Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Blocking celiac antigenicity of the glutamine-rich gliadin 33-mer peptide by microbial transglutaminase†
Lin Zhouabc,
Yujie Wuabc,
Youfei Chengabc,
Jie Wangb,
Jun Luabc,
Jinyan Gaoab,
Juanli Yuanad and
Hongbing Chen *ab
*ab
aState Key Laboratory of Food Science and Technology, Nanchang University, 235 Nanjing Dong Road, Nanchang 330047, China. E-mail: chenhongbing@ncu.edu.cn; Fax: +86 791 88333708; Tel: +86 791 88334552
bSchool of Food Science and Technology, Nanchang University, Nanchang 330031, China
cSino-German Joint Research Institute, Nanchang University, Nanchang 330047, China
dCollege of Pharmaceutical Sciences, Nanchang University, Nanchang 330031, China
First published on 3rd March 2017
Abstract
Celiac disease (CD) is a T cell-mediated enteropathy of the small intestine and caused by the ingestion of wheat gluten and related prolamins in barley and rye. However, there is no effective therapy to alleviate symptoms of celiac disease except for a life-long gluten-free diet. Recent studies showed that modification by microbial transglutaminase (mTG) could reduce the gliadin-specific immune response. In the present study, different acyl-acceptor substrates in combination with mTG were used to modify the model 33-mer peptide (LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF), which is a particular celiac toxic α-gliadin peptide. RP-HPLC and LC-ESI-MS were performed to test the extent of the modifications. R5 ELISA and G12 ELISA were used to analyze the antigenicity of the modified peptide. The shifts of retention time and molecular weight showed great modification of 33-mer peptide after 2 h of incubation with mTG. When acyl-acceptor substrates were crosslinked with 33-mer peptide, the antigenicity of modified peptide forms was decreased compared to its initial level. In summary, it is demonstrated that mTG is active on a variety of chemically acyl-acceptor substrates. Transamidation by mTG with an appropriate amine donor can be used to block the antigenicity of gliadin peptide related to celiac disease. These findings highlight a potential strategy to prevent cereal toxicity in celiac disease.
1. Introduction
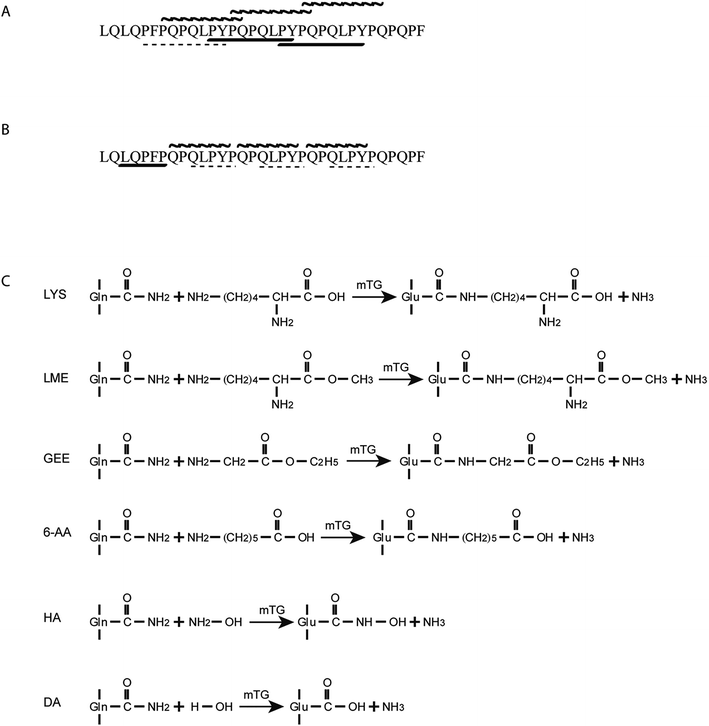
Celiac disease (CD) is an intestinal disorder developed in genetically susceptible individuals and caused by the ingestion of wheat gluten and related prolamins present in barley and rye.1 CD is strongly related to the HLA locus, specifically HLA-DQ2 (A1*0501–B1*0201) and/or HLA-DQ8 (A1*0301–B1*0302).2 The gluten epitopes can induce CD after deamidation by tissue transglutaminase (TG2), which is a crucial step in CD pathogenesis.3,4 The deamidated gluten epitopes have higher affinity to bind to HLA-DQ2 and HLA-DQ8 molecules, leading to stronger T-cell responses.5 Furthermore, the primary toxic components of wheat gluten are gliadins, which are glutamine-rich and proline-rich proteins with resistance to digestion.6,7 Moreover, there exists a particular toxic α-gliadin peptide, LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF, which consists of 33 amino acids including 10 glutamine residues. The 33-mer peptide is naturally formed by digestion with gastric and pancreatic enzymes and is resistant to further degradation by enzymes of the brush-border membrane (BBM).6 The peptide contains three overlapping immunodominant epitopes (Fig. 1A), PFPQPQLYP (one copy), PYPQPQLPY (two copies) and PQPQLPYPQ (three copies) in that their deamidated forms can stimulate T cells to induce adaptive immunity.6,8 However, there is no effective therapy to treat CD except for a life-long gluten-free diet, which is arduous to persist with because of high prices, limited variety and poor sensory properties.9–11In this sense, enzymatic modification of gluten has been utilized to decrease the CD-specific immunological activities without affecting other principal properties. Gluten proteins were modified by binding methionine to the amino lateral groups of glutamine residues with chymotrypsin, which reduced the induction of celiac IgA.12 Furthermore, food-grade microbial transglutaminase (mTG) has been used to reduce the immunological activity of gluten proteins, which typically catalyses pH-dependent transamidation to form an ε-(γ-glutamyl)-lysine bond between glutamine residues (acyl-donor substrate) and lysine residues (acyl-acceptor substrate).13 Detoxification was attributed to the selective