Chen, Y. Wu and X. Cao, Clin. Cancer Res., 2005, 11, 7554–7563 CrossRef CAS PubMed.
- W. Gao, H. Liu, J. Yuan, C. Wu, D. Huang, Y. Ma, J. Zhu, L. Ma, J. Guo, H. Shi, Y. Zou and J. Ge, J. Cell. Mol. Med., 2016, 20, 2318 CrossRef CAS PubMed.
- Y. Wu, W. Deng and D. J. Klinke 2nd, Analyst, 2015, 140, 6631–6642 RSC.
- F. J. Verweij, M. A. van Eijndhoven, E. S. Hopmans, T. Vendrig, T. Wurdinger, E. Cahir-McFarland, E. Kieff, D. Geerts, R. van der Kant, J. Neefjes, J. M. Middeldorp and D. M. Pegtel, EMBO J., 2011, 30, 2115–2129 CrossRef CAS PubMed.
- A. N. Kapustin, M. L. Chatrou, I. Drozdov, Y. Zheng, S. M. Davidson, D. Soong, M. Furmanik, P. Sanchis, R. T. De Rosales, D. Alvarez-Hernandez, R. Shroff, X. Yin, K. Muller, J. N. Skepper, M. Mayr, C. P. Reutelingsperger, A. Chester, S. Bertazzo, L. J. Schurgers and C. M. Shanahan, Circ. Res., 2015, 116, 1312–1323 CrossRef CAS PubMed.
- P. Garnero, E. Sornay-Rendu, M. C. Chapuy and P. D. Delmas, J. Bone Miner. Res., 1996, 11, 337–349 CrossRef CAS PubMed.
- S. Ejiri, M. Tanaka, N. Watanabe, R. B. Anwar, E. Yamashita, K. Yamada and M. Ikegame, J. Bone Miner. Metab., 2008, 26, 409–415 CrossRef CAS PubMed.
- P. Ash, J. F. Loutit and K. M. Townsend, Nature, 1980, 283, 669–670 CrossRef CAS PubMed.
- E. Kelemen, Nature, 1986, 323, 743 CrossRef CAS PubMed.
- B. A. Scheven, J. W. Visser and P. J. Nijweide, Nature, 1986, 321, 79–81 CrossRef CAS PubMed.
- N. Udagawa, N. Takahashi, T. Akatsu, H. Tanaka, T. Sasaki, T. Nishihara, T. Koga, T. J. Martin and T. Suda, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 7260–7264 CrossRef CAS.
- T. Nakashima, M. Hayashi, T. Fukunaga, K. Kurata, M. Oh-Hora, J. Q. Feng, L. F. Bonewald, T. Kodama, A. Wutz, E. F. Wagner, J. M. Penninger and H. Takayanagi, Nat. Med., 2011, 17, 1231–1234 CrossRef CAS PubMed.
- G. Mbalaviele, R. Nishimura, A. Myoi, M. Niewolna, S. V. Reddy, D. Chen, J. Feng, D. Roodman, G. R. Mundy and T. Yoneda, J. Cell Biol., 1998, 141, 1467–1476 CrossRef CAS PubMed.
- D. L. Lacey, E. Timms, H. L. Tan, M. J. Kelley, C. R. Dunstan, T. Burgess, R. Elliott, A. Colombero, G. Elliott, S. Scully, H. Hsu, J. Sullivan, N. Hawkins, E. Davy, C. Capparelli, A. Eli, Y. X. Qian, S. Kaufman, I. Sarosi, V. Shalhoub, G. Senaldi, J. Guo, J. Delaney and W. J. Boyle, Cell, 1998, 93, 165–176 CrossRef CAS PubMed.
- N. Nakano, Y. Nakai, T. B. Seo, Y. Yamada, T. Ohno, A. Yamanaka, Y. Nagai, M. Fukushima, Y. Suzuki, T. Nakatani and C. Ide, Neurosci. Lett., 2010, 483, 57–61 CrossRef CAS PubMed.
- H. Kupcova Skalnikova, Biochimie, 2013, 95, 2196–2211 CrossRef CAS PubMed.
- X. Liang, Y. Ding, Y. Zhang, H. F. Tse and Q. Lian, Cell Transplant., 2014, 23, 1045–1059 Search PubMed.
- G. Camussi, M. C. Deregibus, S. Bruno, V. Cantaluppi and L. Biancone, Kidney Int., 2010, 78, 838–848 CrossRef CAS PubMed.
- J. Li, K. Liu, Y. Liu, Y. Xu, F. Zhang, H. Yang, J. Liu, T. Pan, J. Chen, M. Wu, X. Zhou and Z. Yuan, Nat. Immunol., 2013, 14, 793–803 CrossRef CAS PubMed.
- G. Jalani, R. Naccache, D. H. Rosenzweig, L. Haglund, F. Vetrone and M. Cerruti, J. Am. Chem. Soc., 2016, 138, 1078–1083 CrossRef CAS PubMed.
- J. S. Suk, Q. Xu, N. Kim, J. Hanes and L. M. Ensign, Adv. Drug Delivery Rev., 2016, 99, 28–51 CrossRef CAS PubMed.
- N. Kamaly, B. Yameen, J. Wu and O. C. Farokhzad, Chem. Rev., 2016, 116, 2602–2663 CrossRef CAS PubMed.
- A. M. Roccaro, A. Sacco, P. Maiso, A. K. Azab, Y. T. Tai, M. Reagan, F. Azab, L. M. Flores, F. Campigotto, E. Weller, K. C. Anderson, D. T. Scadden and I. M. Ghobrial, J. Clin. Invest., 2013, 123, 1542–1555 CAS.
- G. Raposo, H. W. Nijman, W. Stoorvogel, R. Liejendekker, C. V. Harding, C. J. Melief and H. J. Geuze, J. Exp. Med., 1996, 183, 1161–1172 CrossRef CAS PubMed.
- W. Sun, C. Zhao, Y. Li, L. Wang, G. Nie, J. Peng, A. Wang, P. Zhang, W. Tian, Q. Li, J. Song, C. Wang, X. Xu, Y. Tian, D. Zhao, Z. Xu, G. Zhong, B. Han, S. Ling, Y. Z. Chang and Y. Li, Cell Discovery, 2016, 2, 16015 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra27931g |
| This journal is © The Royal Society of Chemistry 2017 |