Open Access Article
Open Access ArticleAutomated melting curve analysis in droplet microfluidics for single nucleotide polymorphisms (SNP) genotyping
F.-W. Liua,
S.-T. Dingb,
E.-C. Linb,
Y.-W. Lu *a and
J.-S. R. Jang
*a and
J.-S. R. Jang c
c
aDept. of Bio-Industrial Mechatronics Engineering, National Taiwan University, Taipei, Taiwan, Republic of China. E-mail: yenwenlu@ntu.edu.tw; Tel: +886-2-3366-9748
bDept. of Animal Science, National Taiwan University, Taipei, Taiwan, Republic of China
cDept. of Computer Science and Information Engineering, National Taiwan University, Taipei, Taiwan, Republic of China
First published on 16th January 2017
Abstract
An integrated microchip platform with automated analysis capability for DNA melting curves is developed for Single Nucleotide Polymorphism (SNP) genotyping applications. The microchip contains fluidic channels where genomic DNA samples are encapsulated into a series of droplets and transported through a detection region with a stable temperature gradient. As the temperature is elevated from 60 °C to 85 °C, the DNA strains denature and the associated fluorescence signals decay with this relationship being acquired as the melting curve. The droplets serve as discrete reactors to conduct DNA melting curve analysis in the liquid phase, eliminating the need for immobilization of reagents. They provide the advantage of signal homogeneity, which reduces the signal fluctuations, and thus the signal-to-noise ratio is improved. In addition, a droplet detection and tracking software system which can identify the droplets, records the fluorescence intensities, plots the melting curves, and finds the melting temperatures, is developed making automated SNP genotyping possible. The platform has been verified with genomic DNA from Landrace sows and shown successful SNP discrimination from homozygotes and heterozygotes which demonstrates its potential to conduct on-site SNP genotyping for disease research, medical diagnostics, agriculture, and farm animal reproduction.
1. Introduction
Single nucleotide polymorphism (SNP) – a single base mutation in a genome – is perhaps one of the most common gene variations. It is a useful biomarker to discriminate different genotypes and sometimes to correlate with economic traits of livestock or crops.1,2 While using traditional phenotypic selection has been shown to be insufficient, marker-assisted-selection (MAS) techniques with reliable markers (e.g. SNP markers) have received considerable attention in the livestock industry. In practice, depending on the number of samples and SNP sites, choosing appropriate SNP genotyping techniques for animal breeding involves tradeoffs between reliability, sample preparation, reagent/sample expense, instrument depreciation and procedure complexity. For instance, large throughput SNP methods require a series of complicated steps and special instruments.3 They are high cost, labor intensive and time-consuming. A feasible approach is to utilize the whole-genome-sequence (WGS) of a small population of the individuals and to use a SNP chip for the genotyping of the majority of the individuals should be collectively adapted.4 In particular, lab-on-a-chip technology as a SNP chip, which integrates sample preparation, genome amplification, and detection onto a single chip, can simplify the genotyping process and be very useful for practical animal breeding in small throughput applications.SNP genotyping methods typically include primer extension,5,6 melting curve analysis,7,8 enzymatic ligation,9,10 enzymatic cleavage11 and sequencing.12 Among them, DNA melting curve analysis has increasingly become popular, particularly in lab-on-a-chip devices, because of its accuracy and straightforward procedures. Melting curve analysis usually utilizes the solid or liquid phase as the carrier for the targets and/or reagents. For instance, in solid-phase-based analysis (e.g. microbeads), the mobile microbeads have their surfaces immobilized with DNA targets or probes, and then being genotyped either statically or dynamically.13–15 However, the solid-phase-based platform is limited in reaction rate by diffusion kinetics and requires surface modification for extensive reagent immobilization to ensure effective binding for detection. In contrast, the liquid phase carries the sample in a bulk flow and is introduced for downstream reaction and detection.16,17
One of the most commonly approaches in liquid-phase-based analysis is droplet microfluidics, where samples and reagents are encapsulated inside droplets.18–21 The droplet platforms have many advantages, as they permit enhanced hydrodynamic mixing, eliminate the need for immobilization, and prevent the content inside from contamination and unnecessary channel-wall adsorption.22,23 The droplets provide a micro-environment to allow the samples and reagents to react and to be analyzed as chemical or biological assays.
One typical detection approach during the analysis for the assay is to use fluorescence microscopy with an imaging system to monitor the fluorescent signals inside the analyzed droplets. This approach is labor-intense and time-consuming. Automatic analysis tools thus become inevitable for the droplet microfluidics in practical uses. As a few examples have been shown on studying cell aggregation, enzyme production,24 gene expression of cells,25 and measuring the growth kinetics of microalgae26 in droplet microfluidics, they mostly resort to physical trapping of droplets on chip and observing the images at the certain locations over time. In addition, rapid sensing and detection for chemicals or enzyme screening,27 or automatic target identifying for single cell analysis over time.28–30 There are very few examples to continuously track the droplet motions and monitor the droplet signals over a period of time.
