Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Effects of nanostructured biosilica on rice plant mechanics†
Kanako Sato a,
Noriaki Ozakib,
Kazuki Nakanishic,
Yoshiyuki Sugaharad,
Yuya Oaki
a,
Noriaki Ozakib,
Kazuki Nakanishic,
Yoshiyuki Sugaharad,
Yuya Oaki a,
Christopher Salinase,
Steven Herrerae,
David Kisailuse and
Hiroaki Imai
a,
Christopher Salinase,
Steven Herrerae,
David Kisailuse and
Hiroaki Imai *a
*a
aDepartment of Applied Chemistry, Faculty of Science and Technology, Keio University, 3-14-1 Hiyoshi, Kohoku-ku, Yokohama 223-8522, Japan. E-mail: hiroaki@applc.keio.ac.jp
bDepartment of Biotechnology, Faculty of Bioresource Sciences, Akita Prefectural University, 241-438 Kaidobata-Nishi, Nakano Shimoshinjo, Akita 010-0195, Japan
cDepartment of Chemistry, Graduate School of Science, Kyoto University, Kitashirakawa, Sakyo-ku, Kyoto 606-8502, Japan
dDepartment of Applied Chemistry, School of Advanced Science and Engineering, Waseda University, 3-4-1 Ohkubo, Shinjuku-ku, Tokyo 169-8555, Japan
eDepartment of Chemical and Environmental Engineering, Materials Science and Engineering Program, University of California at Riverside, Bourns Hall B357, Riverside, CA 92521, USA
First published on 24th February 2017
Abstract
Nanostructured amorphous silica in rice plants (biosilica or plant opal) plays an important role in plant growth related to food production. However, the same silica has a structural supporting role as well that has not been uncovered. The current study focuses on the structural design of the two main types of biosilicas in rice plants for the improvement of their mechanical properties. One structural motif is plate-like silicas, which cover most of the surfaces of leaf blades. Another is fan-shaped silicas, which are aligned inside leaf blades, providing a stiff backbone. These biosilica structures consist of 10–100 nm diameter nanoparticles. The mechanical properties, such as hardness and Young's modulus, of the biosilicas are associated with their relative density. Thus, the rice plant mechanics is inferred to be designed by changing the packing of the nanoparticles. Silica plates consisting of loosely packed particles have relatively low density and high flexibility enabling coverage of leaf blade surfaces, while fan-shaped silicas, which consist of tightly packed nanoparticles, are rigid to support the leaf blades as a backbone.
1. Introduction
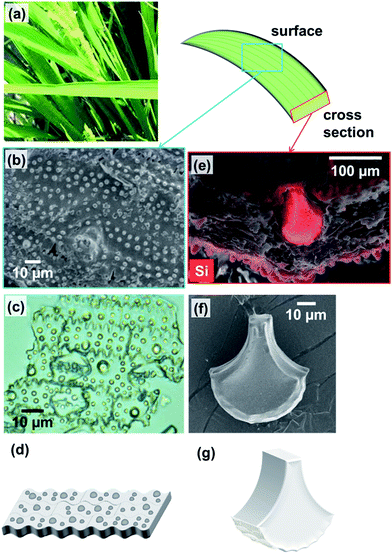
In nature, organisms produce various kinds of materials with precisely controlled in morphologies and a limited selection of elements under ambient conditions.1 The resulting materials demonstrate remarkable properties, with some outperforming synthetic materials.2–5 Amorphous silica (SiO2) is widely used in industry and is one of the essential biominerals that can be produced by organisms. Biogenic amorphous silicas (biosilicas) are widely observed in diatoms,6–10 marine sponges,11–20 and some higher plants.21–30 Although the production of synthetic silica requires significant amounts of energy, biosilicas are produced under ambient temperatures and pressures in aqueous systems near a neutral pH using organic molecules, such as silaffin,9,10,31 silicatein,20,32,33 collagen,34 and long-chain polyamines.35,36 In addition, silica frustules of diatoms and silica skeletons of marine sponges have hierarchical architectures controlled from the nano- to macroscale.8,10,12,13 Moreover, biosilicas are known to provide excellent optical and mechanical properties. For instance, the spicules from a marine sponge are superior in terms of fracture resistance12–14 and show optical properties similar to those of artificial optical fibers.15,16 Thus, many studies have been performed on the biosilicas of diatoms and marine sponges with the aim of revealing their structures, properties, and growth processes.Several kinds of higher plants, such as bamboos,23 equisetums,22,28 and rice plants,25,26 produce biosilicas (called plant opal) by absorbing silicates from soil. Rice plants accumulate a large amount of amorphous silica (20–30 wt%) on the surfaces and inside of leaf blades, stems, and husks.25,27,30 Various shapes of biosilicas, such as plates, fans, prickle hairs, and dumbbells, were observed in rice plants.30,43,44 Since rice plants cultivated on silicon-rich soil were reported to be unlikely to get diseases or fall over,24,25,29 biosilicas would be effective for the prevention against diseases and improve mechanical properties. However, the detailed structures and mechanical properties of biosilicas in rice plants have hardly been investigated.