transamidation of glutamine residues, which blocked the deamidation process by mTG and tTG, which is critical for the development of CD. Although mTG can also catalyse a hydrolysis reaction between glutamine residues and H2O, it has a lower deamidation preference than human tissue transglutaminase (tTG).14 It was found that the reactivity between gluten proteins and R5 mAb was reduced by 42% using R5 ELISA when gluten proteins were modified by mTG and lysine.15 Moreover, previous studies show that the transamidation by mTG/tTG with appropriate acyl-acceptor substrates (lysine or lysine methyl ester) can be used to prevent the immunological activity of gluten proteins, while as it was not sufficient in abolishing the gluten activity.16–19 In addition, recent study highlighted the potential of mTG transamidation to block non-immune mechanisms related to gluten toxicity.20
On this basis, it is of interest to study other compounds that might substitute for lysine as acyl-acceptor substrates with the aim of diversifying biocatalytic applications of mTG in the modification of gluten and the breadth of utility as a green biocatalyst in the food industry. In this experiment, it was the aim to investigate the transamidation of the 33-mer peptide using mTG and different acyl-acceptor substrates, namely L-lysine (LYS), L-lysine methyl ester (LME), glycine ethyl ester hydrochloride (GEE), 6-aminocaproic acid (6-AA) and hydroxylamine (HA), which have different molecular weights (Fig. 1C). Furthermore, in vitro digestions of the 33-mer peptide and its modified forms were executed by trypsin and α-chymotrypsin. Reversed-phase high-performance liquid chromatography (RP-HPLC) and liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) were performed to test the extent of modifications. R5 ELISA and G12 ELISA were used to analyze the possible changes of antigenicity after the transamidation of mTG and digestion of enzyme. R5 ELISA is suggested by Codex Alimentarius for detection and quantification of gluten in food and G12 ELISA is specific for the detection of the 33-mer peptide.
Our hypothesis was that the acyl-acceptor substrates in the experiment can be crosslinked efficiently to glutamine residues of toxic epitopes, resulting in a low level of CD toxic epitopes. Moreover, the isopeptide bonds of modified 33-mer peptide were resistant to the digestion of digestive enzymes, which indicated that all of the transamidated forms would be stable.
2. Materials and methods
2.1 Synthetic peptides and chemicals
The 33-mer peptide was synthesized by GL Biochem Ltd (Shanghai, China). The purity (≥95%) and structure were assessed by reversed-phase high performance liquid chromatography (RP-HPLC) and liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS). mTG was donated by Jiangsu Yiming Biological Products Co., Ltd. (Jiangsu, China) with a declared activity of 1000 units per g. LYS (>98%), GEE (>98%) and 6-AA (>98.5%) were bought from Sangon Biotech (Shanghai, China). LME (98%) and HA (98%) were from Aladdin (Shanghai, China) and Sigma-Aldrich, respectively. Pepsin (≥2500 units per mg protein), trypsin (≥7500 BAEE units per mg), α-chymotrypsin (Type II, lyophilized powder, ≥40 units per mg protein) and bile salt (sodium glycodeoxycholate and sodium taurocholate) were purchased from Sigma-Aldrich. All reagents used in RP-HPLC analysis were of HPLC grade. Other chemicals were of analytical grade.2.2 Determination of mTG activity
An amount of 0.5 g enzyme powder was dissolved in 25 mL 0.01 mol L−1 phosphate buffer, and the mixture was centrifuged at 5000g to remove precipitate.21 The supernatant was dialyzed with a molecular weight cut-off of 3500 Da to remove other components. The resulting enzyme solution was stored at 4 °C, and directly used for subsequent reaction. Enzyme activity was determined by a colorimetric procedure using Z-Gln-Gly as the substrate.22 Z-Gln-Gly reacted with hydroxylamine hydrochloride in the presence of mTG and then the products were combined with ferric ion in acidic conditions. The enzymatic activity was evaluated by UV absorbance at 525 nm. Meanwhile, the calibration curve is established on the basis of the concentration of L-glutamic acid γ-monohydroxamate as standard. One enzyme unit (U) is defined as the formation of 1 μmol L-glutamic acid γ-monohydroxamate per min at 37 °C.Based on the standard curve, the calculated enzyme activity was calculated as about 10 U mL−1.