To have a SNP chip for on-site animal breeding, we construct a droplet-based microchip with an automatic melting analysis tool. Our microchip, which performs spatial melting curve analysis in a series of moving droplets, requires to continuously detect, track the droplet motions and analyze the droplet fluorescent signals. As each droplet encloses the DNA sample and passes through a controlled temperature gradient from low to high temperatures, DNA denaturation occurs. By using a designed sequence of oligonucleotide, the DNA samples with different genotypes have different denaturation temperatures, or melting temperatures (Tm); the genotypes are distinguished. DNA melting curve analysis is then conducted in the droplet format. With our automatic melting curve analysis tool, the functions of target recognition, tracking, image data acquisition and melting analysis are sequentially conducted for each droplet. The tool not only replaces the laborious and time-consuming tasks but also prevents the bias and variations due to manual operations, further improve the efficiency of the assay.
2. Materials and methods
Mechanism and design
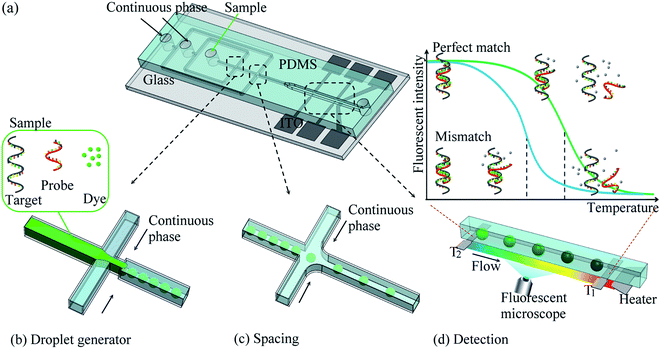
DNA melting curve analysis has been widely utilized in SNP genotyping because neither the enzymatic reaction nor further determination of reaction product is necessary.7 Melting curve analysis employs the denaturation of DNA duplex upon heating. While DNA denatures, the intercalating dye indicates the amount of DNA that still remains in duplex. The relationship between the temperature and the fluorescent intensity of the dye is known as melting curve of this DNA duplex. The sharpest slope in melting curve is where the half of the duplex denatures and its corresponding temperature is DNA melting temperature (Tm). Tm is based on the sequence and conformation of DNA duplex. Therefore, target DNA sequence with SNP mutation can be discriminated by an allele-specific oligonucleotide probe. The probe is designed to be complementary to the target sequence and cover the SNP location.7 When SNP occurs and change the sequence in one nucleotide, the bonding strength between probe and perfect-match target is stronger; therefore the Tm is higher. On the other hand, the probe with a one-mismatched nucleotide has a weaker bonding strength and so the lower Tm.Fig. 1(a) shows the prototype of our droplet-based SNP genotyping microchip, consisted of multifunctional microchannel in PDMS and substrate with on-chip heater/sensors. The microchip enables us to perform (1) droplet generator (2) spacing control and (3) SNP detection region using melting curve analysis in a row. In the first region, a sample solution is dispensed to the microchannel and then squeezed into discrete droplets by continuous phase from the side channels as shown in Fig. 1(b). The droplets are then serially transported into the second region – spacing region – where another stream of continuous phase from the side channels are introduced to control the spacing distance between droplets to prevent the collision of the droplet, as shown in Fig. 1(c). The droplets proceed to enter the third region – detection region – where on-chip heater creates a sharp but steady temperature gradient for melting curve analysis as shown in Fig. 1(d). As the droplet passes through this region, it experiences an increase in temperature and the probe denatures from the target DNA. The fluorescent intensities of the droplets are recorded along with their locations (and temperature), so the melting curves are obtained.
Fabrication of microfluidic device
Photolithography and wet etching technique were used to fabricate the heater/thermometers set on an ITO glass with surface resistance of 16 Ω per square (UNI-WARD Corp. New Taipei City, Taiwan). A master mold of microchannel was first made of negative photoresist of SU8 series (MicroChem Corp. MA, USA). Microchannel was polydimethylsiloxane (PDMS) (Dow Corning Corp. MI, USA) mixture at a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
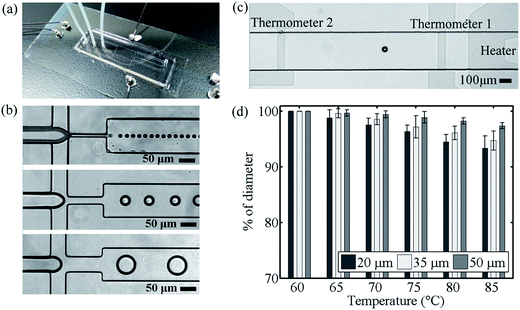
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio which was properly mixed and poured on the mold. After cured, the PDMS was peeled off and punched through at the inlet/outlet position. It was aligned to the region of heater/thermometer on the ITO glass and bonded together by partially-curing bonding method.13 Fig. 2(a) showed the prototype of our droplet-based device with electrical and fluidic connections.
1 ratio which was properly mixed and poured on the mold. After cured, the PDMS was peeled off and punched through at the inlet/outlet position. It was aligned to the region of heater/thermometer on the ITO glass and bonded together by partially-curing bonding method.13 Fig. 2(a) showed the prototype of our droplet-based device with electrical and fluidic connections.
