Open Access Article
Open Access ArticleNovel anti-cancer agents based on germacrone: design, synthesis, biological activity, docking studies and MD simulations†
Lianbao Ye *a,
Jie Wua,
Weiqiang Chenb,
Yu Fenga and
Zhibing Shenc
*a,
Jie Wua,
Weiqiang Chenb,
Yu Fenga and
Zhibing Shenc
aMedicinal Chemistry of Department, Guangdong Pharmaceutical University, Guangzhou 510006, China. E-mail: yelb7909@163.com; Fax: +86-20-39352129; Tel: +86-20-39352139
bSchool of Basic Courses, Guangdong Pharmaceutical University, Guangzhou 510006, China
cSchool of Traditional Chinese Medicine, Guangdong Pharmaceutical University, Guangzhou, Guangdong 510006, China
First published on 13th January 2017
Abstract
Germacrone is a major activity component found in Curcuma zedoaria oil product, which is extracted from Curcuma zedoaria. In the present study, novel germacrone derivatives were first designed based on the theories of bioalkylating agents, synthesized, and then investigated for their inhibition effects on Bel-7402, HepG2, A549, and HeLa cells. Moreover, the study also evaluated the inhibition of these derivatives on c-Met kinase, which is expressed in a number of cancers. The results indicated that most of the compounds showed a stronger inhibitory effect on cancer cells and c-Met kinase than germacrone. Besides these findings, molecular docking studies were also carried out to analyze the results and explain the molecular mechanism of activities to c-Met kinase. Molecular dynamics simulations have been carried out for the further evaluation of binding stabilities between the compounds and their receptors.
Introduction
Curcuma zedoaria (Christm.) Roscoe, also called zedoary, is a common plant grown in tropical countries and it is used as a natural flavor, spice, and herb in Indonesia, India, and China. In China, it is traditionally used for the treatment of dyspepsia, menstrual disorders, flatulence, fever, and cough.1,2 Curcuma zedoaria extract exhibit antitumor, antimicrobial, analgesic, and anti-allergic activities.3–6 Germacrone is a major activity component found in zedoary oil product, which is extracted from Curcuma zedoaria. Germacrone (Fig. 1) has various kinds of biological activities, including antiulcer, anti-bacterial, antifungal, antitumor, antitussive, antifeedant, depressant, anti-inflammatory, hepatoprotector, vasodilator, and choleretic effects.7–10The overexpression and mutation of c-Met and its natural ligand, hepatocyte growth factor (HGF), have been found in different types of tumors including colorectal, lung, pancreatic, breast, gastric, prostate, ovarian, head and neck, glioma, hepatocellular, renal, melanoma, and a number of sarcomas; this overexpression and mutation is correlated with advanced stays and poor prognosis.11–13 Activation of the HGF/c-Met signaling pathway has been shown to lead to a wide array of cellular responses including proliferation, angiogenesis, survival, tissue regeneration, wound healing, scattering, motility, branching morphogenesis and invasion.14–17 Therefore, c-Met has become an attractive target for anti-tumor therapy.
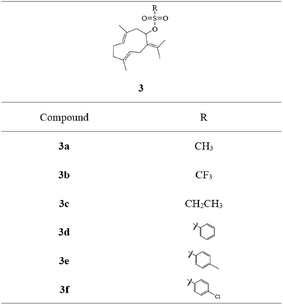
Sulfonates have been widely used as anticancer drugs. In the alkylation reaction of organic synthesis, the existence of the sulfonate group can make the C–O bond active and become a useful alkylation reaction reagent.18 Based on the design theories of bioalkylating agents and combination principles, different sulfonate esters at C-8 were introduced on germacrone to obtain novel derivatives 3a–3f (Fig. 2). These compounds might form intermediates with electrophilic groups and react with the rich electron groups of biomacromolecules (such as DNA, RNA or some important enzymes) to form the covalent binding in vivo resulting in the loss of activity of the DNA molecule, thus playing the role of an antitumor agent.18
In this study, we prepared these compounds and evaluated their inhibitions on Bel-7402, HepG2, A549, and HeLa cells. Simultaneously, the study evaluated the inhibition of these derivatives on c-Met kinase, which has been overexpressed in a number of cancers. Further, molecular docking studies analyzed the results and explained the molecular mechanism of eminent activities to c-Met kinase. Molecular dynamics simulations were applied to carry out further evaluation of the binding stabilities between compounds and their receptors.