2.3 Modification by mTG
In the experiments, five transamidation mixtures and one deamidation mixture were performed. Regarding the transamidation, each acyl-acceptor substrate in Table 1 was used in five different transamidation mixtures, respectively. The reaction solution contained 1 mg of 33-mer peptide, 0.5 U mTG and 0.05 mmol acyl-acceptor substrate in a final volume of 1 mL, as well as the control groups were prepared using 1 mg of 33-mer peptide, 0.05 mmol acyl-acceptor substrate and inactivated mTG. Regarding the deamidation reaction, the mixture was performed with 1 mg of 33-mer peptide, 0.5 U mTG and without the acyl-acceptor substrates. The assay was carried out at 50 °C for 2 h, followed by stopping the enzymatic reaction by heating at 85 °C for 10 min.| Acyl-acceptor substrates | Structure | MW | RTb (min) | Percentagec (%) |
|---|---|---|---|---|
| a LYS: L-lysine; LME: L-lysine methyl ester; GEE: glycine ethyl ester; 6-AA: 6-aminocaproic acid; HA: hydroxylamine; MW: molecular weight.b Retention time (RT) of modified peptide with three acyl-acceptor molecules.c Percentage of modified peptide with three acyl-acceptor molecules. Values are expressed as averages ± standard deviation. | ||||
| 1 LYS | 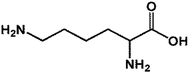 |
146.19 | 14.64 | 90.26 ± 4.43 |
| 2 LME | 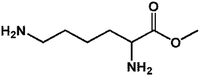 |
160.21 | 15.26 | 47.51 ± 2.47 |
| 3 GEE | 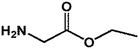 |
103.12 | 20.10 | 42.12 ± 0.33 |
| 4 6-AA | 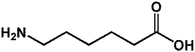 |
131.17 | 18.50 | 23.93 ± 0.49 |
| 5 HA | 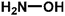 |
33.02 | 16.31 | 36.51 ± 0.31 |
2.4 RP-HPLC analysis and LC-ESI-MS analysis
2.5 In vitro digestions of the 33-mer peptide and its modified forms
The in vitro digestions of each peptide were performed according to previous studies with some modifications.23 For gastric digestions, porcine pepsin was dissolved in simulated gastric fluid with 35 mM NaCl at pH 2.0. Gastric digestions were performed for 60 min at 37 °C with the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (enzyme to peptide, w/w). Aliquots were withdrawn at 60 min. The digestion reactions were stopped by adjusting the pH to 7.0 with NaOH. Duodenal digestion was conducted using the 60 min gastric digests as the starting material. Moreover, the reaction mixtures were supplemented with bile salt, bis–tris, CaCl2 and enzymes (trypsin and α-chymotrypsin). The final composition was 6.15 mM of bile salt, 20.3 mM bis–tris, 7.6 mM CaCl2, trypsin (1
50 (enzyme to peptide, w/w). Aliquots were withdrawn at 60 min. The digestion reactions were stopped by adjusting the pH to 7.0 with NaOH. Duodenal digestion was conducted using the 60 min gastric digests as the starting material. Moreover, the reaction mixtures were supplemented with bile salt, bis–tris, CaCl2 and enzymes (trypsin and α-chymotrypsin). The final composition was 6.15 mM of bile salt, 20.3 mM bis–tris, 7.6 mM CaCl2, trypsin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, enzyme to protein, w/w) and α-chymotrypsin (1
50, enzyme to protein, w/w) and α-chymotrypsin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, enzyme to protein, w/w). The reactions were incubated at 37 °C for 120 min, followed by stopping by heating at 100 °C for 10 min.
200, enzyme to protein, w/w). The reactions were incubated at 37 °C for 120 min, followed by stopping by heating at 100 °C for 10 min.
2.6 The competitive enzyme-linked immunosorbent assay (ELISA)
In the study, both R5 ELISA and G12 ELISA were used to confirm the result of transamidation by mTG. The antigenicity of the 33-mer peptide was estimated by RIDASCREEN® Gliadin Competitive kit (R-Biopharm, Darmstadt, Germany) and GlutenTox®ELISA Competitive (Biomedal, Seville, Spain). For RIDASCREEN® Gliadin Competitive Elisa (R5 ELISA), R5 monoclonal antibody (mAb) was used to recognize potential toxic peptides sequences of gliadins, such as the sequence of QQPFP and LQPFP. GlutenTox®ELISA Competitive (G12 ELISA) is an immunosorbent kit for the detection and quantification of the fraction of gliadin, secalin and barley, based on G12 mAb. G12 ELISA is specific for 33-mer peptide, which can detect QPQLPY, QPQLPF and so on.In order to adjust the absorbance values into the range of the standard curve, samples were appropriately diluted in the buffer solution. The assays were carried out according to the manufacturer's instructions. The absorbance was then measured at 450 nm using a microplate reader (Bio-Rad, USA). The standard curve line of R5 ELISA was done using the cubic spline function and the standard curve line of G12 ELISA was performed by second-order polynomial regression.
