Open Access Article
Open Access ArticleSelection, identification and application of DNA aptamers for the detection of Bifidobacterium breve†
Lujun Hua,
Linlin Wanga,
Wenwei Luab,
Qixiao Zhaiab,
Daming Fanab,
Xiaoming Liuab,
Jianxin Zhaoab,
Hao Zhangab and
Wei Chen *abc
*abc
aState Key Laboratory of Food Science and Technology, School of Food Science and Technology, Jiangnan University, Wuxi 214122, PR China. E-mail: chenwei66@jiangnan.edu.cn; Fax: +86 510 85912155; Tel: +86 510 85912155
bInternational Joint Research Center for Probiotics & Gut Health, Jiangnan University, Wuxi 214122, PR China
cBeijing Innovation Centre of Food Nutrition and Human Health, Beijing Technology and Business University, Beijing 100048, PR China
First published on 16th February 2017
Abstract
In the present study, a single-stranded DNA (ssDNA) aptamer binding to Bifidobacterium breve with high avidity and selectivity was selected through a whole-bacterium-based systemic evolution of ligands using an exponential enrichment (SELEX) process. Following 12 rounds of selection specific for B. breve, three FAM-labeled aptamers were chosen for flow cytometry analysis and the results revealed that all three aptamers possessed a high binding affinity for B. breve. To obtain the optimal sequence, sequence truncation experiments of these three aptamers were conducted. An aptamer variant BB16-11f with high affinity and selectivity was acquired. In addition, the dissociation constant was significantly reduced to 18.66 ± 1.41 nM. Furthermore, an enzyme linked aptamer assay was developed to prove the potential application of the aptamer BB16-11f in the detection of B. breve. The results showed that the colorimetric assay had a linear relationship between the absorbance at 450 nm and the concentrations of B. breve ranging from 103 cfu mL−1 to 107 cfu mL−1 with a correlation coefficient of 0.98. The limit of detection (LOD) of the assay was 1000 cfu mL−1. Additionally, the developed method was also successfully used to detect B. breve in a milk environment. Taken together, we hold that the developed colorimetric bioassay based on the aptamer BB16-11f is a promising method for the detection of B. breve.
Introduction
The contribution of bifidobacteria in maintaining or improving human health has been accepted for quite a while and has brought about the comprehensive development of bifidobacteria as probiotics.1 In particular, Bifidobacterium breve showed immunomodulatory properties and had the potential to prevent intestinal inflammation.2–7 The earlier B. breve was administrated in low birth weight infants, the more helpfully it will promote the colonization of bifidobacteria and the formation of normal intestinal microbiota.8 B. breve also possesses QPS (quality and presumption of safety) status granted by the European Food Safety Authority, and thus, B. breve is often implemented in probiotic products.9–12 However, many reports have proved that the recovery of incorporated probiotic organisms is not always satisfying and the species recovered from these products are not always the same as those indicated on the product labels.13–16 Thus, it is vital to discriminate the different probiotic strains at the species level in food products available on the market.Identification of B. breve is significant for its industrial use, but the conventional methods of detection do not always obtain satisfying results.17,18 The classical culture methods, including isolation and identification of the species, are labor intensive and time consuming. Furthermore, phenotypic characterization is not enough to distinguish B. breve from other strains at the species level in many cases.19 Thus, there is an important need to develop an alternative approach to the identification of B. breve. Fortunately, various molecular methods have been developed to detect B. breve. Molecular techniques such as fluorescence in situ hybridization20 and PCR21 can shorten the detection time, but they increase the analysis cost in that they need sophisticated equipment and well-trained professional.21–23 What is more, these methods are not practical for rapid detection.
Rapid identification methods should supply dependable, low-cost, real time, on-field and amicable detection with same or better sensitivity, selectivity and consistency of the culture-based assays.24 Aptamers, which are screened via an in vitro process, are single-stranded oligonucleotide ligands obtained by SELEX method based on their high binding affinity and selectivity for target biomolecules.25,26 Aptamers have many unique features that make them a better choice than antibodies, including inexpensive, ease of synthesis and modification and comparable stability.27,28 Live bacteria and bacterial surface molecules can be employed as targets during SELEX.29,30 Whole bacteria as target has been implemented to create ssDNA aptamers against Mycobacterium tuberculosis, Lactobacillus acidophilus, Staphylococcus aureus, Campylobacter jejuni, Streptococcus pyogenes and Salmonella typhimurium without knowing a specific target molecule beforehand.31–36 Thus, aptamers may offer an alternative method for the rapid detection of B. breve.
Enzyme linked aptamer assay (ELAA), is a variant of conventional ELISA (enzyme linked immunosorbent assay) using aptamers replacing antibodies. ELAA can realize colorimetric assay based on the catalytic reaction of horseradish peroxidase (HRP) and TMB–H2O2. And colorimetric assays have many advantages such as high-throughput with the help of 96-well microplate and convenience in that the signal readout requires only simple instruments or even no instrumentation when read with the naked eyes. Thus, ELAA has been used in many bioanalytical applications for the target-specific detection of some substances such as ochratoxin A, thrombin and M. tuberculosis.37–39 However, ELAA has not been reported in the detection of B. breve.
In the study, we used an improved whole-bacterium SELEX strategy to screen ssDNA aptamers with high avidity and selectivity for B. breve. Additionally, the truncation experiments were conducted to obtain the optimal aptamer. Furthermore, to confirm the potential application of the candidate aptamer, a colorimetric bioassay was developed for the detection of B. breve, and the developed method was also used to detect B. breve in milk samples.
Materials and methods
Bacterial strains and culture media
B. breve ATCC 15700 was employed as the target during whole-bacterium SELEX. Other bacterial strains adopted in the study (for counter-SELEX and specificity studies) were as follows: B. longum ATCC 15697, B. bifidum ATCC 29521, B. animalis JCM 11658, B. adolescentis ATCC 15705 and L. plantarum ST-III (CGMCC no. 0847). All of the bifidobacterial strains used in the study were supplied by the American Type Culture Collection (ATCC) or the Japan Collection of Microorganisms (JCM). All bifidobacterial strains were grown in MRS broth with addition of 0.05% of L-cysteine hydrochloride monohydrate at 37 °C. The L. plantarum used in this study was cultured in MRS broth at 37 °C (Merck KGaA, Darmstadt, Germany). All bacterial strains were cultured to the logarithmic phase under anaerobic conditions.DNA library and PCR amplification
An 80 nt oligonucleotide ssDNA library was synthesized with the following sequence:5′-AGCAGCACAGAGGTCAGATG-N40-CCTATGCGTGCTACCGTGAA-3′, where the middle N40 represents random nucleic acids with equimolar amounts of A, G, C and T at each position and the fixed primer sequence is at both ends. The initiating ssDNA library and the primers employed to amplify it were supplied by Integrated DNA Technologies (IDT; Coralville, IA).
The PCR amplification methods during SELEX were as follows: 1× PCR amplification buffer, 10 μM forward and reverse primer, 25 mM dNTPs, 5 U μL−1 of Taq DNA polymerase, 2 μL of the template (all reagents were from Invitrogen) and 2.5 μL of DMSO (Sigma) in a total volume of 50 μL. Thermocycling parameters were as follows: pre-denaturation for 6 min at 95 °C, 25 cycles of denaturation at 95 °C for 30 s, annealing for 30 s at 69 °C, elongation for 20 s at 72 °C and final elongation for 6 min at 72 °C (T100 Thermo cycler; Bio-Rad Laboratories, Hercules, CA). PCR amplification products were analyzed by 8% nondenaturing polyacrylamide gel electrophoresis (PAGE) in 1× TBE buffer (Bio-Rad Protean III) at 200 V for 25 min. Qiagen MinElute PCR Purification Kit was used for purifying all PCR amplification products (Qiagen Inc., Valencia, CA).
Aptamer selection
The SELEX procedure for aptamer selection was based on Hamula and colleagues32 with some modifications as follows: B. breve was harvested when it reached logarithmic phase. B. breve cells were centrifuged at 4000 × g and 4 °C for 5 min and then washed twice in binding buffer (50 mM Tris–HCl (pH 7.4), 100 mM NaCl, 5 mM KCl and 1 mM MgCl2). The ssDNA library/pool was heated at 95 °C for 10 min and rapidly chilled for 10 min in an ice bath before incubation. B. breve cells totaling 108 cfu mL−1 were incubated with the 10 μL ssDNA library (200 μM) for initial round or 10 μL aptamer pool (10 μM) for subsequent rounds in binding buffer (600 μL for the initial round, 350 μL for subsequent rounds) for 45 min at room temperature with slight agitation. An excess of tRNA (Sigma) and bovine serum albumin (BSA; Invitrogen) were put into the incubation buffer. Following incubation, B. breve cells were washed three times in binding buffer (from 0.25 to 0.75 mL) with 0.05% BSA. And then, the cells were resuspended in 80 μL of 1× PCR reaction buffer before next heating at 95 °C for 10 min and quickly cooled for 10 min in an ice bath. After centrifugation as described above, the supernatant was separated and designated as the cell-bound aptamer fraction, which was used as template for PCR amplification and the PCR amplification products were adopted in the following round of selection.To exclude aptamers binding to the tube, the resuspended cell solution from each incubation and elution was transferred to a fresh tube. To improve the specificity of aptamer candidates, two rounds (the 9th and 11th rounds) of counter-SELEX were implemented. In brief, the selected aptamer pools were incubated with the pooled counter-SELEX bacteria (B. longum, B. bifidum, B. animalis and B. adolescentis) in binding buffer for 45 min at room temperature with gentle rotation. The aptamer-bound cells were centrifuged and the supernatant were collected for the next round of selection.
Following the 12th round of selection, aptamer pools were cloned (TOPO TA Cloning kit, Invitrogen, Carlsbad, CA) and the plasmid DNA was purified (QIAEX II Gel Extraction Kit; Qiagen, Mississauga, ON, Canada) prior to sequencing. RNA structure 3.0 was adopted for predicting the secondary structure of each sequence, with input conditions of 1 mM MgCl2 and the room temperature. The most likely sequence was selected according to the lowest predicted free energy (ΔG, kcal mol−1).
Binding assays
A FACSCalibur flow cytometer with PowerMacG4 workstation and CellQuest Pro software (BD Biosciences, USA) was used to analyze the binding affinity of the individual sequences to different bacterial cells (B. breve, B. longum, B. bifidum, B. animalis, B. adolescentis and L. plantarum) in separate experiments. The individual sequences with the fluorescent label (5′-FAM) attached were obtained from IDT. The binding analysis were performed by incubating 108 B. breve cells with 100 nM of FAM-labeled aptamer for 45 min at room temperature with gentle rotation to facilitate aptamer binding following heat denaturation. Cells were then washed twice before resuspension in binding buffer for prompt flow cytometric assays. Forward scatter, side scatter and fluorescence intensity were measured, and the gated fluorescence intensity above the background (cells with no aptamers) was quantified. BD CellQuest Pro software was used for analyzing the data from the FACSCalibur and creating the histogram overlays. Binding saturation curves were created to estimate binding dissociation constants (Kd) by altering the aptamer concentration (0–100 nM) with a constant number of B. breve cells (108 cells).32,40 GraphPad Prism 5.0 software was employed to fit a nonlinear regression curve from which Kd values were determined.Aptamer truncation
To determine the minimal sequence necessary for high-affinity binding between the aptamers and B. breve, truncation experiments were conducted.41 Firstly, the specific primer binding sites at both ends of the aptamers were removed. Then, DNA aptamer variants with high binding ability to B. breve were further truncated from the 5′ or 3′ end respectively. In the experiment, the truncated aptamer variants were FAM-labeled at the 5′-end and tested for their binding affinity with B. breve using flow cytometric assays as described above. And the specificity and binding dissociation constants Kd values of the optimized aptamer variants were also determined according to the method described above.Procedure of colorimetric bioassay based on the selected aptamer
A colorimetric sandwich-type assay for detection B. breve was developed according to ELAA. First, avidin was dissolved in NaHCO3 buffer (pH 9.6) and 100 μL diluted avidin (10 mg mL−1) was added into each well of the 96-well plates for incubating overnight at 4 °C. The wells were washed three times with washing buffer (0.01 mol L−1 PBS with 0.05% Tween-20) prior to use. Then, the avidin-coated microplate wells were blocked with the blocking buffer (0.01 mol L−1 PBS with 3% BSA) to prevent the appearance of nonspecific adsorption.Subsequently, 10 μL biotinylated candidate aptamer was added to the wells of the microplates and incubated 30 min at 37 °C after the plates were washed three times with the washing buffer. A series of different concentrations of B. breve cells were then added into each well for incubating at 37 °C for 45 min and the biotinylated aptamer and streptavidin–HRP were mixed at 37 °C for 30 min at the same time. After the microplates were washed three times with the washing buffer, 10 μL samples of the above biotinylated aptamer and streptavidin–HRP complexes were added to the wells of the microplates for reacting for 30 min at 37 °C. Then 200 μL TMB–H2O2 working solutions were added into each well for reacting without direct light exposure after the final wash step. After 15 min, the mixture was terminated with 50 μL 2 M H2SO4, and the OD value at 450 nm was measured with the microplate reader.
Colorimetric detection of B. breve in the milk
Raw milk was ordered from local supermarkets and subjected to centrifugation for 30 min at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm so that protein and fat were removed. Then, the supernatant was filtered with a 0.22 μM filtration membrane. Finally, different amounts of B. breve were added into the treated milk samples to make the spiked samples for colorimetric detection as described above. The plate counting method was also used for detecting B. breve as a control test in the study.
000 rpm so that protein and fat were removed. Then, the supernatant was filtered with a 0.22 μM filtration membrane. Finally, different amounts of B. breve were added into the treated milk samples to make the spiked samples for colorimetric detection as described above. The plate counting method was also used for detecting B. breve as a control test in the study.
Results and discussion
Aptamer selection
To screen aptamers that potentially identify B. breve, a whole-bacterium SELEX including 10 rounds of selection against B. breve and two rounds of counter selection against an incorporative mixture of other bacteria was performed. The whole-bacterium SELEX approach has two advantages for the rapid identification of B. breve. First, it is unnecessary to be aware of the target beforehand and the target is in its native conformation, which inherently increases the possibility of intended application.42 Second, the whole-bacterium SELEX approach is also convenient for both separation based on centrifugation and analysis of binding affinity via flow cytometry.31,43The aptamer pools from the 12th round of SELEX were cloned and sequenced, obtaining 33 sequences totally (data shown in Table S1 in the ESI†). These sequences were then divided into six families on the basis of the homology of the DNA sequence and the similarity of the secondary structure. Analysis of all sequences revealed that many sequences contain high GC content, which indicates the secondary structure formation. Sequences were selected for further characterization according to their predicted secondary structure and free energy of formation.
Determination of affinity and specificity
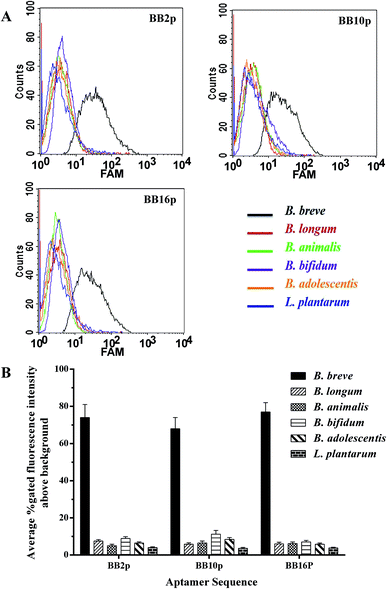
The FAM-labeled aptamers were incubated with B. breve and were then tested via flow cytometry. The results showed that the values of gated fluorescence intensity above the background of the three aptamers BB2p, BB10p and BB16p were approximately 74%, 68% and 77% respectively, which were from the aptamer pool of the 12th round of SELEX. These three aptamer sequences are shown in Table 1. Aptamer BB16p showed the greatest binding affinity with B. breve. In addition, Fig. 1 predicted the secondary structures of the aptamers, and the conditions input were 1 mM MgCl2 and the room temperature.| Name | Sequence (5′ → 3′) |
|---|---|
| a The underlined sequences are the primer binding sites. | |
| BB2p | 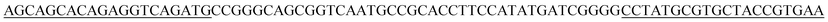 |
| BB10p | 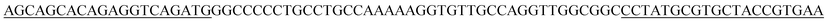 |
| BB16p | 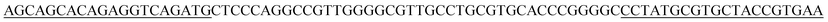 |
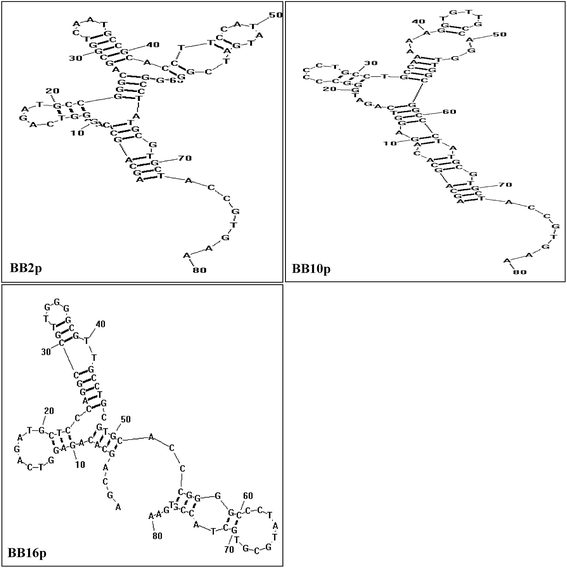 | ||
| Fig. 1 Predicted secondary structures of aptamer sequences that have high binding avidity for B. breve in SELEX. The conditions used were 1 mM MgCl2 and the room temperature. | ||
Fluorescently labeled aptamers BB2p, BB10p and BB16p were analyzed against different species of bacteria, including B. longum, B. bifidum, B. animalis, B. adolescentis and L. plantarum. These three aptamers all showed higher binding affinity for B. breve over the other species of bacteria. Although the percent gated fluorescence intensity above the background was 11.2% for BB10p when incubated with B. bifidum (Fig. 2A and B), the aptamers BB2p and BB16p showed lower binding to B. bifidum with the values of gated fluorescence intensity above the background 8.9% and 7.0% respectively (Fig. 2A and B). Thus, it can be inferred that the aptamers BB2p and BB16p are more specific for B. breve than the aptamer BB10p.
Aptamer optimization
Full-length aptamers, being with fixed primer sequences at each terminus for PCR amplification, were selected through the SELEX process. In general, not all the nucleic acids are necessary for the binding affinity between the aptamers and the targets.44 And longer sequences bring about lower yield and higher cost in synthesis. Moreover, the unnecessary nucleic acids could form intricate secondary structures that destabilize the binding affinity between the aptamers and the targets.41 Thus, in practical usage, it is necessary to obtain the minimal sequence of aptamers which showed the same or higher affinity to the target compared to the full-length aptamer.To obtain the minimal sequence of aptamers, the aptamers BB2p, BB10p and BB16p were truncated to narrow down the sequence region responsible for target binding. All the truncated aptamer variants were shown in Table 2. When the primers of the aptamers were removed, the aptamer variants BB10 and BB16 demonstrated higher binding affinity than the original full-length aptamers respectively, while aptamer variant BB2 revealed lower binding affinity compared to the aptamer BB2p (Fig. 3). Therefore, the results proved that the primer binding sites of BB2p were very important for its binding affinity for the target, while the primer sequences of BB10p and BB16p were unnecessary for their binding abilities to B. breve.
| Name | Sequence (5′ → 3′) | Length (nt) |
|---|---|---|
| BB2 | CCGGGCAGCGGTCAATGCCGCACCTTCCATATGATCGGGG | 40 |
| BB10 | GGCCCCCTGCCTGCCAAAAAGGTGTTGCCAGGTTGGCGGC | 40 |
| BB10-6f | GGCCCCCTGCCTGCCAAAAAGGTGTTGCCAGGTT | 34 |
| BB10-10f | GGCCCCCTGCCTGCCAAAAAGGTGTTGCCA | 30 |
| BB10-14f | GGCCCCCTGCCTGCCAAAAAGGTGTT | 26 |
| BB10-2r | CCCCCTGCCTGCCAAAAAGGTGTTGCCAGGTTGGCGGC | 38 |
| BB10-9r | CCTGCCAAAAAGGTGTTGCCAGGTTGGCGGC | 31 |
| BB10-13r | CCAAAAAGGTGTTGCCAGGTTGGCGGC | 27 |
| BB16 | CTCCCAGGCCGTTGGGGCGTTGCCTGCGTGCACCCGGGGC | 40 |
| BB16-11f | CTCCCAGGCCGTTGGGGCGTTGCCTGCGT | 29 |
| BB16-13f | CTCCCAGGCCGTTGGGGCGTTGCCTGC | 27 |
| BB16-15f | CTCCCAGGCCGTTGGGGCGTTGCCT | 25 |
| BB16-8r | CCGTTGGGGCGTTGCCTGCGTGCACCCGGGGC | 32 |
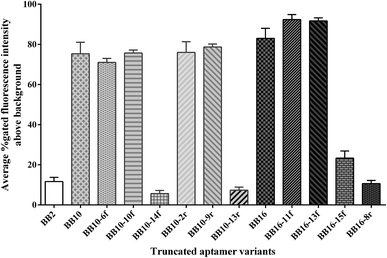 | ||
| Fig. 3 Binding affinity of the truncated aptamer variants for the target B. breve. Binding abilities of these aptamer variants to B. breve were analyzed by flow cytometric. | ||
The aptamer variants BB10 and BB16 were further stepwise truncated 6–15 nt from 3′-end or 5′-end respectively. In other words, when the nucleotides were removed from 3′-end of the aptamer variants, 5′-end remained intact bringing about the further aptamer variants BB10-6f, BB10-10f, BB10-14f, BB16-11f, BB16-13f and BB16-15f, and vice versa, when the sequences of 3′-end were removed resulting in the further aptamer variants BB10-2r, BB10-9r, BB10-13r and BB16-8r. In these ten further truncated variants, one or more G-clusters were removed from the aptamer variants BB10 and BB16 (Table 2). The results revealed that the further aptamer variants BB10-6f, BB10-10f, BB10-2r, BB10-9r, BB16-11f and BB16-13f showed higher binding affinity for B. breve than their corresponding aptamer variants BB10 and BB16 respectively, while the further truncated variants BB10-14f, BB10-13r, BB16-15f and BB16-8r demonstrated lower binding ability to B. breve compared to their corresponding aptamer variants (Fig. 3).
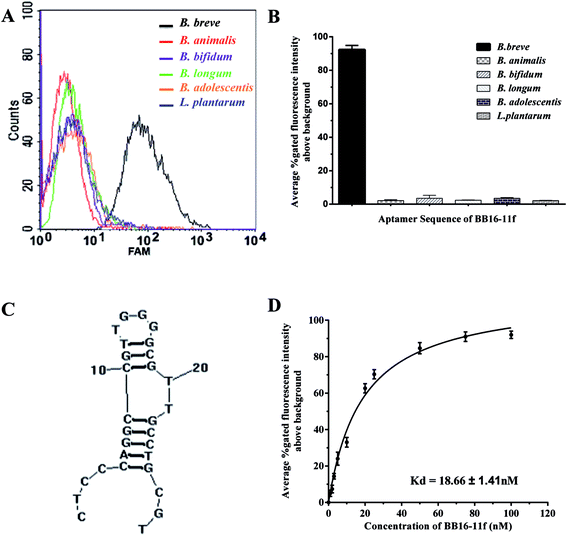
Taken together the results of the truncation experiments, the further truncated variants BB16-11f manifested highest binding affinity for its target B. breve, with the values of gated fluorescence intensity above the background about 92%, in all the aptamer variants. In the flow cytometric analysis, we did not approach 100% of aptamer-labeled B. breve cells, which could be explained in part by variation in target marker expression in the bacterial cell populations on the basis of different growth phase. This phenomenon has been demonstrated when whole-bacterium SELEX was used to select ssDNA aptamers targeted to S. aureus.33
Furthermore, Fig. 4D showed a binding saturation curve from the flow cytometric analysis of the fluorescently labeled aptamer BB16-11f binding to B. breve as described above, and the binding dissociation constant (Kd) of aptamer BB16-11f was measured by nonlinear regression analysis using GraphPad Prism 5.0. The results showed that the Kd value was 18.66 ± 1.41 nM between B. breve and the aptamer BB16-11f. Thus, the candidate aptamer BB16-11f was chosen for the further specificity analysis. The binding saturation curve between the aptamer BB16-11f and B. breve and predicted secondary structures were shown in Fig. 4. The conditions input in the prediction of secondary structures were 1 mM MgCl2 and the room temperature.
To determine the specificity of the candidate aptamer BB16-11f for the target B. breve, FAM-labelled aptamer BB16-11f was measured against a variety of other bacteria, including B. longum, B. bifidum, B. animalis, B. adolescentis and L. plantarum. As shown in Fig. 4, the aptamer BB16-11f showed preferential binding affinity for B. breve over the other bacteria tested. This preferential binding ability demonstrated the excellent specificity between the aptamer BB16-11f and the target B. breve. Therefore, the aptamer BB16-11f was selected for the subsequent practical application.
The calibration curve of B. breve detection
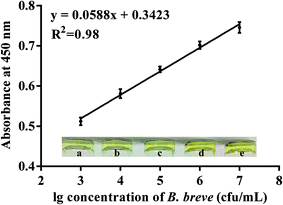
To prove the potential application of the candidate aptamer BB16-11f, the aptamer/target/aptamer sandwich type was developed. In the experiment, when the reaction of the HRP and TMB–H2O2 was terminated with H2SO4, the produced di-imine displayed a yellow color, with an absorbance peak at 450 nm. The results shown in Fig. 5 revealed that the absorbance at 450 nm ranged from 0.512 to 0.746 when the concentrations of B. breve was in the range from 103 cfu mL−1 to 107 cfu mL−1, and a good linear relationship was determined from the data between the OD values at 450 nm and the amounts of B. breve with a regression coefficient of 0.98. The limit of detection of the proposed method was estimated to be 1000 cfu mL−1.Method validation in milk samples
To verify the feasibility of colorimetric assay for practical application, we used the developed method to detect B. breve in milk, and the plate counting method was also used as control in the study. As shown in Fig. 6, we detected six different concentrations of B. breve ranging from 103 cfu mL−1 to 106 cfu mL−1, and the results indicated that the developed method had a good coherence with the plate counting method (R2 = 0.99). Therefore, the colorimetric detection method based on the aptamer BB16-11f can be used for the detection of real samples.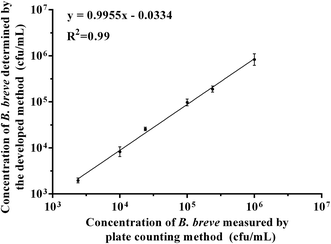 | ||
| Fig. 6 The relationship between the developed method and plate counting method in detecting B. breve in milk samples. | ||
Conclusion
In summary, the ssDNA aptamer BB16-11f with high-affinity binding and excellent specificity for B. breve was identified through a whole-bacterium SELEX process and the truncated experiments. The estimated Kd value was 18.66 ± 1.41 nM between the aptamer BB16-11f and B. breve. Furthermore, we established a colorimetric assay which did not rely on expensive instrumentation for the detection of B. breve based on the aptamer BB16-11f, and the detection method developed is simple and could be adopted to detect B. breve cells as low as 1000 cfu mL−1. In addition, this developed method was also successfully used to detect B. breve in milk samples. Therefore, the high-throughput colorimetric assay has been successfully used for detection of B. breve.Acknowledgements
This work was supported by the Program of National Natural Science Foundation for the Youth of China (No. 31501454), the Program for Changjiang Scholars and Innovative Research Team in University (IRT1249), the Program of Collaborative innovation center of food safety and quality control in Jiangsu Province, and the Program of Introducing Talents of Discipline to Universities (B07029).References
- C. Stanton, G. Gardiner, H. Meehan, K. Collins, G. Fitzgerald, P. B. Lynch and R. P. Ross, Am. J. Clin. Nutr., 2001, 73, 476–483 Search PubMed.
- T. Fujii, Y. Ohtsuka, T. Lee, T. Kudo, H. Shoji, H. Sato, S. Nagata, T. Shimizu and Y. Yamashiro, J. Pediatr. Gastroenterol. Nutr., 2006, 43, 83–88 CrossRef PubMed.
- I. Aloisio, C. Santini, B. Biavati, G. Dinelli, A. Cencič, W. Chingwaru, L. Mogna and D. Di Gioia, Appl. Microbiol. Biotechnol., 2012, 96, 1561–1576 CrossRef CAS PubMed.
- L. Mogna, M. Del Piano and G. Mogna, J. Clin. Gastroenterol., 2014, 48, S37–S39 CrossRef PubMed.
- S. G. Jeon, H. Kayama, Y. Ueda, T. Takahashi, T. Asahara, H. Tsuji, N. M. Tsuji, H. Kiyono, J. S. Ma, T. Kusu, R. Okumura, H. Hara, H. Yoshida, M. Yamamoto, K. Nomoto and K. Takeda, PLoS Pathog., 2012, 8, e1002714 CAS.
- B. Zheng, J. van Bergenhenegouwen, S. Overbeek, H. J. G. van de Kant, J. Garssen, G. Folkerts, P. Vos, M. E. Morgan and A. D. Kraneveld, PLoS One, 2014, 9, e95441 Search PubMed.
- M. Klemenak, J. Dolinšek, T. Langerholc, D. Di Gioia and D. Mičetić-Turk, Dig. Dis. Sci., 2015, 60, 3386–3392 CrossRef CAS PubMed.
- Y. Li, T. Shimizu, A. Hosaka, N. Kaneko, Y. Ohtsuka and Y. Yamashiro, Pediatr. Int., 2004, 46, 509–515 CrossRef PubMed.
- Y. Shimakawa, S. Matsubara, N. Yuki, M. Ikeda and F. Ishikawa, Int. J. Food Microbiol., 2003, 81, 131–136 CrossRef CAS PubMed.
- A. Picot and C. Lacroix, Int. Dairy J., 2004, 14, 505–515 CrossRef CAS.
- L. Masco, G. Huys, E. De Brandt, R. Temmerman and J. Swings, Int. J. Food Microbiol., 2005, 102, 221–230 CrossRef CAS PubMed.
- G. Mazzola, I. Aloisio, B. Biavati and D. Di Gioia, LWT--Food Sci. Technol., 2015, 64, 727–734 CrossRef CAS.
- M. Gueimonde, S. Delgado, B. Mayo, P. Ruas-Madiedo, A. Margolles and C. G. de los Reyes-Gavilán, Food Res. Int., 2004, 37, 839–850 CrossRef CAS.
- M. C. Collado, Y. Moreno, J. M. Cobo and M. Hernández, Eur. Food Res. Technol., 2006, 222, 112–117 CrossRef CAS.
- J. M. T. Hamilton-Miller, S. Shah and J. T. Winkler, Publ. Health Nutr., 1999, 2, 223–229 CAS.
- R. Temmerman, L. Masco, T. Vanhoutte, G. Huys and J. Swings, Appl. Environ. Microbiol., 2003, 69, 6380–6385 CrossRef CAS PubMed.
- G. Klein, A. Pack, C. Bonaparte and G. Reuter, Int. J. Food Microbiol., 1998, 41, 103–125 CrossRef CAS PubMed.
- E. Vlková, J. Nevoral, B. Jencikova, J. Kopečný, J. Godefrooij, I. Trojanová and V. Rada, J. Microbiol. Methods, 2005, 60, 365–373 CrossRef PubMed.
- R. M. Satokari, E. E. Vaughan, H. Smidt, M. Saarela, J. Mättö and W. M. de Vos, Syst. Appl. Microbiol., 2003, 26, 572–584 CrossRef CAS PubMed.
- T. Takada, K. Matsumoto and K. Nomoto, J. Microbiol. Methods, 2004, 58, 413–421 CrossRef CAS PubMed.
- T. Matsuki, K. Watanabe, J. Fujimoto, Y. Kado, T. Takada, K. Matsumoto and R. Tanaka, Appl. Environ. Microbiol., 2004, 70, 167–173 CrossRef CAS PubMed.
- P. S. Langendijk, F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson and G. W. Welling, Appl. Environ. Microbiol., 1995, 61, 3069–3075 CAS.
- E. Torres-Chavolla and E. C. Alocilja, Biosens. Bioelectron., 2009, 24, 3175–3182 CrossRef CAS PubMed.
- E. C. Alocilja and S. M. Radke, Biosens. Bioelectron., 2003, 18, 841–846 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CAS.
- S. M. Nimjee, C. P. Rusconi and B. A. Sullenger, Annu. Rev. Med., 2005, 56, 555–583 CrossRef CAS PubMed.
- A. B. Iliuk, L. Hu and W. A. Tao, Anal. Chem., 2011, 83, 4440–4452 CrossRef CAS PubMed.
- C. L. A. Hamula, J. W. Guthrie, H. Zhang, X. F. Li and X. C. Le, Trends Anal. Chem., 2006, 25, 681–691 CrossRef CAS.
- S. M. Shamah, J. M. Healy and S. T. Cload, Acc. Chem. Res., 2008, 41, 130–138 CrossRef CAS PubMed.
- F. Chen, J. Zhou, F. Luo, A. B. Mohammed and X. L. Zhang, Biochem. Biophys. Res. Commun., 2007, 357, 743–748 CrossRef CAS PubMed.
- C. L. A. Hamula, H. Zhang, L. L. Guan, X. F. Li and X. C. Le, Anal. Chem., 2008, 80, 7812–7819 CrossRef CAS PubMed.
- X. Cao, S. Li, L. Chen, H. Ding, H. Xu, Y. Huang, J. Li, N. Liu, W. Cao, Y. Zhu, B. Shen and N. Shao, Nucleic Acids Res., 2009, 37, 4621–4628 CrossRef CAS PubMed.
- H. P. Dwivedi, R. D. Smiley and L. A. Jaykus, Appl. Microbiol. Biotechnol., 2010, 87, 2323–2334 CrossRef CAS PubMed.
- C. L. A. Hamula, X. C. Le and X. F. Li, Anal. Chem., 2011, 83, 3640–3647 CrossRef CAS PubMed.
- N. Duan, S. Wu, X. Chen, Y. Huang, Y. Xia, X. Ma and Z. Wang, J. Agric. Food Chem., 2013, 61, 3229–3234 CrossRef CAS PubMed.
- L. Barthelmebs, J. Jonca, A. Hayat, B. Prieto-Simon and J. L. Marty, Food Control, 2011, 22, 737–743 CrossRef CAS.
- J. H. Park, Y. S. Cho, S. Kang, E. J. Lee, G. H. Lee and S. S. Hah, Anal. Chem., 2014, 462, 10–12 CAS.
- R. Aimaiti, L. Qin, T. Cao, H. Yang, J. Wang, J. Lu, X. Huang and Z. Hu, Appl. Microbiol. Biotechnol., 2015, 99, 9073–9083 CrossRef CAS PubMed.
- M. Jing and M. T. Bowser, Anal. Chim. Acta, 2011, 686, 9–18 CrossRef CAS PubMed.
- D. Shangguan, Z. Tang, P. Mallikaratchy, Z. Xiao and W. Tan, ChemBioChem, 2007, 8, 603–606 CrossRef CAS PubMed.
- L. Cerchia, F. Ducongé, C. Pestourie, J. Boulay, Y. Aissouni, K. Gombert, B. Tavitian, V. de Franciscis and D. Libri, PLoS Biol., 2005, 3, 0697–0704 CAS.
- D. Shangguan, Y. Li, Z. Tang, Z. C. Cao, H. W. Chen, P. Mallikaratchy, K. Sefah, C. J. Yang and W. Tan, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11838–11843 CrossRef CAS PubMed.
- S. D. Jayasena, Clin. Chem., 1999, 45, 1628–1650 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra27672e |
| This journal is © The Royal Society of Chemistry 2017 |