In the present work, we performed structural and mechanical analyses of biosilicas in rice plants with the aim of clarifying the structure–mechanical property relationships. Here, we focused on two main species contained in leaf blades: silicified long cells, called silica plates, that cover most of the surfaces of leaf blades, and silicified bulliform cells, called fan-shaped silicas, which are aligned inside leaf blade, proving a stiff backbone.37 The nanostructures of biosilicas were characterized using a wide variety of techniques, including scanning electron microscopy (SEM), Fourier transform infrared absorption (FT-IR), silicon-29 solid-state nuclear magnetic resonance (NMR), mercury porosimetry, and nitrogen adsorption. In addition, variations of nanostructure–mechanical properties of biosilicas were investigated by nanoindentation. Thus, we propose the role of these varied biosilica nanostructures within rice plants.
2. Experimental
2.1. Structural characterization of biosilicas in a rice plant (Oryza sativa L.)
Mature leaf blades of rice plants (Oryza sativa L.) were collected from paddy fields in Akita, Japan. The leaf blades were ground with a small pulverizer, and biosilicas in leaf blades, which were heavier than unsilicified cells, were separated by sieving in water. The chemical structures of biosilicas were analyzed with FT-IR using the attenuated total reflection (ATR) method (Bruker Optics Equinox 55), and an energy dispersive spectroscope (EDS; FEI-SIRON) operated at 20 kV, and a fluorescent X-ray analyzer (Horiba XGT-2700) operated at 15 kV. The chemical bonding states of biosilicas were investigated using a silicon-29 solid-state NMR spectrometer (JEOL Lambda-500) at ambient temperature with an inversely gated decoupling pulse sequence. The 29Si π/2 pulses were 4 μs. The relaxation delay was 197 s for silica plates and 170 s for fan-shaped silicas. The number of scans was 1711 for silica plates and 969 for fan-shaped silicas. In order to investigate the nanostructures of biosilicas, the powdery samples of biosilicas were calcined at 300–500 °C for 3 h in air to remove organic compounds after freeze-drying for 24 h. The morphologies of biosilicas before and after the calcination were investigated using field emission SEM (FE-SEM; Hitachi S-4700, FEI-SIRON, JEOL JSM-7600F) at an accelerating voltage of 5 kV and an optical microscope (Keyence VH-Z500R). The pore-size distributions of calcined biosilicas were investigated using mercury porosimetry (Micromeritics AutoPore IV 9500) and the nitrogen adsorption method (Micromeritics 3Flex-3MP).2.2. Mechanical characterization of biosilicas
Nanoindentation maps were performed on polished oblique and transverse sections of leaf blades. Indents were performed at ambient temperature using a nanomechanical testing system (Hysitron TI 950 Triboindenter). Samples (i.e., raw leaf blades) were embedded in epoxy (Spurr's low viscosity resin, Sigma-Aldrich embedding kit). The resin blocks were polished flat using a polisher on 1200 grit silicon carbide paper, and then additional polishing was performed to reveal smoother surfaces using 0.25–6.0 μm polycrystalline diamond suspensions. Displacement-controlled indents were performed to a depth of 250 nm on the polished sections using a diamond cube corner indenter tip. Indents were repeated in a grid with a spacing of 5 μm × 5 μm or 2.5 μm × 2.5 μm. The displacement function consisted of a 5 s loading step, a 2 s hold, and a 5 s unloading step.38 Hardness and elastic modulus were calculated using the Oliver and Pherr method.393. Results
3.1. Structural characterization of biosilicas in a rice plant (Oryza sativa L.)
To investigate the nanostructures of the biosilicas, we removed organic compounds by calcination at 300 °C in air for 3 h. Based on SEM observations (Fig. 2), we found that the biosilicas were comprised of 50–100 nm diameter particles, which were loosely aggregated in silica plates but tightly packed in fan-shaped silicas. The silica plates have a porous body possessing macropores smaller than ∼200 nm. For quantitative evaluation of their porous structures, we explored the micro-, meso-, and macropores of silica plates and fan-shaped silicas using mercury porosimetry and nitrogen adsorption methods.
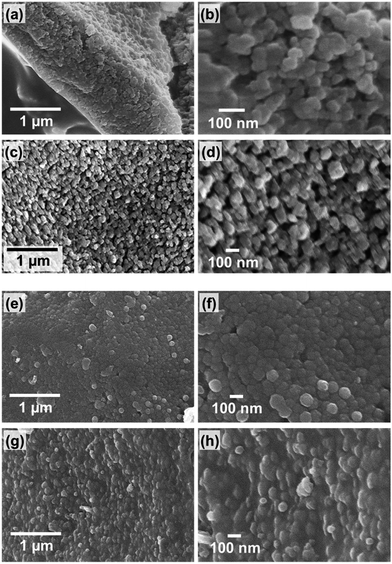 | ||
| Fig. 2 SEM micrographs of the side (a and b) and under surface (c and d) of a silica plate and of the front (e and f) and side surface (g and h) of a fan-shaped silica. | ||
Tables 1 and 2 show the pore structures of biosilicas evaluated by mercury porosimetry and nitrogen adsorption methods, respectively. Since pores larger than 200 nm were not observed in the SEM micrographs in Fig. 2b and d, the volume of pores larger than 200 nm obtained by mercury porosimetry (Fig. S1†) is ascribed to the interparticle spacing in powdered samples. The volume of macro- and mesopores between 5.5 and 200 nm in diameter was 0.19 cm3 g−1 for silica plates and negligible for fan-shaped silicas (pore volume (1) in Table 1). Based on the final amount of mercury infused at 227 MPa, the volumes of meso- and micropores smaller than 5.5 nm in diameter and closed pores were estimated to be 0.52 cm3 g−1 for silica plates and 0.36 cm3 g−1 for fan-shaped silicas (pore volume (2) in Table 1). In order to estimate the pore volume, we assumed that the density of silica matrix is the same as that of silica glass, 2.2 g cm−3. Therefore, the total pore volumes of silica plates and fan-shaped silicas were 0.71 and 0.36 cm3 g−1, respectively. We also investigated the volume of micro- and mesopores in silica plates and fan-shaped silicas using nitrogen adsorption (Fig. S2†). The volumes of open pores smaller than 5.5 nm in diameter were estimated to be 0.18 cm3 g−1 for silica plates and 0.01 cm3 g−1 for fan-shaped silicas (pore volume (3) in Table 2). The closed pore volume was evaluated from the difference of pore volume (2) and (3). Hence, the volumes of closed pores were 0.34 cm3 g−1 for silica plates and 0.35 cm3 g−1 for fan-shaped silicas (pore volume (4) in Table 2). According to adsorption isotherms of nitrogen (Fig. S2†), the Brunauer–Emmett–Teller (BET) specific surface areas of silica plates and fan-shaped silicas were 367 and 28 m2 g−1, respectively.