Materials and methods
All chemicals were purchased from Aladdin or J&K Scientific Ltd. China. Germacrone (99.99%) was obtained from Chengdu Push Bio-technology Co., Ltd. Solvents were purified and dried based on standard procedures and stored over 3 Å molecular sieves. Reactions were tested by TLC using SILG/UV 254 silica-gel plates. Flash chromatography (FC) was carried out by silica gel column chromatography. 1H NMR and 13C NMR spectra were obtained using a Bruker Digital NMR Spectrometer, rep. δ in ppm, J in Hz. EI-MS was obtained using a Waters ZQ4000 instrument. Cells were obtained from the China Center for Type Culture Collection of Wuhan University; c-Met kinase was purchased from Millipore (Billerica, MA). RPMI-1640 culture medium and new-born calf serum were purchased from Gibco (Grand Island, NY), and methyl thiazolyl tetrazolium (MTT) was purchased from Amresco (Solon, OH).Synthesis of compounds
Cell assay
Inhibition effects of the synthesized compounds on Bel-7402, HepG2, A549 and HeLa cells were investigated according to the related ref. 19. The cells were incubated in RPMI 1640 complete culture medium consisting of 10% FBS, 100 U mL−1 penicillin, and 100 µg mL−1 streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. Cell growth was evaluated by MTT assay.10 Cells were inoculated at 1.0 × 104 cells per mL with 200 µL in each well of a 96-well plate and treated with 0–800 µmol L−1 of target products or 0.1% DMSO for 24 and 48 h. 20 µL of MTT was added and the mixture was incubated in the dark at 37 °C for 4 h. After the removal of MTT, cells were treated with 150 µL of DMSO. The absorbances were determined at 590 nm using an automated microplate reader (SpectraMAX190, Molecular Devices, USA).Inhibition of c-Met kinase
The inhibition of c-Met kinase was investigated as described in a reported study.20 The IC50 values were determined using TR-FRET. 6 His-tagged recombinant human c-Met residues (50 nM) 974-end (Millipore) were cultured in a medium containing 2.5 mmol L−1 MnCl2, 10 mmol L−1 MgCl2, 20 mmol L−1 Tris, 2 mmol L−1 DTT, and 0.01% Tween 20 with 5 mmol L−1 ATP and 200 nmol L−1 5FAM-KKK-SPGEYVNIGFG-NH2 at room temperature for 60 min. Compounds were tested with increasing concentrations. Reactions were terminated using IMAP stop solution. Plates were incubated overnight and analyzed using AiphaQuest.Molecular docking
Molecular docking was performed according to the related ref. 11. The docking experiment was carried out using the CDocker program, which was connected with the Accelrys Discovery Studio 2.5.5. The programs were adapted with an empirical scoring function and a patented search engine.21,22 Briefly, ligands were docked into the corresponding protein's binding site in compliance with the protocol, which was generated by ligands from the crystal structure of 3DKC with random hydrogen atoms and Gasteiger–Hückel charges, but not water. The ligands' other parameters were default values except that the threshold was 1. The structure of the receptor was minimized to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cycles using the Powell method in DS 2.5.5. The geometries of all compounds were optimized by the conjugate gradient method of TRIPOS. The convergence criterion was identified as 0.001 kcal mol−1.
000 cycles using the Powell method in DS 2.5.5. The geometries of all compounds were optimized by the conjugate gradient method of TRIPOS. The convergence criterion was identified as 0.001 kcal mol−1.
MD simulations
The MD simulations were performed on the basis of molecular docking using AMBER 10.0 for ligands and AMBER ff03 for proteins. The Gaussian 03 program was used to calculate partial atomic charges at a neutral pH, with histidines 164 and 200 protonated at the δ position. The SHAKE algorithm was used to restrict all the bonds, given the time step of 2 fs and cut-off distance of 8 Å, with long-range electrostatic interactions treated with the particle mesh Ewald (PME) method.23,24 The heating operation was carried out from 0 to 300 K in 50 ps using Langevin dynamics at a constant volume and equilibrated for 100 ps at a constant pressure of 1 atm after four steps of minimizations, which included 2500 cycles of steepest descent minimization, followed by 2500 cycles of conjugated gradient minimization. Heavy atoms of the receptor–ligand complex were restrained to 0, 10, 100, and 500 kcal (mol−1 Å2) and were 10 kcal (mol−1 Å2) during the heating and equilibration steps, whereas solvent molecules were not restricted. Finally, periodic boundary conditions of 8 ns were employed for the whole system with a normal pressure of 1 atm and normal temperature of 300 K in the production step.MM-PBSA estimation of binding free energy ΔGpred
For each system, ΔGpred values were calculated using 100 snapshots from the last 1 ns trajectory with an interval of 10 ps by the molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) method.25–28Results and discussion
Chemistry
The target compounds 2 and 3a–3f were synthesized, as described in Fig. 3. Different sulfonic acids reacted with (3E,7E)-3,7-dimethyl-10-(propan-2-ylidene) cyclodeca-3,7-dienol (2) to obtain (3E,7E)-3,7-dimethyl-10-(propan-2-ylidene) cyclodeca-3,7-dien-1-ylmethanesulfonate (3a), (3E,7E)-3,7-dimethyl-10-(propan-2-ylidene)cyclodeca-3,7-dien-1-yltrifluoromethanesulfonate (3b), (3E,7E)-3,7-dimethyl-10-(propan-2-ylidene)cyclodeca-3,7-dien-1-ylethanesulfonate (3c), (3E,7E)-3,7-dimethyl-10-(propan-2-ylidene)cyclodeca-3,7-dien-1-yl-benzenesulfonate (3d), and (3E,7E)-3,7-dimethyl-10-(propan-2-ylidene)cyclodeca-3,7-dien-1-yl-4-chlorobenzene sulfonate (3f) via acyl chloride esterification reaction with yields of 67%, 61%, 64%, 57%, and 52%, respectively. The esterification reaction provided good yields using dry THF as a solvent and pyridine as an acid-binding agent. The residual target compounds (3E,7E)-3,7-dimethyl-10-(propan-2-ylidene)cyclodeca-3,7-dien-1-yl-4-methylbenzenesulfonate (3e) were obtained via a DMAP/DCC esterification reaction with a yield of 62%. This reaction was carried out using dry DCM or acetonitrile as a solvent, DMAP as a catalyst, and DCC as a dehydrator and can be widely applied.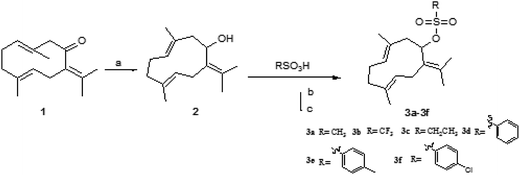 | ||
| Fig. 3 Synthesis of 3a–3f reagents and conditions: (a) LiAlH4, THF, r.t., 1 h; (b) SOCl2, DMF, Py, THF, r.t., 4 h; and (c) DMAP, DCC, r.t., 24 h. | ||
Due to the distinct difference in polarity, compounds could be easily separated. Structural modification of germacrone and a systematic study showed that 8-hydroxyl group may require further optimization studies and a range of substituents could be suitable for this position.
Evaluation of biological activity
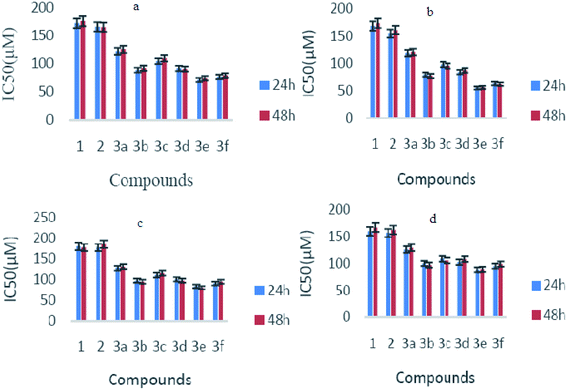
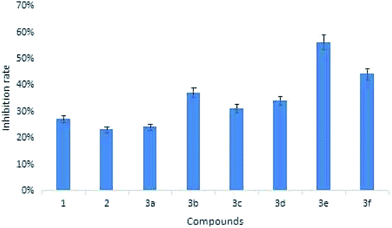
Biological activities were investigated by checking the effect of germacrone and its derivatives on the growth of Bel-7402 cells, HepG2 cells, A549, and HeLa cells. These cells were treated with germacrone and its derivatives at the concentrations of 12.5 µmol L−1, 25 µmol L−1, 50 µmol L−1, 100 µmol L−1, 200 µmol L−1, 400 µmol L−1, and 800 µmol L−1, respectively. An MTT assay was then applied at 24 h and 48 h. As shown in Table 1, the proliferations of HepG2, Bel7402, A549, and Hela cells were inhibited in a dose-dependent manner, and the cytolytic activity was markedly inhibited at the same time. Moreover, the IC50 values of germacrone against Bel-7402 cells, HepG2 cells, A549, and HeLa cells were about 173.54 µM, 169.52 µM, 179.97 µM, and 160.69 µM. The growth-inhibitory effect had no significant differences after incubation with germacrone and derivatives for 24 h and 48 h (Fig. 4); thus, there was no significant influence on the anti-proliferation effect of compounds on Bel-7402 cells, HepG2 cells, A549, and HeLa cells with an extended treatment time after 24 h. This might be caused by the saturation of intracellular drug concentration within 24 hours. Compounds 3e and 3f, in which R was substituted with benzene, showed high inhibition and 3e showed highest activity against HepG2 with an IC50 value of 56.22 µM. The results of inhibition on c-Met showed that these derivatives exhibited a stronger inhibitory effect than germacrone (Fig. 5). Since the results suggested that the derivatives have good activity, we could initially confirm that the designed compounds might be well worth investigation on the basis of molecular docking, molecular dynamics simulations, and preliminary activity tests. In conclusion, our strategy to design germacrone derivatives was successful in obtaining the desired compounds, which demonstrated a stronger inhibitory effect on cancer cells and c-Met kinase. These compounds might be useful as lead structures for the development of anti-cancer agents, and this study could supply important information for building a chemical library and the basis for further research.| Compound | Bel-7402 IC50 (µM) | HepG2 IC50 (µM) | A549 IC50 (µM) | HeLa IC50 (µM) | ||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| 1 | 173.54 | 177.21 | 169.52 | 174.56 | 179.97 | 177.21 | 160.69 | 167.78 |
| 2 | 167.08 | 166.32 | 155.20 | 161.54 | 177.21 | 185.24 | 157.31 | 163.15 |
| 3a | 122.23 | 125.68 | 119.09 | 121.08 | 127.68 | 131.45 | 125.96 | 129.44 |
| 3b | 88.87 | 92.44 | 79.98 | 77.80 | 97.74 | 95.52 | 100.25 | 97.28 |
| 3c | 105.05 | 110.14 | 98.54 | 95.65 | 111.28 | 115.69 | 108.97 | 105.88 |
| 3d | 92.00 | 91.02 | 84.67 | 87.74 | 101.35 | 97.74 | 102.80 | 108.36 |
| 3e | 71.78 | 74.68 | 56.22 | 57.18 | 84.47 | 81.00 | 88.64 | 89.26 |
| 3f | 77.04 | 79.45 | 64.12 | 62.99 | 91.25 | 95.65 | 95.24 | 99.11 |
Docking studies
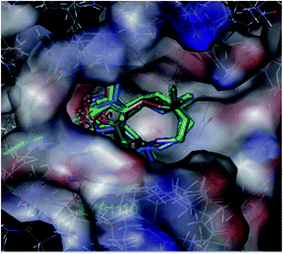
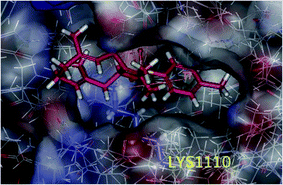