2.7 Statistical analysis
Statistical analysis was performed using Lab Solution, ORIGIN 8.0 and RIDASOFTWin. The data is presented as mean ± standard deviation (S.D.).3. Results
3.1 Modification of the 33-mer peptide by mTG
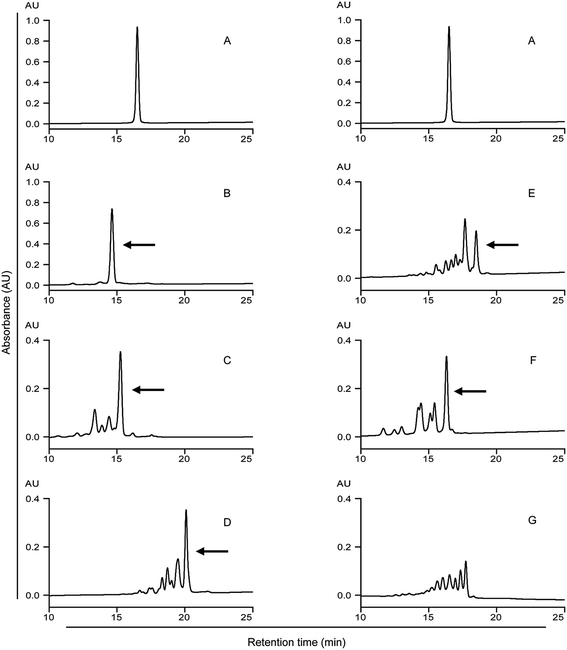 | ||
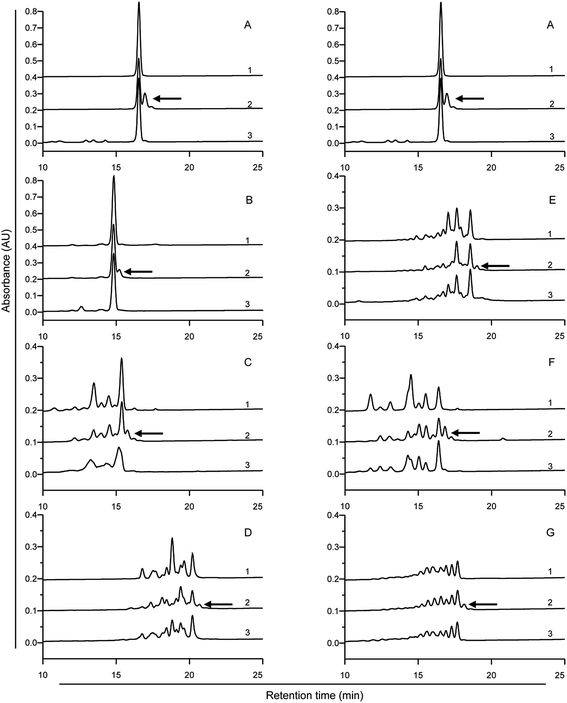
| Fig. 2 HPLC analysis of the 33-mer peptide and its modified forms. Detecting wavelength is 220 nm. (A) Native 33-mer peptide as control; (B) LYS/mTG; (C) LME/mTG; (D) GEE/mTG; (E) 6-AA/mTG; (F) HA/mTG; (G) 1 mg of 33-mer peptide and 0.5 U mTG without acyl-acceptor substrates. Arrows indicate the 33-mer peptide crosslinked with three acyl-acceptor substrate molecules, which percentage is given in the Table 1. LYS: L-lysine; LME: L-lysine methyl ester; GEE: glycine ethyl ester; 6-AA: 6-aminocaproic acid; HA: hydroxylamine. | ||
The native 33-mer peptide is given in Fig. 2A. Fig. 2B to F represent the 33-mer peptide treated with mTG and various acyl-acceptor substrates, while Fig. 2G shows the peptide treated without acyl-acceptor substrates. It was shown that new peaks were present after the enzyme catalysis, suggesting that retention time was different between the native peptide and its modified forms. Moreover, it was indicated that the retention time was reduced when the 33-mer peptide was crosslinked with LYS (Fig. 2B), LME (Fig. 2C), and HA (Fig. 2F). Meanwhile, the crosslinking of GEE (Fig. 2D) and 6-AA (Fig. 2E) had increased the retention time. The proportion of 33-mer peptide crosslinking with three LYS molecules can reach 90.26% (Table 1), while the ratio of 33-mer peptide crosslinking with three LME molecules, three GEE molecules, three 6-AA molecules and three HA molecules were 47.51, 42.12, 23.93 and 36.51%, respectively. When the reaction mixture involved only mTG and 33-mer peptide, several new peaks appeared due to the deamidating activity of mTG (Fig. 2G). This experiment demonstrates the deamidation activity on the 33-mer peptide by mTG as such (Fig. 2G) and the mTG plus acyl-acceptor substrate-specific modifications in the other figures.
