Open Access Article
Open Access ArticleBacterial community compositions of propylene oxide saponification wastewater treatment plants†
Xiangyu Fan‡
 a,
Ying Zhu‡b,
Pengfei Gua,
Yumei Lia,
Guiqing Xiaoc,
Dongxue Songa,
Yiwei Wanga,
Rong Hea,
Huajun Zheng*d and
Qiang Li
a,
Ying Zhu‡b,
Pengfei Gua,
Yumei Lia,
Guiqing Xiaoc,
Dongxue Songa,
Yiwei Wanga,
Rong Hea,
Huajun Zheng*d and
Qiang Li *a
*a
aSchool of Biological Science and Technology, University of Jinan, Jinan, China. E-mail: chm_liq@ujn.edu.cn; Fax: +86 53189736818; Tel: +86 53182767364
bNew Materials Research Institute of Shandong Academy of Sciences, Jinan, China
cCollege of Bioscience and Biotechnology, Hunan Agricultural University, Changsha, China
dKey Laboratory of Reproduction Regulation of NPFPC, SIPPR, IRD, Fudan University, Shanghai, China. E-mail: zhenghj@chgc.sh.cn; Fax: +86 2134617421; Tel: +86 2134617495
First published on 21st April 2017
Abstract
The activated sludge process has been successfully used to treat propylene oxide (PO) saponification wastewater, which has the characteristics of high chlorine contents (22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–26
000–26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg L−1) and high COD (more than 2000 mg L−1). Microorganisms, especially bacteria, play an important role in PO saponification wastewater treatment processes. Analysis of the bacterial composition of the aeration tank and contact oxidation tank, the two main components of PO saponification wastewater treatment plants (SWWTP), revealed their significant community difference in municipal and coking wastewater treatment plants. Interestingly, β-Proteobacteria was almost absent in the PO SWWTP, which was usually abundant in various bio-treatment systems. In the aeration tank of PO SWWTP, the most abundant genera were Marinobacter, Mesorhizobium, Paracoccus, Devosia, Methylophaga and KSA1. In the contact oxidation tank of PO SWWTP, the most abundant genera were Thalassospira, Marinobacter, Owenweeksia, Novispirillum, Mesorhizobium, Sporotomaculum, Pseudidiomarina and KSA1. We also measured the total components and toxicity of PO saponification wastewater in order to establish correlations between bacterial stains, genes and their treatment capacity. The results indicated that most of the bacteria encoded the dehalogenase gene and played an important role in the dechlorinating process of chlorinated organics in the aeration tank. In contrast, most of the bacteria encode the alkJ gene in the contact oxidation tank, which was involved in the degradation of 2,4-dimethyl-2-pentanol or some dechlorinated intermediate products. This study would provide new insight into the microbial community compositions of PO SWWTPs.
000 mg L−1) and high COD (more than 2000 mg L−1). Microorganisms, especially bacteria, play an important role in PO saponification wastewater treatment processes. Analysis of the bacterial composition of the aeration tank and contact oxidation tank, the two main components of PO saponification wastewater treatment plants (SWWTP), revealed their significant community difference in municipal and coking wastewater treatment plants. Interestingly, β-Proteobacteria was almost absent in the PO SWWTP, which was usually abundant in various bio-treatment systems. In the aeration tank of PO SWWTP, the most abundant genera were Marinobacter, Mesorhizobium, Paracoccus, Devosia, Methylophaga and KSA1. In the contact oxidation tank of PO SWWTP, the most abundant genera were Thalassospira, Marinobacter, Owenweeksia, Novispirillum, Mesorhizobium, Sporotomaculum, Pseudidiomarina and KSA1. We also measured the total components and toxicity of PO saponification wastewater in order to establish correlations between bacterial stains, genes and their treatment capacity. The results indicated that most of the bacteria encoded the dehalogenase gene and played an important role in the dechlorinating process of chlorinated organics in the aeration tank. In contrast, most of the bacteria encode the alkJ gene in the contact oxidation tank, which was involved in the degradation of 2,4-dimethyl-2-pentanol or some dechlorinated intermediate products. This study would provide new insight into the microbial community compositions of PO SWWTPs.
Introduction
Propylene oxide (PO), the third derivative of propylene, is an important material in organic chemical engineering.1 PO is widely used in the production of polyether polyol and propene glycol. Two different commercial processes are employed in producing PO: the chlorohydrin process and the hydroperoxide process. In mainland China, the chlorohydrin process is preferred in most PO-producing plants, and saponification wastewater with high chlorine contents (22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–26
000–26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg L−1) and high COD (more than 2000 mg L−1) is generated. Many kinds of chlorinated organic compounds are detected in PO saponification wastewater, such as dichloroisopropyl ether, methyl dichloroacetate, etc., which cause the wastewater to be difficult to treat.1–3 After many years' research and development, the activated sludge process had been successfully used to treat PO saponification wastewater in China mainland.4
000 mg L−1) and high COD (more than 2000 mg L−1) is generated. Many kinds of chlorinated organic compounds are detected in PO saponification wastewater, such as dichloroisopropyl ether, methyl dichloroacetate, etc., which cause the wastewater to be difficult to treat.1–3 After many years' research and development, the activated sludge process had been successfully used to treat PO saponification wastewater in China mainland.4
The performance of activated sludge process largely depends on the activity of bacterial communities in the activated sludge. The microorganisms in municipal activated sludge could only tolerate chloride ions under the concentration of 6000 mg L−1, however the concentration of chloride ions in PO saponification wastewater varies from 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg L−1 to 26
000 mg L−1 to 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg L−1. Accordingly, maybe the microbial community structure in the activated sludge of PO saponification wastewater is different from the municipal activated sludge.
000 mg L−1. Accordingly, maybe the microbial community structure in the activated sludge of PO saponification wastewater is different from the municipal activated sludge.
It is very important to establish the relationship between the bacterial community structures and the degradation efficiency of the pollutants (high chlorine contents) in the saponification wastewater. The performances of activated sludge concerning microbial community structure are widely investigated.5 The investigation of microorganisms in saponification wastewater would develop effective strategies for resources development of activated sludge.6
Many molecular biological methods have been developed to explore the bacterial community and the activated sludge performance.1,2 Among them, high-throughput sequencing technology is an efficient tool to reveal the community structures of activated sludge. In previous studies, attentions were mainly focused on coking wastewater treatment plants7 and municipal wastewater treatment plants,8 lacking an overview of the bacterial community features of high-salinity industry wastewater treatment plants, such as PO saponification wastewater treatment plant (SWWTP). In this study, the bacterial community of the activated sludge in PO wastewater was investigated by Miseq Illumina high-throughput sequencing. The core phyla, class, family and genera of the activated sludge in PO wastewater were all identified, and obvious differences were exhibited comparing with the municipal activated sludge. This study would provide a theoretical basis for resources development of PO SWWTP activated sludge.
Results and discussion
Physico-chemical analysis and toxicity test of PO saponification wastewater
In order to link the structure of the bacterial communities with organics and inorganics of PO saponification wastewater, total components in PO saponification wastewater were measured (Table S1†). The organic components in PO saponification wastewater were 2,4-dimethyl-2-pentanol, dichloroisopropyl ether, 1,7-dichloro-4-oxaheptane and methyl dichloroacetate respectively. The inorganic components were calcium chloride, sodium sulphate, calcium hypochlorite and sodium hydroxide respectively. The results showed that many chlorinated organic compounds existed in PO saponification wastewater. It was inferred that there were many bacteria encoding dehalogenase genes, which played important roles in the degradation process of organic chlorides in PO saponification wastewater. Furthermore, it was incredible that lots of inorganic ion, such as chlorine ion, calcium ion and sodium ion, exist in PO saponification wastewater. It meant that halotolerant or halophilic microorganisms would be found, which adapted to live in high-salinity wastewater. We also monitored the toxicity of PO saponification wastewater. The toxicity test result was ≥0.24 mg L−1 in terms of HgCl2 equivalent concentration, indicating extremely toxicity.9Overview of sequencing revealing microbial community difference between aeration tank and contact oxidation tank
The microbial community diversity and structure of PO SWWTP was investigated by Miseq Illumina high-throughput sequencing. There were a total of 18 samples from six groups (A, C, E, G, I and J). After removing low quality sequences and chimeras, the average number of effective sequences was 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 497, and the average length of sequences was 409 bp. A total of 3812 representative OTUs was obtained for all the samples, and the OTU number of each sample were 568–1083, with average 693 OTUs in aeration tank and 1006 OTUs in contact oxidation tank. The Good's coverage was 97.4–99.4%, indicating the sequencing depth was enough for analysis.