Characteristics of microdevice
Once an ITO glass was first patterned to define the heater and thermometers, the resistances of two thermometers were measured through a data acquisition card (USN-6210, National Instrument Corp., TX, USA) when different voltages were applied to the heater. The temperature distribution generated from the ITO heater was measured by using an infrared camera (P640 Series, Ching Hsing Computer-Tech Ltd, Taipei, Taiwan). The relationships between the temperatures and resistances of the thermometers were obtained before the glass was bonded to PDMS to form our device. As the region between thermometer 1 and 2 was defined as the detection region of the assay, a PID control system was employed to maintain thermometer 1 (T1) at 85 °C and thermometer 2 (T2) at 60 °C during the experiment. The temperature distribution along the detection region was found approximately linear so the temperature at the certain location inside the microchannel could be easily derived.SNP genotyping in droplet-based microchip
Ataxia telangiectasia-mutated (ATM) genes from Landrace sows as the target DNA were tested. ATM genes were proven as an effective biomarker. It played an important roles in swine reproduction—the total number of piglet born, number of piglet born alive and the average birth weight.14 ATM held three SNP genotypes, including homozygous type (e.g. CC and TT), and heterozygous type (e.g. CT). A 73 bp pair fragment of ATM gene containing upstream promoter and exon to intron 2 regions was used as our target DNA sample. A 21 bp oligonucleotide, which was perfectly complementary to CC type of DNA, was used as the probe (Genomics Biotechnology Co., Ltd, Taipei, Taiwan). The sequence of the target and the probe were listed below: ATM-C, 5′-GGC TAC GCT CGA GGG TAG CAG CAT GAT CCA AGC CGC AGG AGT ACC CGC AGT GAG AGA CGA GAC TCA GGT AAA A-3′; ATM-T, 5′-GGC TAC GCT CGA GGG TAG CAG CAT GAT CTA AGC CGC AGG AGT ACC CGC AGT GAG AGA CGA GAC TCA GGT AAA A-3′; probe, 5′-CCT GCG GCT TGG ATC ATG CTG-3′.The sample solution contained the target DNA sample and probe at equal amount in total volume of 10 μL at 30 μM, as well as 10% v/v saturated DNA double strand fluorescent dye, LCGreen. The sample solution was then introduced into the main inlet while the continuous phase (mineral oil and 1.5% wt/wt SPAN 80 as stabilizing surfactant) was introduced from the inner side channels. The flow rates were 2 μL h−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 μL h−1 (sample solution
10 μL h−1 (sample solution![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
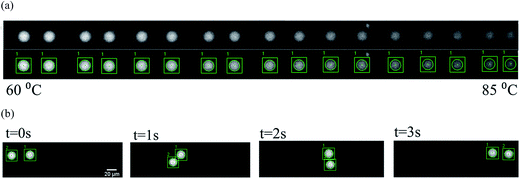
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) continuous phase). A series of droplets were produced at the diameters of 20, 35 and 50 μm by using the nozzles with different widths at 15, 30 and 50 μm respectively, as shown in Fig. 2(b). The spacing flow of the continuous phase was injected at the rate of 10 μL h−1. The droplets were properly isolated and spaced before they proceeded into the detection region.
continuous phase). A series of droplets were produced at the diameters of 20, 35 and 50 μm by using the nozzles with different widths at 15, 30 and 50 μm respectively, as shown in Fig. 2(b). The spacing flow of the continuous phase was injected at the rate of 10 μL h−1. The droplets were properly isolated and spaced before they proceeded into the detection region.
The temperature of thermometer 1 (T1) was set at 85 °C, where was about the downstream exit of the detection region. Meanwhile, the temperature of thermometer 2 (T2) was 60 °C, where was about the upstream entrance of the detection region. After the system reached steady-state, the images of the droplets flowing through the detection region were recorded via the fluorescent microscope (AxioObserver.A1, Carl Zeiss MicroImaging GmbH, Gottingen, Germany) with the excitation light at the wavelength of 425–475 nm and the exposure time at 100 ms.
Automated analysis for droplet image and melting curve
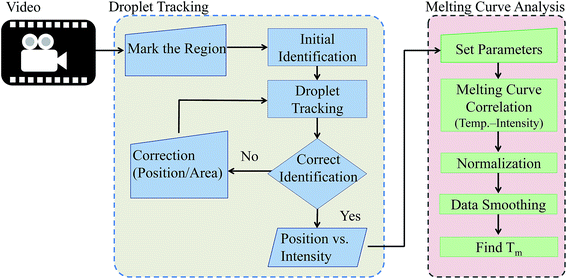
The workflow for the automated analysis was outlined in Fig. 3; it included the two major functions: (1) droplet tracking and (2) melting curve analysis. A sequence of the fluorescent images as the droplet flowing from low to high temperature across the detection region was first obtained and utilized for our analysis. A background image, taken when all light sources were blocked, was subtracted from the image sequence in order to correct the bias within the CCD camera. Each droplet had its fluorescent intensity at different positions along the detection region in every frame. As one of the major hurdles in the melting curve analysis was to correlate the fluorescent intensity and surrounding temperature of the DNA sample, an automatic object detection and tracking based on image processing program was developed. The program identified the droplet, which contained the DNA sample, as well as acquired the information of the fluorescent intensities, droplet locations and surrounding temperatures. It consisted of five parts: (i) user selection for target droplet, (ii) droplet identification, (iii) droplet tracking, (iv) interactive annotation and (v) data acquisition.The region, which contained the droplet of interest, was first selected. The program identified the droplet by following procedures: reducing the noise by median filter, converting to binary image by Otsu's method, creating the mask for the target by dilation and erosion, finding the edge by Sobel edge detection method and fitting a bounding circle to the edge by least squares method since the droplets were mostly spherical. The program tracked the identified droplet by normalized cross-correlation (NCC) matching with linear extrapolation. The bounding circle was used as the template for tracking by comparing the correlation of the location and intensity of each pixel. The area with the highest correlation to the circle in the next frame was recognized as the same droplet and became the template for the next. This process was automatically repeated until the identified droplet image left the field of view. The information of the droplet in its location and image intensity in every frame was obtained.
The position and fluorescent intensity of the droplet was then processed for melting curve analysis. The surround temperature of the droplet was directly related to the droplet position in the channel, so the relationship between the temperature and fluorescent intensity was plotted and obtained as the melting curve for each droplet or sample. The fluorescent data were normalized and the curves were smoothed by Savitzky–Golay (S–G) polynomials. The negative first derivatives of the melting curves were usually taken to find the corresponding melting temperatures of the samples.
3. Results
Droplet preparation
To evaluate the performance of our microchips, the sample solution was introduced to the droplet generator with different widths of the nozzles respectively. The droplets of 20, 35 and 50 μm in diameters were produced and transported through the spacing region into the detection region. Meanwhile, prior to the detection, the microchip has reached temperature equilibrium when thermometer 1 maintaining at 85.0 ± 1.0 °C. The droplets flew steadily at 50 μm per sand results in an overall detection time 25 seconds at the heating rate 1.0 °C s−1. Fig. 2(c) showed the droplets flowing in the detection region between thermometers. The fluorescent intensities of the droplet were recorded as they moving progressively toward higher temperature.During the experiment, the size of droplet was found being affected by the surrounding temperature. As the temperature increased, droplets shrank and their diameters decreased. Droplet became unstable when the temperature exceeded 90 °C in our experiment. At this moment, droplet easily coalesced with others. If the temperature continued to increase, the size would drastically decrease until the interface disappeared. This phenomenon is attributed to the alteration of interfacial tension between water and oil upon heating and directly affects the droplet stability.31 The thermal stability of the droplet can thus be described as the size change with respect to temperature.32,33 To quantify this relationship, the droplet sizes were measured every 5 °C when they flew across the detection region. The average diameter changes were −6.6%, −4.7% and −2.7% for the 20, 35 and 50 μm droplets. The droplet however remained intact, and the interface was confirmed during the entire melting/heating process.
Droplet detection and tracking based on image processing
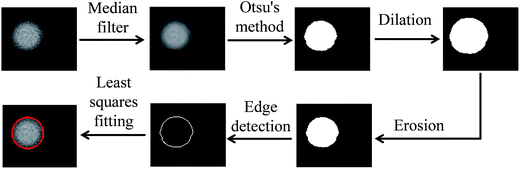
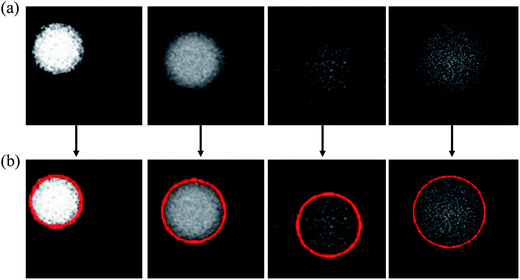
In order to lessen human's effort on labeling droplets during experiment, we developed a highly automatic software system for droplet detection and tracking using techniques from image processing and numerical methods. To be more specific, a user only needed to crop the region containing a specific droplet, our system identified the droplet automatically by putting a bound circle on top of it, and then tracked the droplet all the way until it reached the left end. Whenever there was a mistake in tracking, the user simply clicked the correct droplet and the system restarted the tracking to make sure the user-specified droplet in a specific frame is in the path of tracking. This type of interactive video annotation greatly reduced human's effort on labeling the droplets. Here we explained the methods used in the software system for droplet detection and tracking.34Once the user cropped the region containing the droplet, our system automatically identified the bounding circle. Fig. 4 showed the procedures of droplet detection by putting a bounding circle on top of it, which involves a series of operations:
(1) A median filter to reduce noise.
(2) Otsu's method to find the best threshold for creating a binary image.
(3) Dilation to remove rough edges.
(4) Erosion to reduce to its original size.
(5) Edge detection based on Sobel's method.
(6) Bounding circle to be found, based on least-squares method.
During the bounding cycle procedures, we could simply choose the maximum-area region, if the number of regions are more than one after erosion. Similar procedure was also used to locate the droplet in the next frame once the approximate location was identified. Please note that the proposed method for finding the bounding circle was quite robust, as shown in Fig. 5 where difficult examples of droplets with very weak fluorescent signals or blurred droplet boundaries were successfully handled.
Once the droplet was detected by the bounding circle, the tracking process was invoked to identify the droplet in the next frame. This was accomplished by using normalized cross-correlation matching and forward position extrapolation, which demonstrated the best performance in our experiments.34 The system was highly automatic, in the sense as it reduced the number of frames to only 1.24% of the original frames required by human labeling. Once this efficient and effective process of interactive labeling and tracking was completed, the program returned the correct bounding circles for the underlying droplet in each frame. It became rather easy to collect the locations of the droplet as well as its intensities of all frames, as shown in Fig. 6(a). Moreover, the proposed method could be used to simultaneously track multiple droplets, as shown in Fig. 6(b).