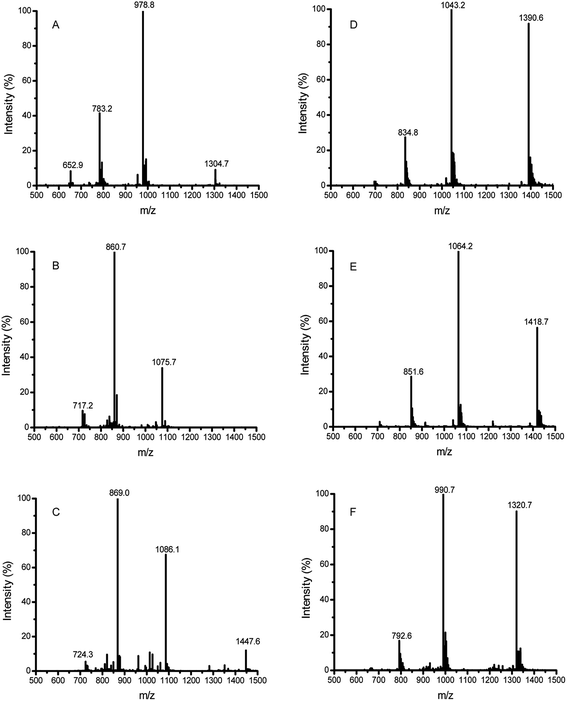
In the LC-ESI-MS (Fig. 3A, Table 2), the m/z value of the 33-mer peptide was 978.8 (4+), indicating that the measured molecular weight (MMW) was 3911.2 daltons. When the LYS molecules were crosslinked to the 33-mer peptide, a strong signal at m/z value 860.7 (5+) was generated (Fig. 3B, Table 2), implying that the detected MW for the new product was 4298.5 daltons. By contrast, there was a shift of 387 daltons from the m/z value 978.8 (4+) (MMW = 3911.2, theoretical average molecular weight (TAMW) = 3911.4) to m/z value 860.7 (5+) (TAMW = 4298.9 daltons), indicating the successful addition of three LYS molecules to the 33-mer peptide. However, the content of 33-mer peptide crosslinking with a single LYS molecule or two LYS molecules was very low. Moreover, the native 33-mer peptide crosslinking with three LME molecules generated a strong signal at m/z value 869.0 (5+) (Fig. 3C, Table 2). The crosslinked 33-mer peptide with one LME molecule (m/z value 1014.9 (4+)) or two LME molecules (m/z value 1050.5 (4+)) were also produced, but they had much lower abundance than the 33-mer peptide with three LME molecules (data not shown). Similarly, the 33-mer peptide crosslinking with three GEE molecules produced a strong signal at m/z value 1043.2 (4+) (Fig. 3D, Table 2), while the m/z values 1000.2 (4+) and 1021.8 (4+) represented the crosslinking of the 33-mer peptide with one GEE molecule and two GEE molecules and the content of two products was low (data not shown). Regarding the 33-mer peptide with three 6-AA molecules, m/z value 1064.2 (4+) was observed (Fig. 3E, Table 2), while the signals of crosslinking with one 6-AA molecule or two 6-AA molecules were very weak (data not shown). In addition, the 33-mer peptide crosslinked with three HA molecules produced the m/z value 990.7 (4+) (Fig. 3F, Table 2), whereas the 33-mer peptide with one HA or two HA molecules had weak signals (data not shown).
| Sample | Precursor ion (m/z) | Charge | MMW | TAMW | No. of added |
|---|---|---|---|---|---|
| a Percentage < 5%; MMW, measured molecular weight; TAMW, theoretical average molecular weight; L-lysine (LYS); L-lysine methyl ester (LME); glycine ethyl ester (GEE); 6-aminocaproic acid (6-AA); hydroxylamine (HA). | |||||
| 33-mer | 978.8 | 4+ | 3911.2 | 3911.4 | |
| LYS | 808.9 | 5+ | 4039.5 | 4040.6 | 1a |
| 834.5 | 5+ | 4167.5 | 4169.8 | 2a | |
| 860.7 | 5+ | 4298.5 | 4298.9 | 3 | |
| LME | 1014.9 | 4+ | 4055.6 | 4054.6 | 1 |
| 1050.5 | 4+ | 4198.0 | 4197.8 | 2 | |
| 869.0 | 5+ | 4340.0 | 4341.0 | 3 | |
| GEE | 1000.2 | 4+ | 3996.8 | 3997.5 | 1 |
| 1021.8 | 4+ | 4083.2 | 4083.6 | 2 | |
| 1043.2 | 4+ | 4168.8 | 4169.7 | 3 | |
| 6-AA | 1007.6 | 4+ | 4026.4 | 4025.6 | 1a |
| 1035.9 | 4+ | 4139.6 | 4139.7 | 2 | |
| 1064.2 | 4+ | 4252.8 | 4253.9 | 3 | |
| HA | 1310.9 | 3+ | 3929.7 | 3927.5 | 1a |
| 986.8 | 4+ | 3943.2 | 3943.5 | 2a | |
| 990.7 | 4+ | 3958.8 | 3959.5 | 3 | |
Regarding the reaction mixture with only mTG and the 33-mer peptide, distinct m/z values were observed (ESI Fig. 1B–E†). According to the ESI Fig. 1B,† the m/z values 979.4 (4+) (MMW 3913.6) and 1305.7 (3+) (MMW 3914.1) arose, indicating a shift of 2–3 daltons from the native 33-mer peptide (MMW 3911.2). Therefore, it was concluded that 2–3 glutamine residues were deamidated to glutamic acid. Interestingly, incubation of 33-mer peptide with mTG yielded different products, such as LQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF (ESI Fig. 1C,† LQ from the native 33-mer peptide), QPFPQPQLPYPQPQLPYPQPQLPYPQPQPF (ESI Fig. 1D,† LQL from the native 33-mer peptide), and LQLQPFPQPQLPYPQPQLPYPQPQLPYPQP (ESI Fig. 1E,† QPF from the native 33-mer peptide).