| Pore volume [cm3 g−1] | |||
|---|---|---|---|
| (1) Meso- & macropores (5.5–200 nm) | (2) Micro-, meso- (<5.5 nm), & closed pores | Total ((1) + (2)) | |
| a “Micro-, meso-, and macropores” are open pores. | |||
| Silica plates | 0.19 | 0.52 | 0.71 |
| Fan-shaped silicas | — | 0.36 | 0.36 |
| Pore volume [cm3 g−1] | ||
|---|---|---|
| (3) Micro- & mesopores (<5.5 nm) | (4) Closed pores ((2) − (3)) | |
| a “Micro- and mesopores” are open pores. | ||
| Silica plates | 0.18 | 0.34 |
| Fan-shaped silicas | 0.01 | 0.35 |
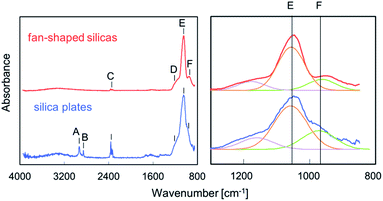
Solid-state nuclear magnetic resonance of biosilicas. Silicon-29 solid-state NMR experiments were carried out to assess the structure of silica (i.e., oxygen bridged silicon as well as silanol groups in biosilicas). Fig. 3 shows that the 29Si NMR spectra of silica plates and fan-shaped silicas are composed of three signals at about −110, −100, and −90 ppm from Q4, Q3 and Q2 structural units, respectively. Q4 stands for three-dimensionally cross-linked Si(OSi)4 units, Q3 for Si(OSi)3(OH) units and Q2 for Si(OSi)2(OH)2 units.23 Based on the NMR spectra, the OH/Si ratio of silica plates and fan-shaped silicas were estimated to be 0.36 and 0.23, respectively. This indicates that the average numbers of silanol groups per one Si atom in silica plates and fan-shaped silicas are much larger than that in artificial silica glasses and similar with other biosilicas.23 Since biosilicas are generally comprised of nanoparticles that have abundant silanol groups on their surfaces, the OH/Si ratio of biosilicas is remarkably higher than those of fused quartz and artificial silica glasses. A smaller value of the OH/Si ratio of fan-shaped silicas than that of silica plates can be explained by the condensation of silanol groups in the surface regions with tightly packing of the silica nanoparticles in their body.
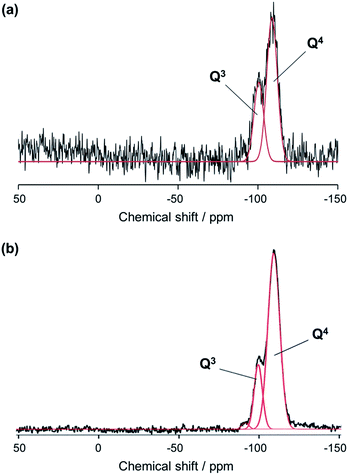 | ||
| Fig. 3 29Si NMR spectra of silica plates (a) and fan-shaped silicas (b) using two Gaussian peaks corresponding to Q4 and Q3 structural units. | ||
Fourier transform infrared absorption of biosilicas. FT-IR absorption experiments were performed to ascertain the chemical bonding state of silica plates and fan-shaped silicas. Fig. 4 shows FT-IR spectra of silica plates and fan-shaped silicas. The bands at 1050 and 950 cm−1 are assigned to the Si–O–Si stretching TO mode and Si–OH stretching mode, respectively. The intensity ratios of Si–OH and Si–O–Si bands were 0.40 for silica plates and 0.26 for fan-shaped silicas. These results agree with those obtained from the 29Si NMR spectra. The absorption bands at 2925 and 2854 cm−1, which are shown in the spectrum of silica plates but not in the spectrum of fan-shaped silica, are assigned as C–H bonds which correspond to the presence of hydrocarbons or lipids. Therefore, silica plates are suggested to contain organics, such as cellulose, in their porous body.
3.2. Mechanical properties of biosilicas
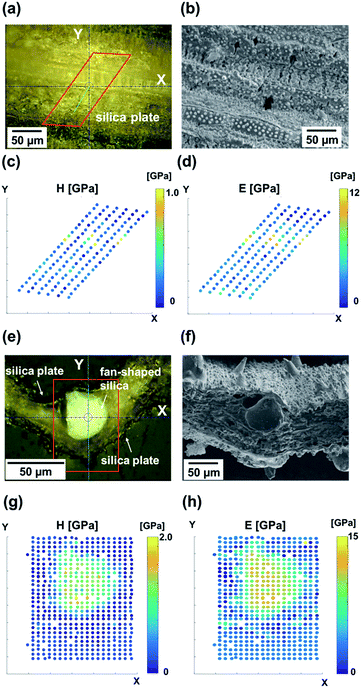
The hardness and modulus of silica plates and fan-shaped silicas were evaluated via nanoindentation experiments. Indents were performed on the oblique (Fig. 5a–c) and transverse (Fig. 5d–f) polished sections of a raw leaf blade embedded in resin.Hardness (Fig. 5c and g) and modulus (Fig. 5d and h) maps of the oblique and transverse sections of a leaf blade reveal that the hardest and stiffest portion of the leaf blade is the fan-shaped silica. On the other hand, silica plates indicated a much lower hardness and modulus than fan-shaped silicas. The presence of dumbbell-shaped silicas is indicated by regions of elevated hardness (c) and stiffness (d). The dumbbell-shaped silicas form a minor portion of the biosilicas present in the leaf blade of the rice plant, therefore we decided to focus only on the silica plates and fan-shaped silicas. Table 3 shows the hardness, Young's modulus and relative density of biosilicas, fused quartz,41 and leaves without biosilica.42 The hardness and modulus of fan-shaped silicas were H = 2.5 ± 1.0 GPa and E = 20 ± 2.4 GPa, and those of silica plates were H = 0.37 ± 0.16 GPa and E = 3.5 ± 1.3 GPa, respectively.
4. Discussion
We demonstrate schematic illustrations representing the summary of the hierarchical pore structures of biosilicas in Fig. 6. Silica plates are comprised of ∼100 nm-sized silica particles. We observed macro- and mesopores (5.5–200 nm) (pore volume (1): 0.19 cm3 g−1) and meso-, micro- (<5.5 nm) and closed pores (pore volume (2): 0.52 cm3 g−1) in the silica plates. Hence, the total pore volume, porosity, and relative density of the silica plate were 0.71 cm3 g−1, 61%, and 0.39, respectively. According to adsorption isotherms of nitrogen (Fig. S2†), the BET specific surface area of silica plates was 367 m2 g−1. Based on the specific surface area, the diameter of silica particles as a building block of the porous framework was estimated to be 7.4 nm for silica plates. This indicates that ∼100 nm silica particles consist of primary particles less than 10 nm in size in silica plates. Moreover, the primary particles contain 0.34 cm3 g−1 of closed pores (pore volume (4)). Fan-shaped silicas are also comprised of ∼100 nm particles. We observed meso-, micro- and closed pores in the fan-shaped silica, and its total pore volume, porosity, and relative density were estimated to be 0.36 cm3 g−1, 44%, and 0.56, respectively. Since the BET specific surface area of fan-shaped silicas was 28 m2 g−1, this indicates that open pores are absent in ∼100 nm silica particles in fan-shaped silicas. However, they contain 0.35 cm3 g−1 of closed pores (pore volume (4)).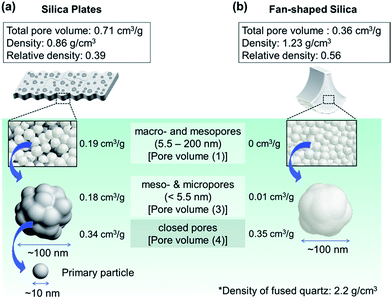 | ||
| Fig. 6 Schematic illustrations of biosilicas and their variously sized pores; (a) silica plates, (b) fan-shaped silica. | ||
Basically, the hierarchically organized biosilicas were reported to be synthesized using organic molecules, such as silaffin,9,10,31 silicatein,20,32,33 and long-chain polyamines35,36 under ambient temperatures and pressures in aqueous systems near a neutral pH. Silicon is taken up by roots in the form of silicic acid and is deposited on cell walls as a polymer of hydrated amorphous silica (i.e., biosilica).26 The unique shapes of biosilicas, such as plates and fans, can be considered to come from the shapes of the plant cells. Although most part of the mechanisms for the biosilica formation in rice plants has yet to be revealed, some sort of organic molecules might be concerned in the formation of biosilica in rice plants as well as diatoms and marine sponges. The interaction between the amino groups in the organic molecules like polyamines and silica sources (i.e., silicic acid derivatives) is considered to promote biosilicification.10,31 In previous works, polyamines were shown to catalyze the polycondensation of silanol groups45 and to act as efficient flocculation agents.46 Nano- and submicrometer-sized porous, core–shell, and hollow silica particles were synthesized using polyamines as a colloidal template.47,48 In addition, our previous work showed that silicates accumulated on a long-chain branched polyethylenimine and then transformed to bunching nanoparticles similar to the biosilica in rice plants.37 However, the production of the upper level of the hierarchical silica structure (i.e., macroscopic framework), which have larger impact on mechanical property, reflecting the shape of organic templates in macro scale has not been achieved using polyamines. Further investigation is required to clarify detailed formation mechanism of the hierarchical architectures of biosilicas from amorphous silica particles.
As mentioned in the previous work,40 Young's modulus of a synthesized amorphous silica decreases with decreasing relative density (Fig. S3†). As discussed in the Structural characterization section above, the relative density of fan-shaped silicas was 0.56, and that of silica plates was 0.39. Therefore, the estimated Young's moduli from the previous work40 (Fig. S3†) are E = 8.2 GPa (silica plates) and E = 17 GPa (fan-shaped silicas). These estimated values are not far from the experimental values: E = 3.5 ± 1.3 GPa (silica plates) and E = 20 ± 2.4 GPa (fan-shaped silicas). From 29Si NMR and FT-IR analyses, the two types of biosilicas, silica plates and fan-shaped silicas, were found to contain large amounts of silanol groups, which would provide the flexibility within the Si–O–Si networks. However, the variation of Young's modulus of the biosilicas can be mainly ascribed to the difference in relative density. This suggests that the structures within the Si–O–Si networks have little influence on their mechanical properties in biosilicas.
The differences between estimated and experimental values are relatively large for silica plates. In hierarchical architectures, the mechanical properties are strongly influenced by the macroscale structures. A relatively large deviation of the mechanical properties from the estimated values is attributed to the presence of meso- and macropores (5.5–200 nm) in the plate body in which silica particles are sparsely packed (Fig. 2b and d).
The mechanical characteristics of silica plates and fan-shaped silicas would contribute to the exhibition of different roles of the biosilicas. Silica plates, which cover all of the surfaces of the leaf blades, protect leaves from viral invasions and insect damage while ensuring flexibility of the leaves. Hence silica plates should have a certain level of hardness and relatively high flexibility (i.e., low Young's modulus). Fan-shaped silicas, which are aligned inside a leaf blade like a backbone, provide structural support to the leaves. Thus, fan-shaped silicas should have a relatively high hardness and modulus. The mechanical properties depending on the relative density of biosilicas are found to be designed by changing the packing state of silica nanoparticles.
5. Conclusions
The microstructures and nanomechanical properties of biosilicas in rice plants were investigated. Both silica plates and fan-shaped silicas consisted of amorphous hydrated silica nanoparticles. The Young's modulus of silica plates was much lower than that of fan-shaped silicas because of the difference in their relative densities originating from their microstructures. These differences found in both the structures and mechanical properties of biosilicas could explain their different roles. The ability of biosilicas to express various functions for any purpose by controlling their microstructures and their ambient synthesis process can provide insights into the design of functional and environmentally friendly silica glass in the future.Acknowledgements
The authors gratefully acknowledge assistance for mercury porosimetry and NMR measurements by Dr Kei Morisato at Kyoto University/GL Sciences Inc. and Takuro Hatanaka at Waseda University, respectively. This work was supported by Grant-in-Aid for Scientific Research (A) (16H02398) from Japan Society for the Promotion of Science.Notes and references
- M. A. Meyers, A. Y. M. Lin, Y. Seki, P. Y. Chen, B. K. Kad and S. Bodde, JOM, 2006, 58, 35 CrossRef CAS.
- U. G. K. Wegst and M. F. Ashby, Philos. Mag., 2007, 84, 2167 CrossRef.
- J. D. Curry, Science, 2005, 309, 253 CrossRef PubMed.
- J. W. C. Dunlop and P. Fratzl, Annu. Rev. Mater. Res., 2010, 40, 1 CrossRef CAS.
- L. K. Grunenfelder, S. Herrera and D. Kisailus, Small, 2014, 10, 3207 CrossRef CAS PubMed.
- T. Coradin and P. J. Lopez, ChemBioChem, 2003, 4, 251 CrossRef CAS PubMed.
- C. W. Foo, J. Huang and D. L. Kaplan, Trends Biotechnol., 2004, 22, 577 CrossRef CAS PubMed.
- R. Gordon, D. Losic, M. A. Tiffany, S. S. Nagy and F. A. S. Sterrenburg, Trends Biotechnol., 2008, 27, 116 CrossRef PubMed.
- K. Kröger and N. Poulsen, Annu. Rev. Genet., 2008, 42, 83 CrossRef PubMed.
- M. Hildebrand, Chem. Rev., 2008, 108, 4855 CrossRef CAS PubMed.
- F. Sandford, Microsc. Res. Tech., 2003, 62, 336 CrossRef CAS PubMed.
- J. C. Weaver, J. Aizenberg, G. E. Fantner, D. Kisailus, A. Woesz, P. Allen, K. Fields, M. J. Porter, F. W. Zok, P. K. Hansma, P. Fratzl and D. E. Morse, J. Struct. Biol., 2007, 158, 93 CrossRef CAS PubMed.
- J. C. Weaver, G. W. Milliron, P. Allen, A. Miserez, A. Rawal, J. P. Garay, J. Thurner, J. Seto, B. Mayzel, L. Friesen, J. B. F. Chmelka, P. Fratzl, J. Aizenberg, Y. Dauphin, D. Kisailus and D. E. Morse, J. Adhes., 2010, 86, 72 CrossRef CAS.
- W. E. G. Müller, X. Wang, K. Kropf, H. Ushijima, W. Geurtsen, C. Eckert, M. N. Tahir, W. Tremel, A. Boreiko, U. Schloßmacher, J. Li and H. C. Schröder, J. Struct. Biol., 2008, 161, 188 CrossRef PubMed.
- V. C. Sundar, A. D. Yablon, J. L. Grazul, M. Ilan and J. Aizenberg, Nature, 2003, 424, 899 CrossRef CAS PubMed.
- J. Aizenberg, V. C. Sundar, A. D. Yablon, J. C. Weaver and G. Chen, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 3358 CrossRef CAS PubMed.
- J. Aizenberg, J. C. Weaver, M. S. Thanawala, V. C. Sundar, D. E. Morse and P. Fratzl, Science, 2005, 309, 275 CrossRef CAS PubMed.
- W. E. G. Müller, X. Wang, B. Sinha, M. Wiens, H. C. Schröder and K. P. Jochum, ChemBioChem, 2010, 11, 1077 CrossRef PubMed.
- W. E. G. Müller, X. Wang, Z. Burghard, J. Bill, A. Krasko, A. Boreiko, U. Schloßmacher, H. C. Schröder and M. Wiens, J. Struct. Biol., 2009, 168, 548 CrossRef PubMed.
- H. C. Schröder, X. Wang, W. Tremel, H. Ushijima and W. E. G. Müller, Nat. Prod. Rep., 2008, 25, 455 RSC.
- D. W. Parry and F. Smithson, Nature, 1958, 181, 1549 CrossRef.
- C. C. Perry and M. A. Fraser, Philos. Trans. R. Soc., B, 1991, 334, 149 CrossRef.
- J. Klinowski, C. Cheng, J. Sanz, J. M. Rojo and A. L. Mackay, Philos. Mag. A, 1998, 77, 201 CAS.
- J. F. Ma, Soil Sci. Plant Nutr., 2004, 50, 11 CrossRef CAS.
- P. J. V. Soest, Anim. Feed Sci. Technol., 2006, 130, 137 CrossRef.
- J. F. Ma, K. Tamai, N. Yamaji, N. Mitani, S. Konishi, M. Katsuhara, M. Ishiguro, Y. Murata and M. Yano, Nature, 2006, 440, 688 CrossRef CAS PubMed.
- H. A. Currie and C. C. Perry, Ann. Bot., 2007, 100, 1383 CrossRef CAS PubMed.
- L. Sapei, N. Gierlinger, J. Hartmann, R. Nöske, P. Strauch and O. Paris, Anal. Bioanal. Chem., 2007, 389, 1249 CrossRef CAS PubMed.
- E. Epstein, Ann. Appl. Biol., 2009, 155, 155 CrossRef CAS.
- S. Neethirajan, R. Gordon and L. Wang, Trends Biotechnol., 2009, 27, 461 CrossRef CAS PubMed.
- N. Kröger, R. Deutzmann and M. Sumper, J. Biol. Chem., 2001, 276, 26066 CrossRef PubMed.
- K. Shimizu, J. Cha, G. D. Stucky and D. E. Morse, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6234 CrossRef CAS.
- W. E. G. Müller, C. Eckert, K. Kropf, X. Wang, U. Schloßmacher, C. Seckert, S. E. Wolf, W. Tremel and H. C. Schröder, Cell Tissue Res., 2007, 329, 363 CrossRef PubMed.
- H. Ehrlich, R. Deutzmann, E. Brunner, E. Cappellin, H. Koon, C. Solazzo, Y. Yang, D. Ashford, J. Thomas-Oates and M. J. Collins, Nat. Chem., 2010, 2, 1084 CrossRef CAS PubMed.
- S. A. Crawford, M. J. Higgins, P. Mulvaney and R. Wetherbee, J. Phycol., 2001, 37, 543 CrossRef.
- N. Poulsen and N. Kröger, J. Biol. Chem., 2004, 279, 42993 CrossRef CAS PubMed.
- T. Hoshino, K. Sato, Y. Oaki, A. S. Narutaki, K. Shimizu, N. Ozaki and H. Imai, RSC Adv., 2016, 6, 1301 RSC.
- L. K. Grunenfelder, E. E. Obaldia, Q. Wang, D. Li, B. Weden, C. Salinas, R. Wuhrer, P. Zavattieri and D. Kisailus, Adv. Funct. Mater., 2014, 24, 6093 CrossRef CAS.
- W. Oliver and G. Pharr, J. Mater. Res., 2004, 19, 3 CrossRef CAS.
- D. Jauffrès, C. Yacou, M. Verdier, R. Dendievel and A. Ayral, Microporous Mesoporous Mater., 2011, 140, 120 CrossRef.
- A. Miserez, J. C. Weaver, P. J. Thurner, J. Aizenberg, Y. Dauphin, P. Fratzl, D. E. Morse and F. W. Zok, Adv. Funct. Mater., 2008, 18, 1241 CrossRef CAS.
- J. Read and G. D. Sanson, New Phytol., 2003, 160, 81 CrossRef.
- S. Yamanaka, H. Takeda, S. Komatsubara, F. Ito, H. Usami, E. Togawa and K. Yoshino, Appl. Phys. Lett., 2009, 95, 123703 CrossRef.
- K. L. Gallagher, A. A. Garcia, J. Sanchez, E. O. Potma and G. M. Santos, Front. Plant Sci., 2015, 6, 753 Search PubMed.
- T. Mitzutani, H. Nagase, N. Fujiwara and H. Ogoshi, Bull. Chem. Soc. Jpn., 1998, 71, 2017 CrossRef.
- R. K. Iler, The Chemistry of Silica, Wiley, New York, 1979 Search PubMed.
- F. Zhou, S. Li, C. D. Vo, J. J. Yuan, S. Chai, Q. Gao, S. P. Armes, C. Lu and S. Cheng, Langmuir, 2007, 23, 9737 CrossRef CAS PubMed.
- M. Pi, T. Yang, J. Yuan, S. Fujii, Y. Kakigi, Y. Nakamura and S. Cheng, Colloids Surf., B, 2010, 78, 193 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra27317c |
| This journal is © The Royal Society of Chemistry 2017 |