497, and the average length of sequences was 409 bp. A total of 3812 representative OTUs was obtained for all the samples, and the OTU number of each sample were 568–1083, with average 693 OTUs in aeration tank and 1006 OTUs in contact oxidation tank. The Good's coverage was 97.4–99.4%, indicating the sequencing depth was enough for analysis.
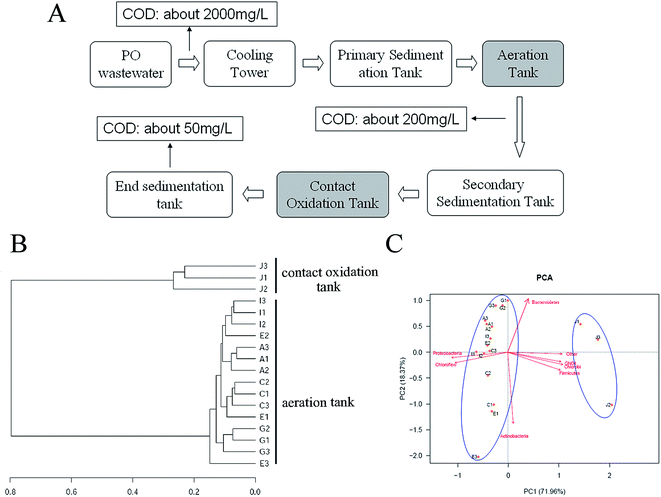
Hierarchical clustering analysis (Fig. 1B) and principal components analysis (PCA) (Fig. 1C) indicated that all samples of aeration tank (A, C, E, G and I) and contact oxidation tank (J) were clustered separately, demonstrating the community structures of aeration tank and contact oxidation tank of PO SWWTP were different. Shannon-Weiner index10 also validated that the species diversity of contact oxidation tank (Shannon-Weiner index: 4.97) was higher than that of aeration tank (Shannon-Weiner index: 3.43).
Bacterial community analysis at high taxonomic levels
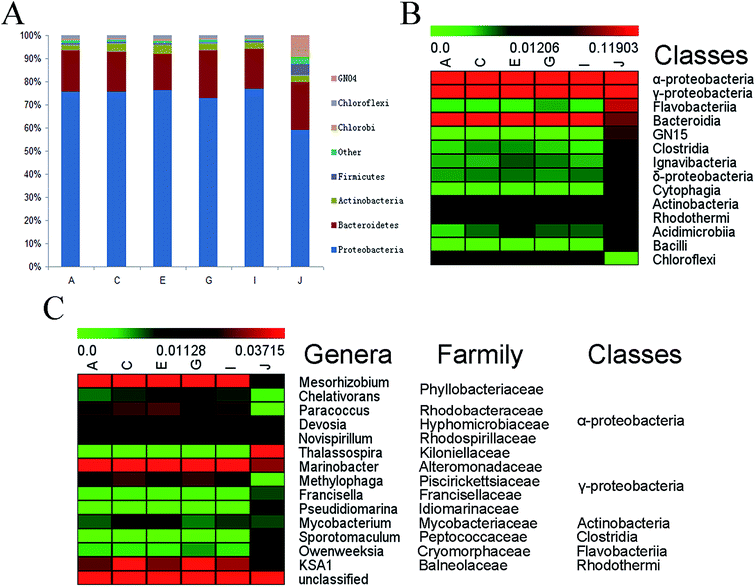
Nearly all the sequences (over 99.99%) can be classified at the phylum level. The major phyla and their proportion for each sampling location are shown in Fig. 2A and Table S2.† No matter in aeration tank or contact oxidation tank, the most abundant phylum was Proteobacteria (minimum 59.13% in contact oxidation tank J and maximum 79.64% in aeration tank I). In aeration tank, the following major phyla were Bacteroidetes (mean 17.77%, each sample 15.84–20.89%), Actinobacteria (mean 3.20%, each sample 2.59–3.94%) and Chloroflexi (mean 1.39%, each sample 1.14–1.61%). In contact oxidation tank, the following major phyla were Bacteroidetes (20.44%), GN04 (5.56%), Firmicutes (4.72%), Chlorobi (3.80%), and Actinobacteria (3.36%). Meanwhile, Metastats analysis11 revealed that 12 phylum showed significant difference of abundance between aeration tank and contact oxidation tank (q-value <1 × 10−5) (Table S2†). Compared with aeration tank, GN04, Firmicutes and Chlorobi occupied higher proportions in contact oxidation tank, and Proteobacteria and Chloroflexi occupied lower proportions (Table S2†). The Proteobacteria was the prominent phylum in PO SWWTP, which was consistent with the former study demonstrating Proteobacteria was the prominent phylum in various wastewater treatment plants.7 As a kind of halotolerant bacteria, GN04 may play an irreplaceable role in the treatment of propylene epoxide saponated sewage.At the class level, 0.049% of the total sequences were not classified. Other sequences belong to 95 identified classes. Bacterial main community compositions of PO SWWTP are shown in Fig. 2B and Table S3.† α-Proteobacteria was the largest class with sequence percentages ranging from 32.49% to 62.40%. The percentages of γ-Proteobacteria were 12.24–22.28%. The percentages of δ-Proteobacteria (0.61–3.35%) were lower than the other proteobacterial groups. Interestingly, β-Proteobacteria was rich in various bio-treatment systems.7,12,13 However, the number of β-Proteobacteria was little in PO SWWTP, suggesting α-Proteobacteria, γ-Proteobacteria and δ-Proteobacteria, rather than β-Proteobacteria, may play important roles in pollutant degradation of PO SWWTP. Except for the subdivisions of Proteobacteria, other classes also functioned in pollutant degradation. Bacteroidia, Actinobacteria, Rhodothermi