Melting curve analysis
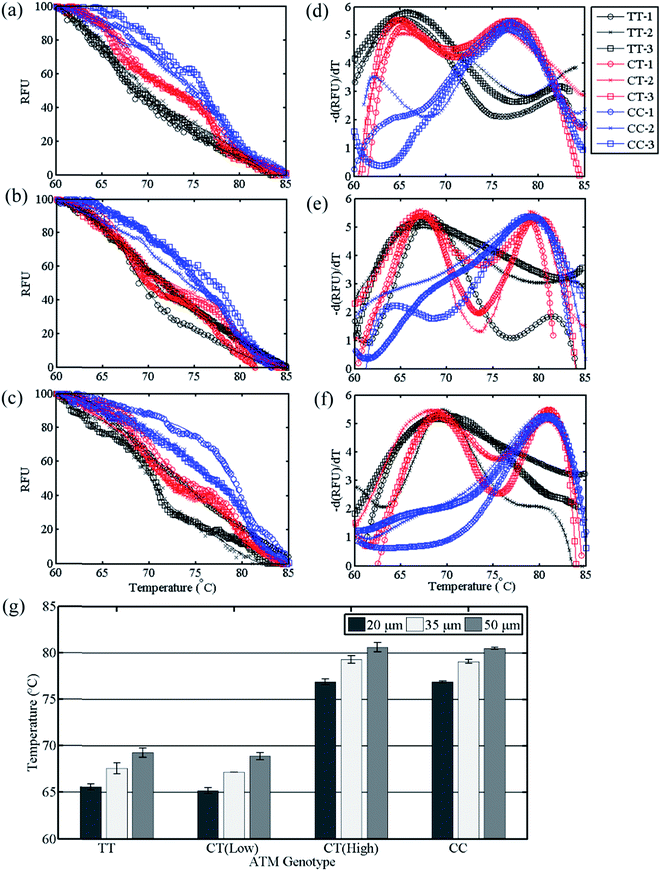
The relationships between the fluorescent intensities and temperatures were obtained as the melting curves shown in Fig. 7(a–c). The total assay time, which included the droplet passed through the detection region, as well as its images were processed for the melting curve and corresponding genotype, was about 220 seconds. As this droplet-based platform allowed us to conduct the analysis one-by-one in series, the average assay time can be greatly reduced, when more droplets were sequentially tested. At least three samples were examined in our preliminary tests for each genotype in each size of the droplets.The melting curves of the same SNP genotype, even in different droplet sizes, had similar tendency. TT type, which had one mismatch in sequence with probe, had high decreasing rate in intensity. Their intensity dropped below 50% before 70 °C due to the weak bonding strength of target–probe duplex. On the other hand, CC type possessed melting curves with lower decreasing rate in intensity at the beginning but higher after 75 °C which was consistent with the stronger bonding of perfect-match target–probe duplex. Heterozygous CT type consisted of both ATM-C and ATM-T as target DNA, which has melting curves lied between CC and TT type.
The negative first derivatives of the melting curves were shown in Fig. 7(d–f). The maxima corresponded to the sharpest slope of the melting curves, where the melting temperatures (Tm) were located. The melting temperatures using the 20 μm droplets were found at 65.6 ± 0.3 °C for TT type, 65.2 ± 0.3 °C and 76.9 ± 0.3 °C for CT type, and 76.9 ± 0.1 °C for CC type. For the 35 μm droplets, the melting temperatures were 67.6 ± 0.6 °C, 67.2 ± 0.0 °C and 79.3 ± 0.4 °C, and 79.1 ± 0.2 °C for TT, CT, and CC types respectively. For 50 μm droplets, the melting temperatures were 69.3 ± 0.5 °C, 68.9 ± 0.4 °C and 80.6 ± 0.5 °C, and 80.5 ± 0.1 °C for TT, CT, and CC types respectively. The average melting temperature using different droplet sizes were listed in Fig. 7(g). The droplet-based platform succeeded in discriminating different SNP genotypes of ATM with variation less than 1.0 °C in Tm. In addition, the difference in melting temperature between two homozygous genotypes (ΔTm) was 11.3 °C in average. It is three folds of the one obtained by using the commercial quantitative Polymerase Chain Reaction (qPCR) machine. The larger ΔTm was beneficial to distinguish different SNP genotypes.
Meanwhile, the droplet size played roles in the melting temperature. For instance, when a 20 μm droplet moved along the temperature gradient region (or detection region), the temperature difference between two ends of a single droplet was estimated as 0.4 °C, assuming the linear temperature distribution along the channel longitudinal direction. Moreover, due to the heat transfer in the latitudinal direction – from the heater on the substrate up to the channel – the temperature difference was approximately 0.5 °C between two ends of a single droplet. Larger droplets would have a more distinct temperature difference across one droplet. Therefore, the melting temperature obtained by using different droplet sizes varied. Larger droplets were found to have higher melting temperatures than smaller droplets did. However, the droplets in different sizes only resulted in minimal changes in the melting curves and melting temperatures.