In our previous study, we performed docking studies using several PDB proteins including 3DKF, 2WGJ, 2RFS, and 3DKC, in which only 3DKC interacted with germacrone and its derivatives owing to sesquiterpene structures of these compounds; therefore, we chose this compound to perform the docking experiments. The 3DKC is the crystal structure of c-Met kinase in complex with ATP and its details can be obtained from http://www.rcsb.org/pdb/explore/materialsAndMethods.do?structureId=3DKC, which is widely used.29–31Molecular docking suggested that these compounds were bound to the active site of the protein 3DKC (shown in Fig. 6). The binding energies of the complexes between each compound (1–3f) and the active site of the receptor are shown in Table 2. The binding energies were higher when R was a benzene ring and the compounds showed higher inhibition, of which a substituted benzene ring had stronger binding energies and inhibition. The interaction between the (E,E)-1,5-cyclodecadiene system and the protein crystal 3DKC seems to be strong, and the binding forces were mainly hydrophobic forces, whereas another clear interaction was the σ–π interaction between the benzene ring with LYS1110 and GLY1087. Compound 3e showed the highest activity owing to the π–π stacking interaction of the benzene ring and hydrophobic interaction of methyl, as shown in Fig. 7.
MD simulations and ΔGpred calculation
Building on the structural data from diverse experimental sources, today's MD simulations permit the exploration of biological processes in an unparalleled detail. Since molecular docking studies reported a probably momentary binding mode, which might be unreasonable and unstable, molecular dynamics simulations were applied to carry out further research of the binding stabilities between the compounds (1–3f) and 3DKC.The crystal structures of the 3DKC complex with 1–3f were used to investigate the reliability of molecular dynamics simulations. The root mean square deviation (RMSD) curve and the surface area curve of the 1 ns MD simulation indicated that the structure of 3e was in a relatively stable state after 1 ns and the MD trajectories were quite smooth (shown in Fig. 8), whereas other compounds were unstable with uneven MD trajectories (as shown in ESI†). The MD parameters were suitable for the MD simulations.
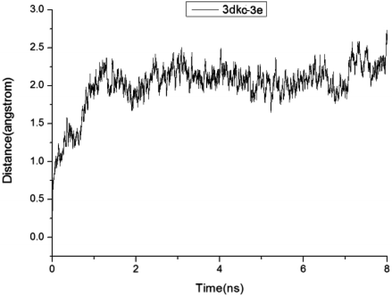 | ||
| Fig. 8 Plots of RMSD for all of the backbone atoms (Å) vs. simulation time (ns) for 3DKC in complex with 3e. | ||
Given identical MD conditions, the two 3DKC–ligand complexes were used to perform the molecular dynamics simulations for eight nanoseconds. The RMSD curves of all compounds were stable during the MD simulations. In the experiment, ΔGpred values of less than −25 kcal mol−1 were selected and used to evaluate 3DKC inhibitory activity. The predicted ΔGpred value of 3e is −28.36 kcal mol−1, whereas the predicted ΔGpred values of other compounds were more than −25 kcal mol−1 (shown in Table 3). To save the computational time and reduce the computational cost of the MD-based screening studies, we did not consider the entropy contribution because of the extremely time-consuming nature of the normal mode calculation of entropy for large systems. Therefore, the calculated ΔGpred values in the present study were inadequate owing to the omission of entropy contribution.
Conclusions
This study designed and synthesized novel bioactivity compounds using the natural product germacrone as the lead compound based on the design theory of alkylating agents. The results showed that the majority of compounds had moderate inhibition on cancer cells and c-Met kinase. These compounds can be further studied as antitumor lead structures to lay the foundation for developing anticancer compounds with clinical value. The results of molecular docking and molecular dynamics simulations were very important in explaining the molecular mechanism of eminent activities to c-Met kinase and binding stabilities between the compounds and their receptors. Moreover, these results can provide a theoretical basis for further research on c-Met inhibitors.Acknowledgements
This project was funded by the Special Fund of Applied Research and Development of Guangdong Provincial Department of Science and Technology (2015B020234009), Special Fund for TCM supported by the State Administration of Traditional Chinese Medicine of China (201507004), Joint Natural Sciences Fund of The Department of Science and Technology and first Affiliated Hospital of Guangdong Pharmaceutical University (No. GYFYLH201317), Guangdong Natural Science Foundation (2015A030313586), and Science and Technology Planning Project of Guangdong Province (Guangdong Branch Regulations [2015]110, 2016A020215159). The authors are grateful to Mr Wei and Lanzhou University for molecular docking experiments and MD Simulations.References
- W. Chen, Y. Liu, M. Gao, J. Wu, A. Wang and R. Shi, Antiangiogenesis effect of essential oil from Curcuma zedoaria in vitro and in vivo, J. Ethnopharmacol., 2011, 133(1), 220–226 CrossRef CAS PubMed.
- C. Chen, Y. Chen and Y. His, et al. Chemical Constituents and Anticancer Activity of Curcuma zedoaria Roscoe Essential Oil against Non-Small Cell Lung Carcinoma Cells in Vitro and in Vivo, J. Agric. Food Chem., 2013, 61(47), 11418–11427 CrossRef CAS PubMed.
- W. Seo, J. Hwang and S. Kang, et al. Suppressive effect of Zedoariae rhizoma on pulmonary metastasis of B16 melanoma cells, J. Ethnopharmacol., 2005, 101(3), 249–257 CrossRef PubMed.
- N. Dde, M. Souza and R. Neto, et al. Phytochemical analysis and analgesic properties of Curcuma zedoaria grown in Brazil, Phytomedicine, 2002, 9(5), 427–432 CrossRef.
- B. Wilson, J. Abraham and V. Manju, et al. Antimicrobial activity of Curcuma zedoaria and Curcuma malabarica tubers, J. Ethnopharmacol., 2005, 99(1), 147–151 CrossRef CAS PubMed.
- H. Matsuda, S. Tewtrakul, T. Morikawa, A. Nakamura and M. Yoshikawa, Antiallergic principles from Thai zedoary: structural requirements of curcuminoids for inhibition of degranulation and effect on the release of TNF-α and IL-4 in RBL-2H3 cells, Bioorg. Med. Chem., 2004, 12(22), 5891–5898 CrossRef CAS PubMed.
- Y. Dang, X. Li, Q. Zhang and Y. Wang, Preparative isolation and purification of six volatile compounds from essential oil of Curcuma wenyujin using high-performance centrifugal partition chromatography, J. Sep. Sci., 2010, 33(11), 1658–1664 CrossRef CAS PubMed.
- W. Cho, J. Nam and H. Kang, et al. Zedoarondiol isolated from the rhizome of Curcuma heyneana is involved in the inhibition of iNOS, COX-2 and pro-inflammatory cytokines via the downregulation of NF-kappa B pathway in LPS-stimulated murine macrophages, Int. Immunopharmacol., 2009, 9(9), 1049–1057 CrossRef CAS PubMed.
- X. Xiao, Y. Zhao and H. Yuan, et al. Study on the effect of Rhizoma curcuma Longa on gastrin receptor, J. Chin. Mater. Med., 2002, 25(3), 184–185 Search PubMed.
- Y. Liu, W. Wang and B. Fang, et al. Anti-tumor effect of germacrone on human hepatoma cell lines through inducing G2/M cell cycle arrest and promoting apoptosis, Eur. J. Pharmacol., 2013, 698(2), 95–102 CrossRef CAS PubMed.
- L. Ye, Y. Tian and Z. Li, et al. Design, synthesis and molecular docking studies of some novel spiro[indoline-3,4′-piperidine]-2(1H)-ones as potential c-Met inhibitors, Eur. J. Med. Chem., 2012, 50, 370–375 CrossRef CAS PubMed.
- P. Ma, T. Kijima and G. Maulik, et al. c-Met mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions, Cancer Res., 2003, 63(19), 6272–6281 CAS.
- P. Ma, S. Jagadeesh and R. Jagadeeswaran, et al. c-Met expression/activation, functions, and mutations in non-small cell lung cancer, Proc. Am. Assoc. Canc. Res., 2004, 45, 1875 Search PubMed.
- D. Renzo, M. Olivero and T. Martone, et al. Somatic mutations of the MET oncogene are selected during metastatic spread of human HNSC carcinomas, Oncogene, 2000, 19(12), 1547–1555 CrossRef PubMed.
- J. Lee, S. Han and H. Cho, et al. A novel germ line juxtamembrane Met mutation in human gastric cancer, Oncogene, 2000, 19(43), 4947–4953 CrossRef CAS PubMed.
- F. Yang, S. Praveena and C. Patrick, Met signaling: novel targeted inhibition and its clinical development in lung cancer, J. Thorac. Oncol., 2012, 7(2), 459–467 CrossRef PubMed.
- M. Li, P. Yang and Y. Liu, et al. Low c-Met expression levels are prognostic for and predict the benefits of temozolomide chemotherapy in malignant gliomas, Sci. Rep., 2016, 6, 1–10 CrossRef PubMed.
- T. L. Lemke, D. A. Williams and V. F. Roche, et al., Foye's principles of medicinal chemistry, Wolters Kluwer: Lippincott William and Wilkins, 7th edn, 2013 Search PubMed.
- W. Chen, Y. Liu and M. Li, et al. Anti-tumor effect of α-pinene on human hepatoma cell lines through inducing G2/M cell cycle arrest, J. Pharmacol. Sci., 2015, 3, 332–338 CrossRef PubMed.
- J. Porter, S. Lumb and R. Franklin, et al. Discovery of 4-azaindoles as novel inhibitors of c-Met kinase, Bioorg. Med. Chem. Lett., 2009, 19(10), 2780–2784 CrossRef CAS PubMed.
- A. Jain, Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine, J. Med. Chem., 2003, 46(4), 499–511 CrossRef CAS PubMed.
- A. Jain, Surflex-Dock2.1: robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search, J. Comput.–Aided Mol. Des., 2007, 21(5), 281–306 CrossRef CAS PubMed.
- Z. Li, Y. Cai and Y. Cheng, et al. Identification of Novel Phosphodiesterase-4D Inhibitors Prescreened by Molecular Dynamics Augmented Modeling and Validated by Bioassay, J. Chem. Inf. Model., 2013, 53(4), 972–981 CrossRef CAS PubMed.
- M. Shuichi and P. Kollman, Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models, J. Comput. Chem., 1992, 13(8), 952–962 CrossRef.
- I. Massova and P. Kollman, Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding, Perspect. Drug Discovery Des., 2000, 18(1), 113–135 CrossRef CAS.
- T. Hou, J. Wang, Y. Li and W. Wang, Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations, J. Chem. Inf. Model., 2011, 51(1), 69–82 CrossRef CAS PubMed.
- M. Liu, M. Yuan and M. Luo, et al. Binding of curcumin with glyoxalase I: molecular docking, molecular dynamics simulations, and kinetics analysis, Biophys. Chem., 2010, 147(1–2), 28–34 CrossRef CAS PubMed.
- L. He, F. He and H. Bi, et al. Isoform-selective inhibition of chrysin towards human cytochrome P450 1A2. Kinetics analysis, molecular docking, and molecular dynamics simulations, Bioorg. Med. Chem. Lett., 2010, 20(20), 6008–6012 CrossRef CAS PubMed.
- S. G. Buchanan, J. Hendle and P. S. Lee, et al. SGX523 is an exquisitely selective, ATP-competitive inhibitor of the MET receptor tyrosine kinase with antitumor activity in vivo, Mol. Cancer Ther., 2009, 8(12), 3181–3190 CrossRef CAS PubMed.
- J. G. Christensen, R. J. Schreck and P. Kuruganti, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo, Cancer Res., 2003, 63(21), 7345 CAS.
- L. Wang, J. Ai and Y. Shen, et al. SOMCL-863, a novel, selective and orally bioavailable small-molecule c-Met inhibitor, exhibits antitumor activity both in vitro, and in vivo, Cancer Lett., 2014, 351(1), 143–150 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra26944c |
| This journal is © The Royal Society of Chemistry 2017 |