Collecting the above results, all acyl-acceptor substrates in this work were active to crosslink with the 33-mer peptide by mTG and the 33-mer peptide has three crosslinking sites. Moreover, it demonstrates the deamidation activity and maybe degradation activity on the 33-mer peptide by mTG without acyl-acceptor substrates. However, it remains unclear which of the three glutamine residues were the modification sites and which site was modified first.
3.2 Modification of the 33-mer peptide by enzymatic digestion
To assess the digestibility of the modified peptides as produced after treatment with mTG plus the various acyl-acceptor substrates by pepsin, trypsin and chymotrypsin, we used HPLC to analyse the digested products. There were no significant differences in digestibility between the modified peptides and the native 33-mer peptide (Fig. 4). After 1 h of in vitro gastric digestion, some new peaks appeared. As indicated in the Fig. 4A2–G2, some peaks disappeared after 2 h of intestinal digestion. However, most of the initial peaks still remained. In summary, it is demonstrated that the enzymes from gastrointestinal tract failed to degrade the isopeptide.3.3 Inhibition of antigenicity by mTG
4. Discussion
In this study, the obtained results show that the gliadin 33-mer peptide can be transamidated with mTG combined with different acyl-acceptor substrates. These results were in agreement with the previous study that the chemical and steric distinction of acyl-acceptor substrates gave rise to the different properties and reactivity.26 The most extensive modification of the 33-mer occurred when enzyme treatment was given using 1 mg of 33-mer, 0.5 U mTG and 0.05 mmol LYS in a volume of 1 mL (Fig. 2B). That may be due to the reaction conditions being optimum for the reactivity of LYS. Interestingly, the crosslinking output level of the 33-mer peptide with three acyl-acceptor molecules was much higher than that of the 33-mer peptide with one acyl-acceptor molecular or two acyl-acceptor molecules in the same condition, demonstrating the 33-mer peptide has three transamidation sites. This work is in agreement with Mazzeo et al. that three Q sites were modified in the sequence of the 33-mer peptide (displayed in bold, LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF).20 Moreover, it has been shown that most of the modifications by mTG could be explained by mentioned patterns in the previous study that the Q is not a target of mTG in QP and QXXP sequences, while the Q is a target of mTG in the sequences QXP, QXX(FYWMLIV) and QXPF(YWMLIV).27 Actually, based on this mechanism, three Q modification sites were deduced in the 33-mer peptide, which are displayed in bold (33-mer: LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF). However, the occurrence of some other peaks in the HPLC spectrum remains unclear, except the peaks of the 33-mer peptide plus three acyl-acceptor substrates (Fig. 2). These may be due to the multifunction of mTG, including transamidation activity, deamidation activity and hydrolysis activity. Nevertheless, in view of our data, it has been shown that transamidation activity of mTG played a more important role compared to deamidation activity in the presence of acyl-acceptor substrates, which is consistent with the previous study that the transamidation rate was about 10 times higher than the deamidation rate in the presence of lysine.14Furthermore, it indicated that all these peptides were degraded slightly after 1 h of in vitro gastric digestion and 2 h of in vitro intestinal digestion, which is consistent with a previous study in which the 33-mer peptide appeared to be highly resistant to digestion, even when exposed to enzymes of the brush-border membrane (BBM).6 After 1 h of gastric digestion, some new peaks appeared because amino-terminal leucine residue was cut by pepsin.6 However, most of the initial peaks still existed. Actually, these results were the same as found earlier showing that digestive enzymes were ineffective to degrade the gluten epitopes.28 In summary, it has been demonstrated that the enzymes from the gastrointestinal tract failed to degrade the isopeptide, which confirms that acyl-acceptor substrates remained linked to the peptide after in vitro digestion by pepsin and subsequently by pancreatin.29
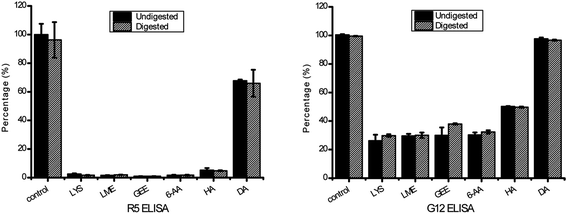
The transamidation activity of mTG greatly decreased the affinity of the 33-mer peptide binding to R5 mAb. The sequence of LQPFP in the N-terminus of the 33-mer peptide can be recognized by the R5 mAb, however, the sequence of QLPYP remains a weak reactivity to the antibody.24 The structural modification of three Q sites could be a reason for the reduction of R5 mAb recognition for the 33-mer peptide.24 Moreover, after being deamidated by mTG, the reactivity of the antibody declined to 67.70% of its initial level. This is in accordance with the low antibody binding observed in previous studies showing that the substitution of glutamine with glutamic acid in the epitopes may decrease the recognition of R5 mAb.3,24
Similarly, the transamidated forms had a low affinity to G12 mAb in comparison with the native 33-mer peptide. A previous study described that the G12 mAb strongly recognizes the sequence of QPQLPY, which is repeated three times in the 33-peptide.25 Therefore, the decreased antigenicity was presented because acyl-acceptor substrates were crosslinked to Q site, which changed the structure of 33-mer peptide. However, there was no obvious difference in the antigenicity between the untreated 33-mer peptide and the deamidated peptide, which was different from the previous study that the affinity of G12 mAb with the peptide QPQLPYPQP was much higher than for that of G12 mAb with the deamidated peptide QPELPYPQP.25 It may be due to incomplete deamidation of the 33-mer peptide in 2 h leading to the modified peptide have the similar immunological activity to the untreated peptide.
Additionally, the purity of 33-mer peptide crosslinked with three LYS molecules can reach to 90.26% (Table 1). When the 33-mer peptide was crosslinked with three LME molecules, three GEE molecules, three 6-AA molecules and three HA molecules, the purity was 47.51, 42.12, 23.93 and 36.51%, respectively. Nevertheless, there was no obvious difference between them in the binding activity to R5 and G12 mAb. It can be explained that 33-mer peptide crosslinked with three LYS molecules was more stable, while some of 33-mer peptide crosslinked with three LME molecules, three GEE molecules, three 6-AA molecules and three HA molecules was broken down because of the hydrolysis activity of mTG. For instance, amino-terminal LQL or carbon-terminal QPF was broken down by mTG from LQLQPF(PQPQLPY)3PQPQPF with three acyl-acceptor molecules as deamidation mixtures. Moreover, the Q sites were still crosslinked with these acyl-acceptor molecules. Therefore, most of transamidated forms had similar affinity R5 and G12 mAb.
Regarding the test of the digested product of the crosslinked 33-mer, it was concluded that there were only slight changes on the antigenicity according to the results of R5 ELISA and G12 ELISA and it is in agreement with the result of HPLC analysis that both native peptide and modified peptide could not be digested completely. Actually, it is also consistent with the observation that the L-lysine of ε-(y-L-glutamy1)-L-lysine was not released even after much prolonged incubation with the mucosal scraps.29 To summarize, it was suggested that mTG combined with acyl-acceptor substrates can modify the 33-mer peptide, resulting in decreasing its antigenicity.
In conclusion, it is suggested that a model 33-mer peptide related to CD can be modified in vitro by different acyl-acceptor substrates with mTG. After 2 h of transamidation, the antigenicity of the peptide decreased greatly compared to its initial level. The formation of isopeptide bonds probably prevents the presenting process by antigen presenting cells and the recognition by gluten-specific T cells. Moreover, this finding broadens the application of mTG to catalyse the gluten modification for reducing their immunogenicity in celiac disease. Additionally, in food processing, the crosslinking of various macromolecules to proteins strengthen food resistance to luminal protease, resulting in low digestibility and absorption. Nevertheless, the crosslinking of small acyl-acceptor molecules to proteins might exert only a weak impact on their digestibility and absorption, meanwhile inactivating their immunogenicity. Strikingly, the small acyl-acceptor molecules would be cost-effective for applications of the food industry, in comparison with the macromolecules. Overall, the results might provide a new sight to develop new types of food with better nutritional, structural characteristics and being safe for people suffering from celiac disease.
Conflict of interest
The authors declare that there are no conflicts of interest.Acknowledgements
The work was supported by the International Science & Technology Cooperation Program of China (No. 2013DFG31380), National High Technology Research and Development Program of China (863 Program; No. 2013AA102205), and the Research Program of State Key Laboratory of Food Science and Technology (No. SKLF-ZZA-201612).References
- J. F. Ludvigsson, D. A. Leffler, J. C. Bai, F. Biagi, A. Fasano, P. H. Green, M. Hadjivassiliou, K. Kaukinen, C. P. Kelly and J. N. Leonard, Gut, 2013, 62, 43–52 CrossRef PubMed.
- V. Abadie, L. M. Sollid, L. B. Barreiro and B. Jabri, Annu. Rev. Immunol., 2011, 29, 493–525 CrossRef CAS PubMed.
- C. Kristiansen, L. Madsen, L. Fugger, H. Scott, O. Norén, P. Roepstorff, K. E. Lundin, H. Sjöström and L. M. Sollid, Nat. Med., 1998, 4, 713–717 CrossRef.
- Y. van de Wal, Y. Kooy, P. van Veelen, S. Peña, L. Mearin, G. Papadopoulos and F. Koning, J. Immunol., 1998, 161, 1585–1588 CAS.
- S. Tollefsen, H. Arentz-Hansen, B. Fleckenstein, Ø. Molberg, M. Ráki, W. W. Kwok, G. Jung, K. E. Lundin and L. M. Sollid, J. Clin. Invest., 2006, 116, 2226 CAS.
- L. Shan, Ø. Molberg, I. Parrot, F. Hausch, F. Filiz, G. M. Gray, L. M. Sollid and C. Khosla, Science, 2002, 297, 2275–2279 CrossRef CAS PubMed.
- J. Van de Kamer and H. Weijers, Acta Paediatr., 1955, 44, 465–469 CrossRef CAS PubMed.
- S.-W. Qiao, E. Bergseng, Ø. Molberg, J. Xia, B. Fleckenstein, C. Khosla and L. M. Sollid, J. Immunol., 2004, 173, 1757–1762 CrossRef CAS.
- B. Meresse, G. Malamut and N. Cerf-Bensussan, Immunity, 2012, 36, 907–919 CrossRef CAS PubMed.
- V. Capriles and J. Arêas, Food Funct., 2016, 7, 1266–1272 CAS.
- M. Bustos, G. Perez and A. Leon, RSC Adv., 2015, 5, 30780–30792 RSC.
- F. Cabrera-Chávez, A. R. Islas-Rubio, O. Rouzaud-Sández, N. Sotelo-Cruz and A. M. C. de la Barca, J. Cereal Sci., 2010, 52, 310–313 CrossRef.
- J. Folk and S. I. Chung, Methods Enzymol., 1985, 113, 358–375 CAS.
- A. Heil, J. Ohsam, C. Büchold, R. Pasternack, K. Yokoyama, Y. Kumazawa and M. Hils, J. Cereal Sci., 2016, 70, 47–56 CrossRef CAS.
- N. G. Heredia-Sandoval, A. R. Islas-Rubio, F. Cabrera-Chávez and A. M. C. de la Barca, Food Funct., 2014, 5, 1813–1818 CAS.
- C. Gianfrani, R. A. Siciliano, A. M. Facchiano, A. Camarca, M. F. Mazzeo, S. Costantini, V. M. Salvati, F. Maurano, G. Mazzarella and G. Iaquinto, Gastroenterology, 2007, 133, 780–789 CrossRef CAS PubMed.
- L. Elli, L. Roncoroni, M. Hils, R. Pasternack, D. Barisani, C. Terrani, V. Vaira, S. Ferrero and M. T. Bardella, Hum. Immunol., 2012, 73, 992–997 CrossRef CAS PubMed.
- G. Mazzarella, V. M. Salvati, G. Iaquinto, R. Stefanile, F. Capobianco, D. Luongo, P. Bergamo, F. Maurano, N. Giardullo and B. Malamisura, Clin. Dev. Immunol., 2012, 2012, 329150 Search PubMed.
- E. Lombardi, P. Bergamo, F. Maurano, G. Bozzella, D. Luongo, G. Mazzarella, V. R. Aufiero, G. Iaquinto and M. Rossi, J. Leukocyte Biol., 2013, 93, 479–488 CrossRef CAS PubMed.
- M. F. Mazzeo, R. Bonavita, F. Maurano, P. Bergamo, R. A. Siciliano and M. Rossi, Biochim. Biophys. Acta, Gen. Subj., 2013, 1830, 5166–5174 CrossRef CAS PubMed.
- C.-H. Tang, X. Sun, S.-W. Yin and C.-Y. Ma, Food Res. Int., 2008, 41, 941–947 CrossRef CAS.
- J. Folk and P. Cole, Biochim. Biophys. Acta, Enzymol. Biol. Oxid., 1966, 122, 244–264 CrossRef CAS.
- L. Shan, S.-W. Qiao, H. Arentz-Hansen, Ø. Molberg, G. M. Gray, L. M. Sollid and C. Khosla, J. Proteome Res., 2005, 4, 1732–1741 CrossRef CAS PubMed.
- F. Kahlenberg, D. Sanchez, I. Lachmann, L. Tuckova, H. Tlaskalova, E. Méndez and T. Mothes, Eur. Food Res. Technol., 2006, 222, 78–82 CrossRef CAS.
- B. Morón, M. T. Bethune, I. Comino, H. Manyani, M. Ferragud, M. C. López, Á. Cebolla, C. Khosla and C. Sousa, PLoS One, 2008, 3, e2294 Search PubMed.
- M. T. Gundersen, J. W. Keillor and J. N. Pelletier, Appl. Microbiol. Biotechnol., 2014, 98, 219–230 CrossRef CAS PubMed.
- E. Dekking, P. Van Veelen, A. De Ru, E. Kooy-Winkelaar, T. Gröneveld, W. Nieuwenhuizen and F. Koning, J. Cereal Sci., 2008, 47, 339–346 CrossRef CAS.
- G. Janssen, C. Christis, Y. Kooy-Winkelaar, L. Edens, D. Smith, P. van Veelen and F. Koning, PLoS One, 2015, 10, e0128065 Search PubMed.
- K. Iwami and K. Yasumoto, J. Sci. Food Agric., 1986, 37, 495–503 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra27893k |
| This journal is © The Royal Society of Chemistry 2017 |