and Chloroflexi were rich in aeration tank, while Flavobacteriia, GN15, Clostridia, Ignavibacteria, Cytophagia, Actinobacteria, Rhodothermi, Acidimicrobiia and Bacilli were rich in contact oxidation tank (Table S3†). Obviously, the percentages of Flavobacteriia and GN15 in contact oxidation tank were far higher than aeration tank, and there was no Chloroflexi in contact oxidation tank. Chloroflexi, a kind of bacterium including dehalogenase coding genes, populations abundant in anaerobic sludge digesters,14 was known to be important in reductive dechlorination for chlorinated pollutants.15–17 The result bore out our speculation that there were dechlorinating bacteria in PO saponification wastewater. The absence of Chloroflexi in contact oxidation tank may indicate that dechlorinating process mainly occurred in aeration tank.
Identification of the core genera
The major genera in each group are summarized in Fig. 2C and Table S4.† Metastats analysis11 revealed that 37 genera showed significantly different of abundance between aeration tank and contact oxidation tank (q-value <1 × 10−5) (Table S4†). Marinobacter, belonged to Alteromonadaceae, was a major component (about 7%) of activated sludge in aeration tank of PO SWWTP. The species of Marinobacter were reported as a kind of extremely halotolerant hydrocarbon-degrading bacteria and facultatively anaerobic.18 This bacterium could survive at high salt contents and degrade various environmental organic contaminants in PO saponification wastewater. Mesorhizobium of Phyllobacteriaceae encoded dehalogenases, which were key enzymes for the degradation of halogenated pollutants.19 Therefore, the occurrences of Mesorhizobium related to high concentrations of dichloroisopropyl ether, methyl dichloroacetate and other chlorinated hydrocarbon. Other core genus in aeration tank included Paracoccus, Devosia, Methylophaga and KSA1. Paracoccus, belonging to the family Rhodobacteraceae, is deemed to degrade pyridine,20 chlorpyrifos21 and acetone,22 which might be the dechlorinated product of dichloroisopropyl ether, one component in PO saponification wastewater. Devosia is a kind of mycotoxin deoxynivalenol – degrading bacterium.23 Methylophaga is a dimethylsulfide degrader.24In contact oxidation tank of PO SWWTP, the most abundant genera were Thalassospira, Marinobacter, Owenweeksia, Novispirillum, Mesorhizobium, Sporotomaculum, Pseudidiomarina and KSA1. It was reported that a novel species of Thalassospira could degrade environmental organic contaminants, including polycyclic aromatic hydrocarbon.25 The ring structures of dibenzo-p-dioxin, dibenzofuran (DF), and related compounds can be degraded by aerobic bacteria containing aromatic hydrocarbon dioxygenases with broad substrate specificity.15 Owenweeksia honkongenesis was identified as species capable of degrading fenvalerate and benzene.26,27 Novispirillum, a kind of purple non-sulfur bacteria, has been used to treat wastewater.28 Sporotomaculum is a novel benzoate-degrading bacterium which has been found to take part in the process of wastewater treatment.29 Pseudidiomarina was found to be polycyclic aromatic hydrocarbon degraders.30 They all have the potential to degrade dechlorinated products of chlorinated hydrocarbon in PO saponification wastewater.
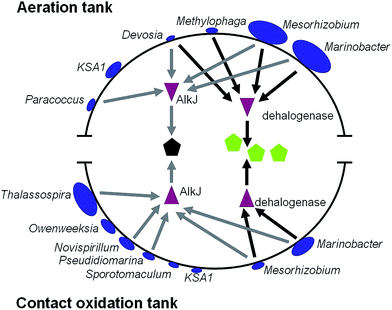
To establish relationship between bacterial stains and their treatment capacity, we detected the association of bacterial stains, genes and organic components in PO saponification wastewater. We found that three kinds of chlorinated organic compounds (dichloroisopropyl ether, 1,7-dichloro-4-oxaheptane and methyl dichloroacetate) and 2,4-dimethyl-2-pentanol existed in PO saponification wastewater. Dehalogenase is a key enzyme responsible for the degradation of halogenide.19 Moreover, AlkJ (alcohol dehydrogenase) is a key enzyme for the degradation of alcohols.31 In such a case we performed a search of the two proteins against all bacteria genomes in the core genera. The results showed that four kinds of bacteria in aeration tank can encode dehalogenase gene, including Marinobacter and Mesorhizobium that were the two largest number of core genera (Fig. 3). As for contact oxidation tank, there were only two kinds of bacteria encoding dehalogenase gene and five kinds of bacteria encoding alkJ gene (Fig. 3). It indicated that dechlorinating process mainly occurred in aeration tank, while the degradation of 2,4-dimethyl-2-pentanol or some dechlorinated products occurred in contact oxidation tank.
It was a remarkable fact that Rhodobacteraceae was the richest in aeration tank, and Rhodospirillaceae was the richest in contact oxidation tank at the family level (Table S5†). However, at the genera level, the great mass of these bacteria can not be identified, and 70% of the sequences were not assigned to any genera in both aeration tank and contact oxidation tank. We analyzed 16S rRNA sequence of the major unidentified bacteria using BLAST-N and listed the closest bacteria of each OTU (Table S6†). Among them, OTU0001 occupied 27% of all the bacterial population in aeration tank and showed 96% identify with genera Roseovarius, which played an important role in degrading low molecular weight polycyclic aromatic hydrocarbons (PAHs), followed by degradation of the 4- and 5-ring PAHs by Gammaproteobacteria, Marinobacter and Methylophaga.32 The high proportion of these novel bacteria, especially in aeration tank, indicated they played important roles in PO saponification wastewater treatment processes. Further research such as metagenome sequencing technologies will be needed for unidentified genera.
Bacterial community compositions of several wastewater treatment systems have been already analyzed. The core genera in municipal wastewater treatment plants (such as Zoogloea, Dechloromonas, Ferruginibacter, Prosthecobacter and Gp6)8,12 and in coking wastewater treatment plants (such as Thiobacillus, Comamonas, Thauera, Azoarcus and Rhodoplanes)7 were almost undetectable in the PO SWWTP. Differences in the bacterial community compositions among several wastewater treatment plants can be partly explained by physicochemical characteristics of wastewater samples. We found that there were many bacteria which can degrade organic chlorides and dechlorinated product in PO SWWTP. In addition, most strains were halotolerant or halophilic organisms in PO SWWTP. These data show the correlation between the structure of the bacterial communities and the abiotic process parameters in PO saponification wastewater. Our study shows the community structures of PO SWWTP and explains why the activated sludge can be successfully used to treat the PO saponification wastewater. However, no one knows how the different bacteria communicate in the treatment. We will focus on this and use the processes to improve the degradation efficiency of wastewater.
Materials and methods
Physico-chemical analysis and toxicity test
Total components were measured in PO saponification wastewater. All analytical data were provided by Science Standard Institute of Testing (Qingdao, China) using Energy Dispersive X-ray Spectroscopy (FEI Quanta 200FEG), PY-GC/MS (QP2010 Ultra), HS-GC/MS (Agilent GC6890-5975I MS), GC/MS (Trace-1310/Thermo ISQ MS detector), Fourier transform infrared spectroscopy (Nicolet iS5), Karl Fischer Titrator (V20), Thermogravimetric Analysis Instrument (TA Q500) and X-ray diffraction (Bruker D8 Advanced XRD). The toxicity was assessed by a bioassay based on the light emission of Photobacterium phosphoreum T3 (the national standard method of China, GB/T 15441-1995).33 The toxicity test was provided by Science Standard Institute of Testing (Qingdao, China).PO SWWTP and sample collection
The PO saponification wastewater used in this study was collected from treatment plant in Befar Group Co., Ltd, Binzhou, Shandong, China. The process chart of PO saponification wastewater treatment is shown in Fig. 1A. And the treatment process of PO saponification wastewater mainly occurs in aeration tank and contact oxidation tank. Samples of activated sludge were collected from aeration tank (A, C, E, G and I) and contact oxidation tank (J) at SWWTP of Shandong Befar Group Co., Ltd in China. Triplicate samples for every tank were collected and stored at −80 °C before use.DNA extraction, PCR and high-throughput sequencing data analysis
Activated sludge samples were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min. And the supernatant was removed. For the genomic DNA, 1.0 g sediment was used. Community DNA of samples were extracted using E.Z.N.A.™ Soil DNA Kit (Omega Bio-tek, Inc.) according to E.Z.N.A.™ Soil DNA Protocol in the kit. Primers for sequencing were 343F (5′-TACGGRAGGCAGCAG-3′) and 798R (5′-AGGGTATCTAATCCT-3′), with different barcodes for the V3 and V4 regions of 16S rRNA gene.34 The PCR was performed at 95 °C for 3 min; and 25 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s; and a final extension at 72 °C for 10 min. PCR products were pooled and purified using the AMPure XP Beads (Beckman). Finally, the DNA library was constructed and run on the MiSeq (Illumina) at Shanghai Hanyu Biotech Co., Ltd (Shanghai, China).
000g for 5 min. And the supernatant was removed. For the genomic DNA, 1.0 g sediment was used. Community DNA of samples were extracted using E.Z.N.A.™ Soil DNA Kit (Omega Bio-tek, Inc.) according to E.Z.N.A.™ Soil DNA Protocol in the kit. Primers for sequencing were 343F (5′-TACGGRAGGCAGCAG-3′) and 798R (5′-AGGGTATCTAATCCT-3′), with different barcodes for the V3 and V4 regions of 16S rRNA gene.34 The PCR was performed at 95 °C for 3 min; and 25 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s; and a final extension at 72 °C for 10 min. PCR products were pooled and purified using the AMPure XP Beads (Beckman). Finally, the DNA library was constructed and run on the MiSeq (Illumina) at Shanghai Hanyu Biotech Co., Ltd (Shanghai, China).
The low quality sequences and chimera were detected and removed by Mothur (version 1.32.01) and UCHIME (version 4.2.40), respectively. Operational taxonomic units (OTUs) were generated by Mothur (version 1.32.01) at the 97% sequence similarity threshold. Tree construction was performed using the hierarchical clustering algorithm in R and visualized using TREEVIEW. Principal components analysis was carried out by SPSS software (version 13.0). The taxonomic assignment was performed using the Ribosomal Database Project Classifier program with a confidence cutoff of 80%. The bacterial taxa at the phylum, class, family and genus level was analyzed respectively. Heatmaps were generated with Excel and MeV (version 4.9.0).
Conclusions
In this study, the community structures of PO SWWTP were explored for the first time. We measured total components and toxicity in PO saponification wastewater in order to establish the correlation between the abiotic process parameters and the structure of the bacterial communities. In aeration tank and contact oxidation tank, the main phyla, class, family, and genera were identified. In aeration tank of PO SWWTP, the most abundant genera were Marinobacter, Mesorhizobium, Paracoccus, Devosia, Methylophaga and KSA1. In contact oxidation tank of PO SWWTP, the most abundant genera were Thalassospira, Marinobacter, Owenweeksia, Novispirillum, Mesorhizobium, Sporotomaculum, Pseudidiomarina and KSA1. We detected the association of bacterial stains, genes and treatment capacity. Therefore, we found that dechlorinating process mainly occurs in aeration tank, while the degradation of 2,4-dimethyl-2-pentanol or some dechlorinated products occurs in contact oxidation tank. We also found that contact oxidation tank harbors more microbial diversity than aeration tank, and both of the community structures were quite different from the usual activated sludge. Interestingly, β-Proteobacteria was not appeared in PO SWWTP, which was usually rich in various bio-treatment systems. The special community structure of PO SWWTP endows the activated sludge to treat the PO saponification wastewater. The understanding of PO SWWTP bacterial communities and how to facilitate the coexistence of different physiological groups of bacteria, will be helpful to develop efficient strategies for the degradation of PO saponification wastewater.Acknowledgements
This work was supported by National Natural Science Foundation of China (Project No. 31100088, 51208290, 31300045, 31600148, 31600066), and Shandong Province Science and Technology Development Plan (Grant No. 2013GSF12006), and Shandong Excellent Young Scientist Award Fund (BS2014YY031), and Shandong Provincial Natural Science Foundation (ZR2016CL02, ZR2016CB20), and Foundation of University of Jinan (XBS1519, XKY1633). The authors declare that there are no conflicts of interest.Notes and references
- Q. A. Li, H. L. Liu, Q. S. Qi, F. S. Wang and Y. Z. Zhang, New Biotechnol., 2010, 27, 789–794 CrossRef CAS PubMed.
- B. K. Singh, S. Munro, E. Reid, B. Ord, J. M. Potts, E. Paterson and P. Millard, Eur. J. Soil Sci., 2006, 57, 72–82 CrossRef CAS.
- H. M. Moreno, L. A. Garbe, R. Tressl, L. Adrian and H. Görisch, Arch. Microbiol., 2003, 179, 234–241 CrossRef PubMed.
- Y. Wang, Y. Zhu, P. Gu, Y. Li, X. Fan, D. Song, Y. Ji and Q. Li, Int. J. Biol. Macromol., 2017, 98, 34–38 CrossRef CAS PubMed.
- C. Yang, W. Zhang, R. H. Liu, Q. Li, B. B. Li, S. F. Wang, C. J. Song, C. L. Qiao and A. Mulchandani, Environ. Sci. Technol., 2011, 45, 7408–7415 CrossRef CAS PubMed.
- R. Margesin and F. Schinner, Extremophiles, 2001, 5, 73–83 CrossRef CAS PubMed.
- Q. Ma, Y. Qu, W. Shen, Z. Zhang, J. Wang, Z. Liu, D. Li, H. Li and J. Zhou, Bioresour. Technol., 2015, 179, 436–443 CrossRef CAS PubMed.
- T. Zhang, M. F. Shao and L. Ye, ISME J., 2012, 6, 1137–1147 CrossRef CAS PubMed.
- G. Na, Y. Zhang, T. Chen, L. Zhang, C. Liu, J. Gu, Z. Lin and Z. Yao, Environ. Monit. China, 2010, 26, 61–64 Search PubMed.
- A. P. Martin, Appl. Environ. Microbiol., 2002, 68, 3673–3682 CrossRef CAS PubMed.
- J. R. White, N. Nagarajan and M. Pop, PLoS Comput. Biol., 2009, 5, e1000352 Search PubMed.
- X. Wang, M. Hu, Y. Xia, X. Wen and K. Ding, Appl. Environ. Microbiol., 2012, 78, 7042–7047 CrossRef CAS PubMed.
- E. L. Figuerola and L. Erijman, Environ. Microbiol., 2007, 9, 1780–1789 CrossRef CAS PubMed.
- H. D. Ariesyady, T. Ito and S. Okabe, Water Res., 2007, 41, 1554–1568 CrossRef CAS PubMed.
- N. Yoshida, N. Takahashi and A. Hiraishi, Appl. Environ. Microbiol., 2005, 71, 4325–4334 CrossRef CAS PubMed.
- J. Yan, B. A. Rash, F. A. Rainey and W. M. Moe, Environ. Microbiol., 2009, 11, 833–843 CrossRef CAS PubMed.
- H. Liu, J. W. Park and M. M. Haggblom, Environ. Pollut., 2014, 184, 222–230 CrossRef CAS PubMed.
- W. Gao, Z. Cui, Q. Li, G. Xu, X. Jia and L. Zheng, Anton. Leeuw. Int. J. G., 2013, 103, 485–491 CrossRef CAS PubMed.
- Y. Sato, M. Monincova, R. Chaloupkova, Z. Prokop, Y. Ohtsubo, K. Minamisawa, M. Tsuda, J. Damborsky and Y. Nagata, Appl. Environ. Microbiol., 2005, 71, 4372–4379 CrossRef CAS PubMed.
- Y. Bai, Q. Sun, C. Zhao, D. Wen and X. Tang, Biodegradation, 2008, 19, 915–926 CrossRef CAS PubMed.
- G. Xu, W. Zheng, Y. Li, S. Wang, J. Zhang and Y. Yan, Int. Biodeterior. Biodegrad., 2008, 62, 51–56 CrossRef CAS.
- H. Siller, F. A. Rainey, E. Stackebrandt and J. Winter, Int. J. Syst. Bacteriol., 1996, 46, 1125–1130 CrossRef CAS PubMed.
- I. Sato, M. Ito, M. Ishizaka, Y. Ikunaga, Y. Sato, S. Yoshida, M. Koitabashi and S. Tsushima, FEMS Microbiol. Lett., 2012, 327, 110–117 CrossRef CAS PubMed.
- R. Boden, D. P. Kelly, J. C. Murrell and H. Schäfer, Environ. Microbiol., 2010, 12, 2688–2699 CAS.
- B. Zhao, H. Wang, R. Li and X. Mao, Int. J. Syst. Evol. Microbiol., 2010, 60, 1125–1129 CrossRef CAS PubMed.
- H. Boricha and M. Fulekar, Rom. Biotechnol. Lett., 2010, 15, 5104–5110 CAS.
- K. W. Lau, C. Y. Ng, J. Ren, S. C. Lau, P.-Y. Qian, P.-K. Wong, T. Lau and M. Wu, Int. J. Syst. Evol. Microbiol., 2005, 55, 1051–1057 CrossRef CAS PubMed.
- L. Zhang, Y. Sun, D. Guo, Z. Wu and D. Jiang, Afr. J. Microbiol. Res., 2012, 6, 6444–6453 CAS.
- Y. L. Qiu, Y. Sekiguchi, H. Imachi, Y. Kamagata, I. C. Tseng, S. S. Cheng, A. Ohashi and H. Harada, Arch. Microbiol., 2003, 179, 242–249 CrossRef CAS PubMed.
- Z. Shao, J. Yuan, Q. Lai and T. Zheng, Front. Microbiol., 2015, 6, 853 Search PubMed.
- J. B. van Beilen, M. G. Wubbolts and B. Witholt, Biodegradation, 1994, 5, 161–174 CrossRef CAS PubMed.
- S. Gallego, J. Vila, M. Tauler, J. M. Nieto, P. Breugelmans, D. Springael and M. Grifoll, Biodegradation, 2014, 25, 543–556 CrossRef CAS PubMed.
- X. Wang, R. Qu, Z. Wei, X. Yang and Z. Wang, Ecotoxicol. Environ. Saf., 2014, 104, 231–238 CrossRef CAS PubMed.
- C. W. Nossa, W. E. Oberdorf, L. Yang, J. A. Aas, B. J. Paster, T. Z. DeSantis, E. L. Brodie, D. Malamud, M. A. Poles and Z. Pei, World J. Gastroenterol., 2010, 16, 4135 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra27808f |
| ‡ Xiangyu Fan and Ying Zhu contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |