Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Silica sub-microspheres induce autophagy in an endocytosis dependent manner†
Dengtong Huang,
Hualu Zhou,
Xuanqing Gong and
Jinhao Gao *
*
The Key Laboratory for Chemical Biology of Fujian Province, State Key Laboratory of Physical Chemistry of Solid Surfaces, iChEM, Department of Chemical Biology, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, China. E-mail: jhgao@xmu.edu.cn
First published on 21st February 2017
Abstract
The health risk of exposure to manufactured nano- and submicro-materials leads to an increasing effort to explore their biological effects and potential toxicity in detail. Here, we show that silica sub-microspheres (0.1 to 2.1 μm in diameter), the major component of dust or particulate matter less than 2.5 μm (PM 2.5), induce autophagy depending on the levels of cellular endocytosis. Due to the suitable size for cellular endocytosis, 0.5–0.7 μm silica particles induce the highest levels of autophagy among particles from 0.1 to 2.1 μm in diameter. Changes in cellular endocytosis of silica sub-microspheres lead to alteration of autophagy levels. Furthermore, dephosphorylation of FOXO3A and subsequent translocation to the nucleus may be associated with this autophagy process. Our results reveal the manner in which silica sub-microspheres induce autophagy, emphasize the potential risk of endocytosis of fine particles or other non-degradable materials, and suggest a new signaling pathway involved with autophagy induced by sub-micromaterials.
1. Introduction
A large variety of functional materials have been developed for better life. Due to the small size, submicro- and nano-materials possess unique physical and chemical properties that differ from the corresponding bulk materials, which endow them with great potential applications in biomedicine.1–3 By the same token, submicro- and nano-materials also display unexpected toxicity, especially the latter, which have a higher surface-to-volume ratio. An emerging discipline “nanotoxicology” is developing rapidly to cope with potential hazards of nanomaterials.4 In contrast, sub-micromaterials are considered less hazardous due to their larger size and lower surface-to-volume ratio than nanomaterials. Nevertheless, sub-micromaterials can be harmful to humans as well. For example, particulate matter less than 2.5 μm (PM 2.5), which is one of the major airborne pollutants, can cause many chronic diseases.5 Moreover, nanomaterials tend to aggregate to sub-microscale due to the destruction of electrostatic stabilization by electrolytes.6,7 We hypothesize that the results are highly implicated in sub-microscale aggregates rather than individual nanoparticles. Therefore studying the biological effects of sub-micromaterials in depth is of great importance.Autophagy is a basic biological process by which cells maintain normal functions, respond to stress, and protect survival, through degrading unnecessary and harmful inner components.8 A great diversity of nanomaterials can stimulate autophagy induction due to their toxicity.9 In contrast, the autophagic effect of sub-micromaterials receives little attention for a long time. This may be due to a habitual thought that sub-micromaterials have little toxicity compared to nanomaterials so that aggregation of nanomaterials would reduce the cytotoxicity.10,11 However, iron oxide and silica nanomaterials, which have good biocompatibility and wide applications in biomedicine fields,12–14 are able to induce high level of autophagy when they aggregate to sub-microscale size.15 This phenomenon suggests that sub-micromaterials with low cytotoxicity may induce autophagy as well.
To verify this assumption, we select silica sub-microspheres as the testing material because they are widely used in biomedical fields due to their good biocompatibility.16–19 In addition, silica is one of the primary components of PM 2.5 and widely distributed in nature. It is also very important that silica particles with diameters in a range from nanometer to micrometer are easy to synthesize. In this study, we fabricated a series of uniform silica sub-microspheres with diameters from 0.1 to 2.1 μm for investigation of autophagic effect. We found that silica sub-microspheres with various sizes induced autophagy to different extents depending on the levels of endocytosis.
2. Experimental
Synthesis and modification of silica sub-microspheres
Silica sub-microparticles were synthesized through hydrolysis of tetraethylorthosilicate (TEOS) in aqueous solution containing arginine. The resulting silica nanoparticles were used as seeds to regrow via Stöber process.20 Surface modification with carboxylate or PEG was performed in alkaline solution while modification with amino groups was performed in acid solution. The detailed procedure is provided in ESI.†Cell culture
HeLa cells were purchased from cell bank of Chinese Academy of Sciences. HeLa-GFP-LC3 cells stably expressing GFP-LC3 were constructed using lentivirus as described previously.15 All the cells were cultured with Minimum Essential Medium (MEM) supplemented 10% fetal bovine serum (FBS) and maintained in a humidified atmosphere with 5% CO2 at 37 °C.Western blot
After treated, cells were harvested by trypsin and centrifugation. Proteins were extracted using radio immune precipitation assay (RIPA) lysis buffer containing 1% protein inhibitor cocktail. After separated in SDS-PAGE, proteins were transferred to 0.45 μm PVDF membrane. The PVDF membrane was blocked in 5% milk for 30 min and then incubated with primary antibody for 2 h and secondary antibody for 1 h. Signal was developed with chemiluminescence substrate and acquired in chemiluminescence imaging systems (C-DiGit, Licor). For detailed procedure, please refer to our previous publication.15 The antibodies are listed in ESI.†Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde, permeabilized with methanol and blocked with goat serum. Then cells were stained with primary antibody at 4 °C overnight and secondary antibody at room temperature for 1 h. Images were acquired at confocal microscope (Leica TCS SP5). For more detailed procedure, please refer to our previous publication.15 Antibodies are listed as follows: anti-LAMP1 (1/100, Cell Signaling Technology, #9091), anti-FOXO3A (1/100, Abcam, ab53287), goat anti-rabbit IgG (H + L) DyLight 633 Conjugated (1/200, Pierce, Thermo scientific).Flow cytometry measurement
Cells were collected by trypsin and centrifugation after treatment and staining. Subsequently, cells were re-suspended in cold PBS and placed in ice bath until measurement. Data were acquired in BD FACSAria II cytometry. 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were recorded for analysis. The first sample was re-measured at the end of measurement to confirm the stability of signal. Cyto-ID Autophagy Detection Kit (Enzo Life Science) was used to measure autophagic vacuoles according to the instruction manual from manufacturer. To avoid interference, we used HeLa cells that did not express GFP-LC3.
000 events were recorded for analysis. The first sample was re-measured at the end of measurement to confirm the stability of signal. Cyto-ID Autophagy Detection Kit (Enzo Life Science) was used to measure autophagic vacuoles according to the instruction manual from manufacturer. To avoid interference, we used HeLa cells that did not express GFP-LC3.
Statistical methods
Mean value and standard deviation (S.D.) were calculated from at least three independent repeated experiments. P values were calculated using student's t test methods. P value < 0.05 was considered statistically significant.3. Results and discussion
Size effect of silica sub-microspheres in autophagy induction
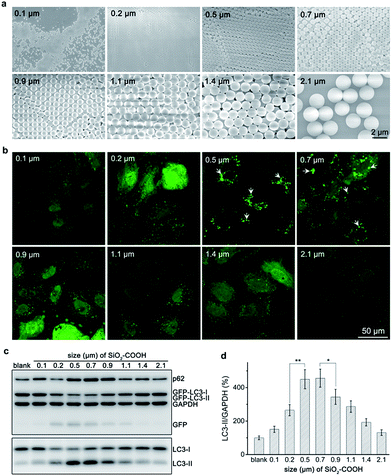
Carboxylated silica sub-microspheres with diameters ranging from 0.1 μm to 2.1 μm (Fig. 1a) were synthesized using seed-mediated growth and Stöber method.20,21 XRD spectra confirmed their amorphous structure (Fig. S1†). After sterilized and purified, silica sub-microspheres were tested their autophagic effect. We found that particles with low dose (60 μg mL−1) can induce considerable accumulation of LC3-II, a marker of autophagosome,22 until 12–24 h (Fig. S2†). Thus, we chose 60 μg mL−1 and 24 h as the test condition and found that particles with different sizes showed distinct ability to induce autophagy. Cells treated with 0.5–0.7 μm silica particles displayed many GFP-LC3 (green fluorescent protein fused with microtubule-associated proteins 1 light chain 3) fluorescent puncta (Fig. 1b) and high expression levels of autophagy related proteins (e.g. LC3-II, GFP-LC3-II and GFP)23 (Fig. 1c and d), indicating high-level induction of autophagy. However, when the particle size was smaller than 0.5 μm or larger than 0.7 μm, autophagic level decreased. Cells treated with 0.1 μm or 2.1 μm silica particles had negligible change in GFP-LC3 puncta or expression levels of autophagy related proteins compared to the control group. These results confirm the ability of silica sub-microspheres to stimulate autophagy induction and suggest 0.5–0.7 μm as a suitable size for high-level induction of autophagy.When keep mass concentration the same, particles with various sizes would have dramatically different number concentrations, and vice versa. Thus defining the dose of particles by mass concentration or number concentration is controversial,24,25 which makes it difficult to standardize the test conditions. Mass concentration is widely accepted for its convenient usage. Nevertheless, we also tested the size effect of silica sub-microspheres at the same number concentration. Western blot revealed that 0.7 μm silica particles induced more LC3-II accumulation (Fig. S3†) than 0.9 μm or larger particles. As the numbers of particles were the same, so the sizes of sub-microscale particles play a key role in causing autophagy to various extents. Considering both mass concentration and number concentration, we suggest 0.5–0.7 μm silica particles as a suitable size for high-level induction of autophagy. Due to the wide usage of mass concentration, we still used 0.5 μm silica sub-microspheres as the typical sample and 60 μg mL−1 as the testing dose.
Positive correlation relationship between autophagic level and endocytic level
We propose that cellular endocytosis may be an important factor because the formation of sub-microscaled aggregation facilitates the endocytosis of nanomaterials and subsequent autophagy induction.15 Transmission electron microscope (TEM) imaging revealed that 0.5 μm silica particles had the highest endocytic level by number (Fig. 2a and b), followed by 0.2 μm, 0.7 μm, and 0.9 μm particles. 1.1 μm or larger sub-microspheres had a low endocytic level. This is in accordance with their autophagic effects except for 0.2 μm particles, which may be explained by their low endocytic level by weight. Scanning electron microscope (SEM) imaging results further confirmed the different endocytosis levels of various-sized silica sub-microspheres. 0.5–0.7 μm silica particles had the highest cellular endocytic levels, followed by 0.2 μm and 0.9 μm particles, while 1.1 μm or 0.1 μm particles had little endocytosis (Fig. S4†). Despite the slight difference, TEM and SEM images suggest that cells internalize more 0.5–0.7 μm silica particles than silica sub-microspheres with other sizes. This may be the reason that these particles induce a high level of autophagy. Ideally, it is better to quantify the endocytic level accurately. The common practice is to wash the cells with phosphate buffered saline (PBS) for three times, detach and then digest, followed by inductively coupled plasma mass spectrometry (ICP-MS) analysis. The resulting data, however, are derived from extracellular particles as well as intracellular particles. The extracellular particles, whether adhered on the surface of cells or the substrate of culture dishes, are difficult to wash out, and thus make a contribution to final results. Hence we cannot get the reliable endocytic level of particles in this way. Fluorescent labeling and quantitation can be useful for comparison among particles of the same size, but not particles with different sizes. So we chose to estimate the endocytic levels of different sized silica sub-microspheres by TEM imaging and found 0.5–0.7 μm was the suitable size for cellular endocytosis (Fig. 2a). This result suggests the positive correlation relationship between autophagic level and endocytic level.To explore the detailed relationship between endocytosis and autophagy, we investigated their induction processes. Endocytic levels at different time points could be quantified by flow cytometry using fluorescent silica sub-microspheres. Since there is only one type of particles, and the fluorescent particles in solution have negligible interference with cells (Fig. S5†), a reliable quantitative result of endocytic level is attainable. Autophagic level was quantified by flow cytometry using Cyto-ID™ Autophagy Detection Kit.15 This commercial kit is special for autophagic vacuoles with negligible staining of lysosomes.26 Quantitative analysis revealed that autophagy induction was quite slowly when cells had a low endocytic level at first 6 h (Fig. 2c). After the internalized particles accumulated to a certain amount at 6–12 h, cells accelerated autophagy induction. However, when the uptake of particles was slow at 12–24 h, autophagy induction was decelerated (Fig. 2c). This result indicates that the endocytic level of silica sub-microspheres may be decisive to the autophagic level.
Tuning autophagy by modulation of endocytosis
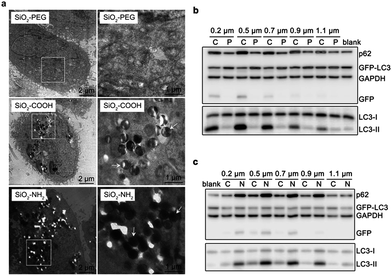
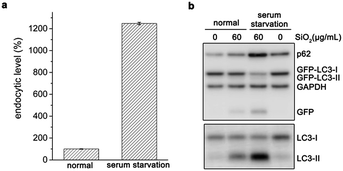
To further confirm their relationship, we varied the cellular endocytic levels of silica sub-microspheres by surface modification. Polyethylene glycol (PEG) can reduce the nonspecific interaction between materials and cells.27,28 It was difficult for silica sub-microspheres coated with PEG to enter cells (Fig. 3a and S6†). Subsequently, the autophagic effect was diminished considerably (Fig. 3b). In contrast, surface modification with amino groups promoted endocytosis of silica sub-microspheres (Fig. 3a and S6†) and then substantially enhanced autophagy induction (Fig. 3c). Furthermore, 0.5–0.7 μm silica particles still had the highest autophagic levels. We speculate that 0.5–0.7 μm may be a critical size for silica sub-microspheres to induce autophagy or other biological effects.Besides surface modification, serum diminishes the interaction between particles and cells, and thus reduces uptake and cytotoxicity of nanoparticles29–31 or sub-microspheres.32 When cells underwent serum starvation for 4 h, the endocytic level of silica sub-microspheres dramatically increased (Fig. 4a). During the following period when cells were cultured in fresh complete medium to eliminate the effects of starvation, the large amount of intracellular silica sub-microspheres substantially enhanced autophagic effect (Fig. 4b and S7†). It is noted that short-term serum starvation alone induced negligible autophagic effect (Fig. 4b). These results strongly indicate that autophagic level is dependent on the endocytic level of silica sub-microspheres.
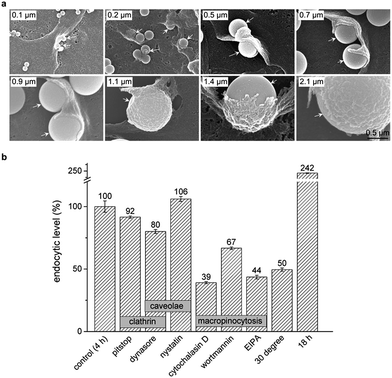
To clarify the detailed process that internalized silica sub-microspheres induce autophagy, we further explore the endocytic pathway and cellular trafficking of silica sub-microspheres. The most extensively studied endocytic pathways are phagocytosis, clathrin mediated endocytosis, caveolin mediated endocytosis and macropinocytosis.33 Phagocytosis is typically restricted to special cells such as phagocytes. The cargos are usually less than 0.3 μm in clathrin mediated endocytosis and less than 80 nm in caveolae mediated endocytosis. Cargos larger than 1 μm can be internalized by non-phagocytic cells via macropinocytosis.34 We used SEM to monitor the endocytic process of silica sub-microspheres and observed the typical structure of macropinocytosis. Cells extended the membrane ruffles to engulf silica sub-microspheres. Then the ruffles fused back with the plasma membrane and encapsulated silica sub-microspheres into macropinosomes (Fig. 5a).
We further found that the endocytosis of silica sub-microspheres could be effectively impeded by low temperature (30 °C) and inhibitors of macropinocytosis such as wortmannin, cytochalasin D, and 5-(N-ethyl-N-isopropyl) amiloride (EIPA) (Fig. 5b). In particular, EIPA is usually used as a symbolic inhibitor of macropinocytosis.35 In contrast, inhibitors of clathrin or caveolae mediated endocytosis such as pitstop, dynasore, and nystatin did not sufficiently inhibit endocytosis of silica. These results provide strong evidences that cells internalize silica sub-microspheres via macropinocytosis. Thus inhibitors of macropinocytosis may be used to modulate endocytosis of sub-microscale particles. However, cells cannot sustain these inhibitors for a long time due to their high cytotoxicity and disturbance to autophagy. For this reason, we adopted epidermal growth factor (EGF), an activator of macropinocytosis,36 to test the autophagic level after treatment. EGF facilitated endocytosis of silica sub-microspheres and subsequent increased autophagy induction (Fig. S8†). This result further confirms the positive correlation between endocytosis and autophagy.
We utilized 0.5 μm fluorescent silica to monitor the cellular trafficking of particles. After incubated with cells for 24 h, most internalized 0.5 μm silica particles co-localized with GFP-LC3 and lysosomal-associated membrane protein 1 (LAMP1) (Fig. S9†), revealing that they located in autophagosomes and lysosomes.23 1.4 μm and 2.1 μm silica particles are large enough to be individually identified in optical imaging without fluorescent labelling. These 1.4 μm and 2.1 μm silica particles all located in LAMP1 labelled lysosomes after internalized by cells (Fig. S10†). Among these internalized particles, few located in GFP-LC3 and LMAP-1 double-labelled autolysosomes.23 However, no particles located in GFP-LC3 single-labelled autophagosomes. These results suggest that silica sub-microspheres may be captured into lysosomes directly after internalized. Considering that autophagosomes fuse with lysosome quickly after formation,37 silica sub-microspheres may be engulfed into autophagosomes and then fuse with lysosomes. The details of this process need to be further elucidated.
Possible involved signalling pathway
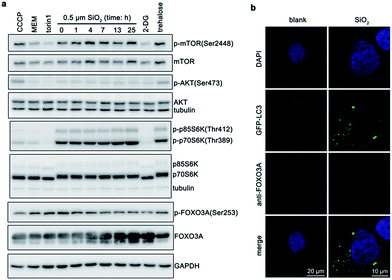
Finally, we also explored signaling pathways that may be involved in this process. Excess reactive oxygen species (ROS) will cause oxidative stress, damage proteins and organelles (e.g., mitochondria and lysosomes), and thus induce autophagy.38,39 Silica nanoparticles can cause ROS generation.40,41 Autophagy induced by silica nanoparticles is also usually regulated by ROS.42 However, silica sub-microspheres did not increase cellular level of ROS (Fig. S11†). Moreover, N-acetyl-cysteine (NAC), a ROS scavenger, could not diminish the autophagic effect induced by silica sub-microspheres, suggesting that ROS did not regulate this process. Due to the low surface-to-volume ratio and high chemical inertness, it is not surprising that silica sub-microspheres do not stimulate ROS generation. Another popular pathway by which nanomaterials regulate autophagy is mammalian target of rapamycin (mTOR),9 which is also the major regulator of autophagy.43 When cells undergo serum deprivation or nutrient starvation, mTOR is de-phosphorylated and inhibited, resulting in autophagy induction. After cells were treated with silica sub-microspheres, mTOR was slightly phosphorylated and activated (Fig. 6a), leading to the activation of its downstream p70S6 kinase (p70S6K). These results indicate that autophagy induced by silica sub-microspheres may not be closely related to the inhibition of mTOR.Instead, we found FOXO3 may be involved in the endocytosis of silica sub-microspheres and subsequent induction of autophagy. FOXO3 belongs to the superfamily of Forkhead box O (FOXO) transcription factors, which modulate the expression of genes involved in apoptosis, cell cycle, DNA damage repair, oxidative stress, and other cellular functions.44 When inactivated, FOXO3 is phosphorylated and locates in cytoplasm. Once activated, FOXO3 is dephosphorylated and translocated to nucleus, which may facilitate the expression of autophagy-related proteins. Recently, FOXO3 has been found to regulate autophagy in skeletal muscle cells45 and haematopoietic stem cells46 by increasing the transcription activity of autophagy-related genes. It also plays a crucial role in autophagy induced by alcohol or glutamine.47,48 After treated for 1 h, cells internalized some silica sub-microspheres (Fig. 2c). At this time point, we found that FOXO3A also began to be dephosphorylated (Fig. 6a). As expected, when cells internalized more and more silica sub-microspheres, an increasing amount of FOXO3A was dephosphorylated. The dephosphorylated FOXO3A was translocated from cytosol to nucleus (Fig. 6b), which would promote the transcription activity of autophagy-related genes and enhance autophagy induction.48 This result suggests that FOXO3A is involved in the regulation of autophagy induced by silica sub-microspheres. However, Akt/PKB, an upstream of FOXO3A,49 was not dephosphorylated after treated (Fig. 6a), indicating that the upstream of FOXO3A in this process is to be determined. The details of FOXO3A-involved pathway need to be further clarified.
The main reason is that cells ingest particles of 0.5–0.7 μm the most among particles of 0.1–2.1 μm. We speculate that internalization of larger particles is more difficult than smaller particles. Previous reports also support that cells ingest more particles of <1 μm than particles of >1 μm.50,51 Moreover, autophagosomes forming at normal conditions are usually less than 1 μm.52,53 It may be more difficult for cells to form larger autophagosomes. Besides silica sub-microspheres, submicro-size aggregation of gold nanoparticles also shows size effect in autophagy induction (Fig. S12†). Small aggregation of nanoparticles considerably facilitated autophagy induction, while large aggregation did not.
Our results support that autophagic level is determined by endocytic level. It may lead to a hypothesis that cells initialize autophagy induction to cope with accumulation of foreign materials which are difficult to degrade. We further confirmed that many other insoluble and persistent submicro materials such as calcium phosphate precipitates,54,55 polystyrene,56 titanium dioxide, carbon, barium sulfate, copper sulphide, calcium carbonate and alumina can also induce autophagy (Fig. S13†). These foreign materials can be endocytosed by cells and locate in lysosomes or autolysosomes ultimately. However, they cannot be degraded in lysosomes. Although cells can eliminate these indigestible materials via division or exocytosis over the long term, accumulation of indigestible materials in a short term may stress cells and trigger autophagy. For static cells in vivo, indigestible materials may accumulate for a long term and impair lysosomes and autophagy. This may be a reason for defective autophagy in silicosis patients or smokers, whose pulmonary cells may ingest many silica sub-microspheres or carbon particles.
Autophagy induced by silica sub-microspheres has something different from that induced by typical small molecules and nanoparticles. It involves FOXO3A-related pathway but not canonical mTOR or ROS. Is it related to cellular immune response to foreign matter or pathogen like bacteria or virus? We propose that endocytosis of silica sub-microspheres triggers cellular recognition of foreign materials and subsequent induction of autophagy. To remove or degrade these foreign materials by autophagy, cells initiate dephosphorylation of FOXO3A to enhance autophagy activity. However, as silica sub-microspheres are enwrapped by biomembrane and there is not special spatial structure on the surface, how do cells recognize them? This may be interesting but needs further exploration. The involvement of FOXO3A could be a good clue although its upstream in this process remains unknown temporally.
4. Conclusions
In summary, we demonstrate that silica sub-microspheres induce endocytosis-dependent autophagy. 0.5–0.7 μm is the suitable size for silica sub-microspheres to induce autophagy due to the high level of endocytosis. Particles with the same size but different surface can induce different levels of autophagy. When predicting the ability of particles to induce autophagy, we should pay more attention to endocytic level instead of particle size. This will lead to a warning that endocytosis of fine particles or other non-degradable materials may be harmful as well. Moreover, the FOXO3A-involved signalling pathway will offer a new clue for research in cell–particle interactions.Acknowledgements
This work was supported by the National Key Basic Research Program of China (2013CB933901, 2014CB744502, and 2014CB932004), National Natural Science Foundation of China (21521004, 81370042, and 81430041), and Fok Ying Tung Education Foundation (142012).Notes and references
- P. Couvreur, Adv. Drug Delivery Rev., 2013, 65, 21–23 CrossRef CAS PubMed.
- Y. Cai, Y. Chen, X. Hong, Z. Liu and W. Yuan, Int. J. Nanomed., 2013, 8, 1111–1120 Search PubMed.
- Z. Fang, L.-Y. Wan, L.-Y. Chu, Y.-Q. Zhang and J.-F. Wu, Expert Opin. Drug Delivery, 2015, 12, 1943–1953 CrossRef CAS PubMed.
- S. J. Soenen, W. J. Parak, J. Rejman and B. Manshian, Chem. Rev., 2015, 115, 2109–2135 CrossRef CAS PubMed.
- M. A. G. Leiva, D. A. Santibañez, S. E. Ibarra, P. C. Matus and R. Seguel, Environ. Pollut., 2013, 181, 1–6 CrossRef PubMed.
- S. Aryal, B. K. C. Remant, N. Bhattarai, C. K. Kim and H. Y. Kim, J. Colloid Interface Sci., 2006, 299, 191–197 CrossRef CAS PubMed.
- Y. Zhang, Y. Chen, P. Westerhoff and J. Crittenden, Water Res., 2009, 43, 4249–4257 CrossRef CAS PubMed.
- Z. Yang and D. J. Klionsky, Nat. Cell Biol., 2010, 12, 814–822 CrossRef CAS PubMed.
- K. Peynshaert, B. B. Manshian, F. Joris, K. Braeckmans, S. C. De Smedt, J. Demeester and S. J. Soenen, Chem. Rev., 2014, 114, 7581–7609 CrossRef CAS PubMed.
- J. M. Zook, R. I. MacCuspie, L. E. Locascio, M. D. Halter and J. T. Elliott, Nanotoxicology, 2011, 5, 517–530 CrossRef CAS PubMed.
- Q. He, Z. Zhang, Y. Gao, J. Shi and Y. Li, Small, 2009, 5, 2722–2729 CrossRef CAS PubMed.
- Z. Zhou, L. Wang, X. Chi, J. Bao, L. Yang, W. Zhao, Z. Chen, X. Wang, X. Chen and J. Gao, ACS Nano, 2013, 7, 3287–3296 CrossRef CAS PubMed.
- N. Lee, D. Yoo, D. Ling, M. H. Cho, T. Hyeon and J. Cheon, Chem. Rev., 2015, 115, 10637–10689 CrossRef CAS PubMed.
- X. Wu, M. Wu and J. X. Zhao, Nanomedicine, 2014, 10, 297–312 CAS.
- D. Huang, H. Zhou and J. Gao, Sci. Rep., 2015, 5, 14361 CrossRef CAS PubMed.
- Y. Zhang, B. Y. W. Hsu, C. Ren, X. Li and J. Wang, Chem. Soc. Rev., 2015, 44, 315–335 RSC.
- A. Bitar, N. M. Ahmad, H. Fessi and A. Elaissari, Drug Discovery Today, 2012, 17, 1147–1154 CrossRef CAS PubMed.
- Z. Zhao, X. Wang, Z. Zhang, H. Zhang, H. Liu, X. Zhu, H. Li, X. Chi, Z. Yin and J. Gao, ACS Nano, 2015, 9, 2749–2759 CrossRef CAS PubMed.
- Z. Zhang, H. Liu, H. Zhou, X. Zhu, Z. Zhao, X. Chi, H. Shan and J. Gao, Nanoscale, 2016, 8, 4373–4380 RSC.
- K. D. Hartlen, A. P. T. Athanasopoulos and V. Kitaev, Langmuir, 2008, 24, 1714–1720 CrossRef CAS PubMed.
- R. Watanabe, T. Yokoi, E. Kobayashi, Y. Otsuka, A. Shimojima, T. Okubo and T. Tatsumi, J. Colloid Interface Sci., 2011, 360, 1–7 CrossRef CAS PubMed.
- Y. Kabeya, N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi and T. Yoshimori, EMBO J., 2000, 19, 5720–5728 CrossRef CAS PubMed.
- D. J. Klionsky, K. Abdelmohsen, A. Abe, M. J. Abedin, H. Abeliovich, A. Acevedo Arozena, H. Adachi, C. M. Adams, P. D. Adams and K. Adeli, et al., Autophagy, 2016, 12, 1–222 CrossRef PubMed.
- J. G. Teeguarden, P. M. Hinderliter, G. Orr, B. D. Thrall and J. G. Pounds, Toxicol. Sci., 2007, 95, 300–312 CrossRef CAS PubMed.
- B. Kong, J. H. Seog, L. M. Graham and S. B. Lee, Nanomedicine, 2011, 6, 929–941 CrossRef CAS PubMed.
- L. L.-Y. Chan, D. Shen, A. R. Wilkinson, W. Patton, N. Lai, E. Chan, D. Kuksin, B. Lin and J. Qiu, Autophagy, 2012, 8, 1371–1382 CrossRef CAS PubMed.
- J. Xie, C. Xu, N. Kohler, Y. Hou and S. Sun, Adv. Mater., 2007, 19, 3163–3166 CrossRef CAS.
- A. S. Karakoti, S. Das, S. Thevuthasan and S. Seal, Angew. Chem., Int. Ed., 2011, 50, 1980–1994 CrossRef CAS PubMed.
- D. Guarnieri, A. Guaccio, S. Fusco and P. Netti, J. Nanopart. Res., 2011, 13, 4295–4309 CrossRef CAS.
- Z.-J. Zhu, T. Posati, D. F. Moyano, R. Tang, B. Yan, R. W. Vachet and V. M. Rotello, Small, 2012, 8, 2659–2663 CrossRef CAS PubMed.
- H. Cai and P. Yao, Colloids Surf., B, 2014, 123, 900–906 CrossRef CAS PubMed.
- R. E. Serda, J. Gu, R. C. Bhavane, X. Liu, C. Chiappini, P. Decuzzi and M. Ferrari, Biomaterials, 2009, 30, 2440–2448 CrossRef CAS PubMed.
- G. Sahay, D. Y. Alakhova and A. V. Kabanov, J. Controlled Release, 2010, 145, 182–195 CrossRef CAS PubMed.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC.
- M. A. Krzyzaniak, M. T. Zumstein, J. A. Gerez, P. Picotti and A. Helenius, PLoS Pathog., 2013, 9, e1003309 CAS.
- D. M. Bryant, M. C. Kerr, L. A. Hammond, S. R. Joseph, K. E. Mostov, R. D. Teasdale and J. L. Stow, J. Cell Sci., 2007, 120, 1818–1828 CrossRef CAS PubMed.
- L. Jahreiss, F. M. Menzies and D. C. Rubinsztein, Traffic, 2008, 9, 574–587 CrossRef CAS PubMed.
- R. Scherz-Shouval and Z. Elazar, Trends Cell Biol., 2007, 17, 422–427 CrossRef CAS PubMed.
- G. Filomeni, D. De Zio and F. Cecconi, Cell Death Differ., 2015, 22, 377–388 CrossRef CAS PubMed.
- M. Ahamed, Hum. Exp. Toxicol., 2013, 32, 186–195 CAS.
- J. Ahmad, M. Ahamed, M. J. Akhtar, S. A. Alrokayan, M. A. Siddiqui, J. Musarrat and A. A. Al-Khedhairy, Toxicol. Appl. Pharmacol., 2012, 259, 160–168 CrossRef CAS PubMed.
- Y. Yu, J. Duan, Y. Yu, Y. Li, X. Liu, X. Zhou, K.-f. Ho, L. Tian and Z. Sun, J. Hazard. Mater., 2014, 270, 176–186 CrossRef CAS PubMed.
- C. H. Jung, S.-H. Ro, J. Cao, N. M. Otto and D.-H. Kim, FEBS Lett., 2010, 584, 1287–1295 CrossRef CAS PubMed.
- H. Huang and D. J. Tindall, J. Cell Sci., 2007, 120, 2479–2487 CrossRef CAS PubMed.
- C. Mammucari, G. Milan, V. Romanello, E. Masiero, R. Rudolf, P. Del Piccolo, S. J. Burden, R. Di Lisi, C. Sandri and J. Zhao, et al., Cell Metab., 2007, 6, 458–471 CrossRef CAS PubMed.
- M. R. Warr, M. Binnewies, J. Flach, D. Reynaud, T. Garg, R. Malhotra, J. Debnath and E. Passegue, Nature, 2013, 494, 323–327 CrossRef CAS PubMed.
- K. E. van der Vos, P. Eliasson, T. Proikas-Cezanne, S. J. Vervoort, R. van Boxtel, M. Putker, I. J. van Zutphen, M. Mauthe, S. Zellmer and C. Pals, et al., Nat. Cell Biol., 2012, 14, 829–837 CrossRef CAS PubMed.
- H.-M. Ni, K. Du, M. You and W.-X. Ding, Am. J. Pathol., 2013, 183, 1815–1825 CrossRef CAS PubMed.
- S. P. Tenbaum, P. Ordonez-Moran, I. Puig, I. Chicote, O. Arques, S. Landolfi, Y. Fernandez, J. R. Herance, J. D. Gispert and L. Mendizabal, et al., Nat. Med., 2012, 18, 892–901 CrossRef CAS PubMed.
- K. A. Foster, M. Yazdanian and K. L. Audus, J. Pharm. Pharmacol., 2001, 53, 57–66 CrossRef CAS PubMed.
- W. Zauner, N. A. Farrow and A. M. R. Haines, J. Controlled Release, 2001, 71, 39–51 CrossRef CAS PubMed.
- S. Jäger, C. Bucci, I. Tanida, T. Ueno, E. Kominami, P. Saftig and E.-L. Eskelinen, J. Cell Sci., 2004, 117, 4837–4848 CrossRef PubMed.
- N. Mizushima, A. Yamamoto, M. Matsui, T. Yoshimori and Y. Ohsumi, Mol. Biol. Cell, 2004, 15, 1101–1111 CrossRef CAS PubMed.
- W. Gao, W.-X. Ding, D. B. Stolz and X.-M. Yin, Autophagy, 2008, 4, 754–761 CrossRef CAS PubMed.
- S. Sarkar, V. I. Korolchuk, M. Renna, A. R. Winslow and D. C. Rubinsztein, Autophagy, 2009, 5, 307–313 CrossRef CAS PubMed.
- S. Kobayashi, T. Kojidani, H. Osakada, A. Yamamoto, T. Yoshimori, Y. Hiraoka and T. Haraguchi, Autophagy, 2010, 6, 36–45 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra26649e |
| This journal is © The Royal Society of Chemistry 2017 |