4. Discussion
The genotyping results obtained by using our droplet platform (droplet microfluidic, spatial melting analysis) and Rotor-Gene Q system (tube-based, temporal melting analysis) were consistent. The ΔTm of 11.3 °C by this droplet device was about 3.2 times larger than the ΔTm of 3.5 °C by Rotor-Gene Q system. Larger ΔTm was beneficial to distinguish the perfectly-matched and the mismatched duplex. There were two main reasons for this advantage: (1) high heating rate and (2) enhanced thermal stability. First, given the flow rate and the temperature gradient in our device, the heating rate was about 1.0 °C s−1, which was higher than 0.1 °C s−1 used in the tube-based system. Meanwhile, since the droplet has a very small heating capacity, a high heating rate during DNA melting procedure was achieved and thus promoted the Tm shifted to a higher value.14,35 The perfectly-matched DNA duplex shifted more than the mismatched one did, resulting in the increase of ΔTm. Secondly, the enhancement of thermal stability due to microchannel laminar configuration shifted the Tm and improved the discrimination between perfectly-matched and mismatched DNA duplex, compared to that in bulk solution.36The size change of the droplet would affect the fluorescent intensity. As discussed earlier, the droplets shrank, as they moved from low temperature to high temperature. The droplet volume decreased and the fluorescent dye became concentrated, so the intensity increased. The 20 μm droplets were found to have about an 18.5% decrease in volume, the 35 μm droplets had about 13.4%, and the 50 μm droplets had about 7.9% when they moved across the detection region (from 60 °C to 85 °C). The fluorescent intensities then could be corrected, by taking account of the relationships of the droplet volume and temperature in Fig. 2(d). The fluorescent intensity varied linearly to the concentration of the fluorescent dye, which was inversely proportional to the droplet volume. In consequent for the 20 μm droplets, which had the most volume changes during the experiment, the melting temperatures became 65.9 °C for TT type and 77.2 °C for CC type after the correction. The differences in the melting temperature due to the droplet volume change in the detection region were relatively small. The thermal stability of the droplets had minimal impact on SNP genotype discriminations. The thermal stability effect on the melting temperatures was even more minor for the droplets with diameters of 35 and 50 μm, or larger droplets.
Meanwhile, although only droplets at 20, 35 and 50 μm in diameter have been tested and demonstrated the functionality of the device, it is possible to have smaller droplets. It is found that the fluorescent intensities of the droplets become very weak when the droplet becomes smaller than 20 μm. It will require an image acquisition system with higher sensitivity to acquire the fluorescent signals from the droplets.
Using droplet as mobile carriers permits the DNA melting curve analysis in spatial-heating configuration, which is more beneficial than in temporal-heating configurations.7,13 Spatial-heating usually resorts to a temperature gradient which only require a minimum area being heated; rapid thermal response and less power consumption are achieved due to small thermal capacity. In contrast, the temporal-heating usually requires the reagent tethered on the fixed surfaces and has larger thermal capacity. Thus, when adapted in spatial-heating, the droplets have an advantage as the faster thermal response can result in larger melting temperature differences (ΔTm).
Compared to the bead-based platform,14 the droplet-based platform was beneficial in potential on-site SNP genotyping by taking small reagent consumption and laboratory procedure automation – the advantages of microfluidics over traditional tube-based approach. The droplet volume in nanoliter scale greatly reduced the consumption of reagent, as well as the sample preparation procedures became simple and straight forward. The DNA sample, probe and intercalating dye could be all injected into the devices without thorough mixing beforehand, as the droplets allowed rapid mixing while they were moving through the microchannel. The immobilization of the target DNA onto solid-phase supports was not necessary, so was the following cleaning process to remove the unbinding targets during the sample preparation.
Discrepancies were found between this droplet-based and bead-based platforms because of their natures. First of all, the droplets required elevated concentrations of the reagents, while the microbeads concentrate the DNA samples from dilute ones through biotin–streptavidin reactions. At low concentrations, the droplets require to be in a larger size to enhance the signal-to-noise ratios, while the microbeads can still maintain stronger signals than the droplets. The DNA concentration used in this study is a little higher than usual to accommodate the small droplets of 20 μm. The DNA samples with lower concentrations at 17 μM, directly from PCR product, have been tested at 35 μm and 50 μm droplets and showed the feasibility of our platform.
In addition, droplet-based analysis took advantage over bead-based analysis for the signal homogeneity and rapid mixing effect. Though the bead-based analysis permitted extraordinary contrast of the signals on the microbeads to the background, it had a larger variation across the whole bead surface. The microbeads had notable fluorescent intensity variation on the surfaces, while the droplets had uniform distribution. The fluorescent signals on the droplets thus decayed more smoothly than the ones on the microbeads during the melting curve analysis. As the results, the melting curves by using droplet-based platform had less fluctuation than the one using bead-based platform.
The automatic droplet image analysis software provided a standard protocol for droplet identification, instead of manual labeling frame by frame. It allowed us to distinguish the droplet images even when the fluorescent signals were very weak. This program automated the image processing process, reduced the human errors and greatly improved the accuracy of the melting curve analysis.
5. Conclusions
A droplet-based platform that can perform melting curve analysis for sensitive and rapid SNP genotyping is successfully demonstrated. The droplets, encapsulate DNA samples and reagents, not only provides better isolation and hydrodynamic mixing to achieve effective genotyping analysis, but more importantly eliminates the need of the surface modification and reagent immobilization; thereby the sample preparation and genotyping procedures are greatly simplified. The platform is able to discriminate homozygotes and heterozygotes genotypes from genomic samples, with the results consistent to the ones from the commercial qPCR machine. In addition, we have developed a highly automatic software system that can detect and track droplets with minimum human intervention to find the corresponding melting temperature, making automation of SNP genotyping possible. All the above shows our droplet-based genotyping platform with great potential to be applied to the on-site genotyping for breeder's use.Acknowledgements
The authors thank Prof. J. C. Hsu in the Department of Entomology and Dr Y. C. Lin in Department of Bio-Industrial Mechatronics Engineering (BIME) at National Taiwan University (NTU) for the assistance in qPCR technique, as well as the Center of Biotechnology at NTU for providing samples and SNP genotyping technique. The authors also thank Ms T. Kirk for manuscript editing. This project has been supported by Career Development Project at NTU and Ministry of Science and Technology, Taiwan (MOST 105-2221-E-002-111-). A part of the microfabrication was conducted in BIME clean room and NEMS Research Center at NTU.References
- B. J. Hayes, H. A. Lewin and M. E. Goddard, The future of livestock breeding: genomic selection for efficiency, reduced emissions intensity, and adaptation, Trends Genet., 2013, 29(4), 9 CrossRef PubMed.
- W. Yang, X. Kang, Q. Yang, Y. Lin and M. Fang, Review on the development of genotyping methods for assessing farm animal diversity, J. Anim. Sci. Biotechnol., 2013, 4(1) DOI:10.1186/2049-1891-4-2.
- M. Revilla, Y. Ramayo-Caldas, A. Castelló, J. Corominas, A. Puig-Oliveras and N. Ibáñez-Escriche, et al., New insight into the SSC8 genetic determination of fatty acid composition in pigs, Genet., Sel., Evol., 2014, 46(1), 28 CrossRef PubMed.
- C.-W. Huang, Y.-T. Lin, S.-T. Ding, L.-L. Lo, P.-H. Wang and E.-C. Lin, et al., Efficient SNP Discovery by Combining Microarray and Lab-on-a-Chip Data for Animal Breeding and Selection, Microarrays, 2015, 4(4), 570 CrossRef PubMed.
- K. Takatsu, T. Yokomaku, S. Kurata and T. Kanagawa, A FRET-based analysis of SNPs without fluorescent probes, Nucleic Acids Res., 2004, 32(19), e156 CrossRef PubMed.
- J. Zhu, C. Qiu, M. Palla, T. Nguyen, J. J. Russo and J. Ju, et al., A microfluidic device for multiplex single-nucleotide polymorphism genotyping, RSC Adv., 2014, 4(9), 4269–4277 RSC.
- W. M. Howell, M. Jobs, U. Gyllensten and A. J. Brookes, Dynamic allele-specific hybridization, Nat. Biotechnol., 1999, 17, 2 CrossRef PubMed.
- Y. C. Chung, Y. C. Lin, M. Z. Shiu and W. N. T. Chang, Microfluidic chip for fast nucleic acid hybridization, Lab Chip, 2003, 3(4), 228–233 RSC.
- C.-W. Liu, Y.-W. Lin, C.-C. Huang and H.-T. Chang, Fluorescence detection of single-nucleotide polymorphisms using a thymidine-based molecular beacon, Biosens. Bioelectron., 2009, 24(8), 2541–2546 CrossRef CAS PubMed.
- K. Knez, D. Spasic, F. Delport and J. Lammertyn, Real-time ligation chain reaction for DNA quantification and identification on the FO-SPR, Biosens. Bioelectron., 2015, 67, 394–399 CrossRef CAS PubMed.
- V. Lyamichev, A. L. Mast, J. G. Hall, J. R. Prudent, M. W. Kaiser and T. Takova, et al., Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes, Nat. Biotechnol., 1999, 17, 292–296 CrossRef CAS PubMed.
- Q. B. Li, Z. W. Liu, H. Monroe and C. T. Culiat, Integrated platform for detection of DNA sequence variants using capillary array electrophoresis, Electrophoresis, 2002, 23, 1499–1511 CrossRef CAS PubMed.
- P.-C. Kao, S.-T. Ding, E.-C. Lin, K.-C. Li, L. Wang and Y.-W. Lu, A bead-based single nucleotide polymorphism (SNP) detection using melting temperature on a microchip, Microfluid. Nanofluid., 2014, 17, 477–488 CrossRef CAS.
- K. C. Li, S. T. Ding, E. C. Lin, L. Wong and Y.-W. Lu, Melting analysis on microbeads in rapid temperature-gradient inside microchannels for single nucleotide polymorphisms detection, Biomicrofluidics, 2014, 8(6), 064109 CrossRef PubMed.
- J.-B. Fan, A. Oliphant, R. Shen, B. G. Kermani, F. Garcia and K. L. Gunderson, et al., Highly Parallel SNP Genotyping, Cold Spring Harbor Symp. Quant. Biol., 2003, 68, 69–78 CrossRef CAS PubMed.
- S. O. Sundberg, C. T. Wittwer, J. Greer, R. J. Pryor, O. Elenitoba-Johnson and B. K. Gale, Solution-phase DNA mutation scanning and SNP genotyping by nanoliter melting analysis, Biomed. Microdevices, 2007, 9, 159–166 CrossRef CAS PubMed.
- N. Crews, C. T. Wittwer, J. Montgomery, R. Pryor and B. Gale, Spatial DNA melting analysis for genotyping and variant scanning, Anal. Chem., 2009, 81(6), 2053–2058 CrossRef CAS PubMed.
- M. T. Guo, A. Rotem, J. A. Heymanab and D. A. Weitz, Droplet microfluidics for high-throughput biological assays, Lab on Chip, 2012, 12, 10 Search PubMed.
- H. N. Joensson, M. L. Samuels, E. R. Brouzes, M. Medkova, M. Uhlén and D. R. Link, et al., Detection and analysis of low-abundance cell-surface biomarkers using enzymatic amplification in microfluidic droplets, Angew. Chem., Int. Ed., 2009, 48(14), 4 CrossRef PubMed.
- Z. Guan, Y. Zou, M. Zhang, J. Lv, H. Shen and P. Yang, et al., A highly parallel microfluidic droplet method enabling single-molecule counting for digital enzyme detection, Biomicrofluidics, 2014, 8(1), 014110 CrossRef PubMed.
- Q. Tian, B. Yu, Y. Mu, Y. Xu, C. Ma and T. Zhang, et al., An integrated temporary negative pressure assisted microfluidic chip for DNA isolation and digital PCR detection, RSC Adv., 2015, 5(100), 81889–81896 RSC.
- Y. Zhu and Q. Fang, Analytical detection techniques for droplet microfluidics—A review, Anal. Chim. Acta, 2013, 787, 12 CrossRef PubMed.
- W.-L. Chou, P.-Y. Lee, C.-L. Yang, W.-Y. Huang and Y.-S. Lin, Recent Advances in Applications of Droplet Microfluidics, Micromachines, 2015, 6, 23 CrossRef.
- A. Huebner, D. Bratton, G. Whyte, M. Yang, A. J. deMello and C. Abell, et al., Static microdroplet arrays: a microfluidic device for droplet trapping, incubation and release for enzymatic and cell-based assays, Lab Chip, 2009, 9(5), 692–698 RSC.
- J.-U. Shim, L. F. Olguin, G. Whyte, D. Scott, A. Babtie and C. Abell, et al., Simultaneous Determination of Gene Expression and Enzymatic Activity in Individual Bacterial Cells in Microdroplet Compartments, J. Am. Chem. Soc., 2009, 131(42), 15251–15256 CrossRef CAS PubMed.
- A. Dewan, J. Kim, R. H. McLean, S. A. Vanapalli and M. N. Karim, Growth kinetics of microalgae in microfluidic static droplet arrays, Biotechnol. Bioeng., 2012, 109(12), 2987–2996 CrossRef CAS PubMed.
- J. J. Agresti, E. Antipov, A. R. Abate, K. Ahn, A. C. Rowat and J.-C. Baret, et al., Ultrahigh-throughput screening in drop-based microfluidics for directed evolution, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(9), 4004–4009 CrossRef CAS PubMed.
- Y. Yan, D. Boey, L. T. Ng, J. Gruber, A. Bettiol and N. V. Thakor, et al., Continuous-flow C. elegans fluorescence expression analysis with real-time image processing through microfluidics, Biosens. Bioelectron., 2016, 77, 428–434 CrossRef CAS PubMed.
- E. Zang, S. Brandes, M. Tovar, K. Martin, F. Mech and P. Horbert, et al., Real-time image processing for label-free enrichment of Actinobacteria cultivated in picolitre droplets, Lab Chip, 2013, 13(18), 3707–3713 RSC.
- M. A. Khorshidi, P. K. P. Rajeswari, C. Wahlby, H. N. Joensson and H. Andersson Svahn, Automated analysis of dynamic behavior of single cells in picoliter droplets, Lab Chip, 2014, 14(5), 931–937 RSC.
- S. M. S. Murshed, S.-H. Tan and N.-T. Nguyen, Temperature dependence of interfacial properties and viscosity of nanofluids for droplet-based microfluidics, J. Phys. D: Appl. Phys., 2008, 41(8), 085502 CrossRef.
- T. Sharma, G. S. Kumar, B. H. Chon and J. S. Sangwai, Thermal stability of oil-in-water Pickering emulsion in the presence of nanoparticle, surfactant, and polymer, J. Ind. Eng. Chem., 2015, 22, 324–334 CrossRef CAS.
- G. Bolognesi, A. Hargreaves, A. D. Ward, A. K. Kirby, C. D. Bain and O. Ces, Microfluidic generation of monodisperse ultra-low interfacial tension oil droplets in water, RSC Adv., 2015, 5(11), 8114–8121 RSC.
- W. C. Lei, An effective and interactive approach to particle tracking for DNA melting analysis, National Taiwan University, Taipei, 2015 Search PubMed.
- C. N. Gundry, J. G. Vandersteen, G. H. Reed, R. J. Pryor, J. Chen and C. T. Wittwer, Amplicon melting analysis with labeled primers: A closed-tube method for differentiating homozygotes and heterozygotes, Clin. Chem., 2003, 49(3), 396–406 CAS.
- M. P. B. Nagata, K. Yamashita, M. Miyazaki, H. Nakamura and H. Maeda, Enhanced thermal stability and mismatch discrimination of mutation-carrying DNA duplexes and their kinetic and thermodynamic properties in microchannel laminar flow, Anal. Biochem., 2009, 390(1), 38–45 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |