Open Access Article
Open Access ArticleDelivery of VP1 siRNA to inhibit the EV71 virus using functionalized silver nanoparticles through ROS-mediated signaling pathways†
Yinghua Liac,
Zhengfang Lina,
Tiantian Xua,
Changbing Wanga,
Mingqi Zhaoa,
Misi Xiaoa,
Hanzhong Wangb,
Ning Deng*c and
Bing Zhu *ac
*ac
aCenter Laboratory, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou, 510120, China
bState Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, China
cGuangdong Province, Key Laboratory of Molecular Immunology and Antibody Engineering, Jinan University, Guangzhou, China. E-mail: zhubing2016@hotmail.com; DengNing123@hotmail.com
First published on 5th January 2017
Abstract
Enterovirus 71 (EV71) is the primary causative agent of hand, foot, and mouth disease (HFMD). There is no effective drug therapy for EV71 at present. Small interfering RNA (siRNA), as a new therapeutic modality, provides a promising antiviral treatment, but it is unable to cross cell membranes. To overcome this limitation, nanotechnology has been proposed to mediate siRNA transfection. The antiviral activity of silver nanoparticles (AgNPs) has attracted increasing attention in recent years and can be employed in biomedical interventions. In this study, a simple method to prepare surface decorated AgNPs using polyethylenimine (PEI) and antiviral siRNA has been demonstrated. The development of AgNPs and PEI co-delivery of siRNA was designed to be antiviral. MTT assays and TEM images showed that PEI and siRNA-modified AgNPs (Ag@PEI@siRNA) have remarkable inhibition against EV71 infection and less toxicity to Vero cells. The mechanistic investigations revealed that Ag@PEI@siRNA could block EV71 from infecting host cells and prevent DNA fragmentation, chromatin condensation and activation of caspase-3. Ag@PEI@siRNA effectively inhibited the accumulation of reactive oxygen species (ROS) by the EV71 virus and activation of AKT and p53. Taken together, this study demonstrates that Ag@PEI@siRNA is a novel promising efficient virucide for EV71.
1 Introduction
As one of the major causative agents of hand, foot, and mouth disease (HFMD), enterovirus 71 (EV71) may lead to severe nervous system damage and even fatalities.1–3 EV71 has caused numerous casualties and a large socioeconomic burden to endemic countries and regions.4,5 To date, no effective vaccine or treatment is available, partly due to the fact that the molecular mechanism by which EV71 induces infection remains elusive.6,7 The virus is a member of the enterovirus genus in the family picornaviridae, which includes echoviruses and coxsackieviruses.8,9 The genome of EV71 consists of a single stranded positive-sense RNA of approximately 7400 nucleotides and contains 5′- and 3′-untranslated regions (UTRs) that are essential for viral RNA replication.10–12 The genome is translated as a single large polyprotein that is composed of four capsid proteins, VP1 to VP4, and seven nonstructural proteins, 2A, 2B, 2C, 3A, 3B, 3C and 3D.13–15 There is a surface depression, called “canyon”, around the 5-fold-related plateaus of both the non-infectious empty particle and the infections mature virion.16 VP1 proteins are located around the icosahedral fivefold axes and form the “northern” rim of the canyon. The most prominent surface loop is the “puff”, which forms the “southern rim” of the canyon in VP2. The largest protrusion on the surface of the virus from VP3 is the “knob”. VP4 proteins are attached to the inner surface of the capsid.17 VP1 has been considered a critical site of positive selection in the molecular evolution of EV71.18Small interfering RNA (siRNA) could induce sequence-specific degradation of mRNA and be widely used to knockdown gene expression.19 SiRNA holds great promise as a powerful research tool and represents a novel way for gene therapy.20 Nevertheless, the therapeutic applications of siRNA have been hampered by its strong negative charge, instability in the presence of nucleases and low transfection efficiency, which lead to quick enzymatic degradation and inability to cross cell membranes.21,22 Recently, different types of gene vectors, including lipids, peptide, iron oxide, silica, gold nanoparticles and cationic polymers, have been investigated as carriers for siRNA delivery.23–27 The major obstacles of an effective delivery system are to avoid potentially toxic side effects and low transfection efficiency.28 As one of the most traditional polycationic polymers, polyethylenimine (PEI) has been widely reengineered to protect the siRNA release from endosomes to achieve gene delivery.29
Nanomaterials, as new antimicrobials, have been widely used owing to their unique chemical and physical properties.30,31 Silver nanoparticles (AgNPs) have attracted considerable public attention for a wide range of biomedical applications compared to other patterns of silver sources and have an easy access to cells and tissues because of their unique antimicrobial activities.32–34 AgNPs have been extensively explored as antibacterial, anti-biofilm, anti-fungi, anti-inflammation and anti-thrombosis agents.35 Furthermore, AgNPs have also been developed to contact with HIV, HBV, HSV and inhibit the virus multiplication inside the host cell.36,37 Nanoparticle-mediated siRNA delivery is a promising therapeutic approach.38 Recently, biofunctionalized surface-modified silver nanoparticles for gene delivery were reported.39–42 In this study, we wish to expose novel silver nanoparticles that can interfere with the interaction between the EV71 virus and host cells. Based on the particular properties of both VP1 siRNA and AgNPs, we aimed to verify that PEI and VP1 siRNA-modified AgNPs (Ag@PEI@siRNA) have excellent inhibition of EV71 virus infection. Reactive oxygen species (ROS) play an important role in several physiological processes and encompass highly reactive molecules.30 Oxidative stress, which has a marked imbalance, is often explicated between consumption and cellular defense mechanisms of ROS.43 The redox imbalance is contacted with much pathology, such as diabetes, cancer and other diseases.44 Previous research groups have described the antimicrobial activity of AgNPs, but little is reported about the antiviral mechanisms of AgNPs. Therefore, this study was to ascertain how PEI and siRNA-modified AgNPs in redox balance will antagonize EV71 virus-induced Vero cell apoptosis.
2 Experimental
2.1 Materials
Vero cells were purchased from ATCC®CCL-81. The enterovirus 71 (EV71) virus was subtype C4 (GenBank number FJ360545.1), isolated from the throat swab of a child patient in Guangzhou China. EV71 VP1 siRNA was chemically synthesized by Sangon Biotech (China). AgNO3, vitamin C (VC), branched polyethylenimine (PEI) with a MW of 25 kDa, thiazolyl blue tetrazolium bromide (MTT), 4′,6-diamidino-2-phenyindole (DAPI), 2′,7′-dichlorofluorescein diacetate (DCFH-DA), 6-coumarin and propidium iodide (PI) were obtained from Sigma. LysoTracker Deep Red was obtained from Invitrogen. Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), penicillin and streptomycin were obtained from Gibco. Caspase-3 was obtained from Cell Signaling Technology.2.2 Preparation and characterization of Ag@PEI@siRNA
Ag@PEI@siRNA nanoparticles were synthesized as follows: 0.1 ml 400 μg ml−1 vitamin C was added into 4 ml 400 μg ml−1 AgNO3 drop by drop under magnetic stirring for 2 hours. The excess AgNO3 and VC were eliminated by dialysis against Milli-Q water overnight. Then, 2 ml of a 10 μg ml−1 AgNPs nanoparticles solution was added to 3.792 mg PEI and 12 μl of 10 μM siRNA at room temperature. After stirring for 1 h, the Ag@PEI@siRNA complex was purified by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and re-dispersion in Milli-Q water. The morphology, particle zeta potential and size distribution of AgNPs, Ag@PEI and Ag@PEI@siRNA were characterized by transmission electron microscopy (TEM) and a Zetasizer Nano ZS particle analyzer.
000 rpm for 10 min and re-dispersion in Milli-Q water. The morphology, particle zeta potential and size distribution of AgNPs, Ag@PEI and Ag@PEI@siRNA were characterized by transmission electron microscopy (TEM) and a Zetasizer Nano ZS particle analyzer.
2.3 Determination of siRNA complexation by a gel retardation assay
The encapsulation degree between siRNA and Ag@PEI was assessed by agarose gel electrophoresis.45 Ag@PEI@siRNA nanoparticles were loaded onto 1% agarose gels with Gold View at 100 V for 30 min in TBE buffer. Retardation of siRNA mobility was observed via UV light and gel photography.2.4 In vitro siRNA release
Ag@PEI@siRNA nanoparticles were dispersed in pH 5.3 and pH 7.4. The release rate of siRNA was detected by an Eppendorf Biophotometer Plus (the wavelength of siRNA was 260 nm). In vitro siRNA release of Ag@PEI@siRNA nanoparticles was measured as previously reported.46,472.5 Cell viability assay
The cytotoxicity of Ag@PEI@siRNA nanoparticles was detected with a cell viability assay as previously reported.48 Briefly, Vero cells were seeded in 96-well culture plates at a density of 4 × 104 cells per well at 37 °C for 24 h and then treated with the EV71 virus for 2 h. After incubation with the virus for 2 h, Vero cells were thoroughly washed with PBS to eliminate viruses that were not internalized. The indicated concentrations of Ag@PEI@siRNA were added and incubated with Vero cells for 24 h. After treatment, 20 μl per well of MTT solution (5 mg ml−1 in PBS) was added and incubated for another 5 h. The formazan crystals were dissolved by adding 150 μl per well DMSO. The cell viability was detected at 570 nm using a microplate spectrophotometer. This study was conducted in accordance with the Declaration of Helsinki and with approval from the Ethics Committee of Guangzhou Women and Children's Medical Center, Guangzhou Medical University.2.6 Intracellular trafficking of Ag@PEI@siRNA
To examine the intracellular translocation of the nanoparticles, the localization of 6-coumarin-labeled Ag@PEI@siRNA in Vero cells was monitored via DAPI and LysoTracker using the method previously reported.49 Vero cells were cultured in 6 cm cell culture dishes and incubated with of 6-coumarin-loaded Ag@PEI@siRNA for various time periods. The localization of 6-coumarin labeled Ag@PEI@siRNA in Vero cells was monitored using LysoTracker and DAPI. The cells were washed with PBS three times and were detected by fluorescence microscopy. The mechanism of cellular uptake of Ag@PEI@siRNA in Vero cells was investigated using endocytosis inhibitors.502.7 Evaluation of mitochondrial membrane potential (ΔΨm)
JC-1 was used to assess the status of ΔΨm in Vero cells exposed to Ag@PEI@siRNA, as previously described.51 Cells cultured in 6-well plates were trypsinized and re-suspended in PBS buffer containing 10 μg ml−1 JC-1 and then incubated at 37 °C for 10 min. The cells were then harvested by centrifugation, re-suspended in PBS and analyzed by flow cytometry. The green fluorescence from JC-1 monomers was used to represent the cells that lost ΔΨm.2.8 Annexin-V-FITC staining assay
Translocation of phosphatidylserine in Vero cells treated with Ag@PEI@siRNA was detected using the Annexin-V-FITC staining kit, as previously described.52 Briefly, the cells were treated with Ag@PEI@siRNA for 24 h and stained with Annexin-V-FITC solution for 15 min. Then, the cells were washed with PBS three times and directly analyzed by flow cytometry.2.9 Flow cytometry analysis
The effect of Ag@PEI@siRNA on the cell cycle distribution was detected through flow cytometry, as previously reported.53 The cells incubated with Ag@PEI@siRNA were collected and centrifuged at 1500 rpm for 10 min. The Vero cells were harvested and fixed with pre-cooled 70% ethanol at −20 °C overnight, followed by propidium iodide. The apoptotic cells were measured by quantifying the sub-G1 peak in the cell cycle pattern.2.10 TUNEL–DAPI co-staining assay
DNA fragmentation was detected with fluorescence staining by the TUNEL apoptosis detection kit, as previously described.54 Briefly, the Vero cells were labeled with the TUNEL reaction mixture for 1 h. Vero cells were incubated with 1 μg ml−1 of DAPI for 15 min at 37 °C, washed with PBS three times, and observed under a fluorescence microscope.2.11 Caspase-3 activity
Caspase-3 activity was detected as previously described.55 Vero cells were harvested and suspended in cell lysis buffer and incubated on ice for 1 h. Cell lysates were added into 96-well plates and then incubated with specific caspase-3 substrates (Ac-DEVD-AMC) for 1 h at 37 °C. The fluorescence intensity was detected under fluorescence microscope with excitation at 380 nm and emission at 460 nm.2.12 The efficacy of silencing the VP1 gene by real-time quantitative PCR
The expression of VP1 was quantitatively detected using the real-time quantitative PCR (Q-PCR) technique.56 The primers used in the Q-PCR experiment were as follows: VP1 siRNA (forward 5′GGCAUCAUCAAAUGCUAGUTT3′, reverse 5′ACUAGCAUUUGAUGAUGCCTT3′) and scramble siRNA (forward 5′UUCUCCGAACGUGUCACGUTT3′, reverse 5′ACGUGACACGUUCGGAGAATT3′). The β-actin primers were as follows: (forward 5′GATATCGCCGCGCTCGCTGTC3′, reverse 5′TTCTGACCCATGCCCACCATCAC3′).2.13 TEM image of thin sections of Vero cells
After incubation with EV71 virus for 2 h, Vero cells were rinsed with PBS and incubated with Ag@PEI@siRNA for a period time. TEM analysis of MDCK cells was performed in situ without displacement from the Petri dish, as previous described.57 Briefly, Vero cells were fixed in PBS solution containing 2.5% gluteraldehyde and 4% paraformaldehyde for 1 h. Ultra-thin sections, sliced with an LKB ultramicrotome, were stained with 2% aqueous uranyl acetate and 2% aqueous lead citrate and then imaged under TEM.2.14 Determination of reactive oxygen species (ROS) generation
ROS accumulation inhibited by Ag@PEI@siRNA treated Vero cells was determined as previously described.58 Vero cells were harvested by centrifugation and suspended in PBS containing 10 mM of DCFH-DA. The ROS level was determined by detecting the fluorescence intensity of DCF with excitation at 500 nm and emission at 529 nm.2.15 Western blotting analysis
The effects of Ag@PEI@siRNA treated with Vero cells on the expression levels of various proteins were determined by Western blotting, as previously reported.59 The total proteins were obtained after Vero cells with Ag@PEI@siRNA treatment for 24 h and incubated with lysis buffer. BCA assay was used to quantify the protein concentration. Protein bands were visualized using an enhanced chemiluminescence detection reagent (ECL kit).2.16 Statistical analysis
Experiments were performed and repeated at least three times. All the data are presented as mean ± SD. The differences between two groups were analyzed by two-tailed student's t test. One-way analysis of variance was used in multiple group comparisons. These analyses were carried out using SPSS 13.0 for windows.3 Results and discussion
3.1 Synthesis and characterization of Ag@PEI@siRNA
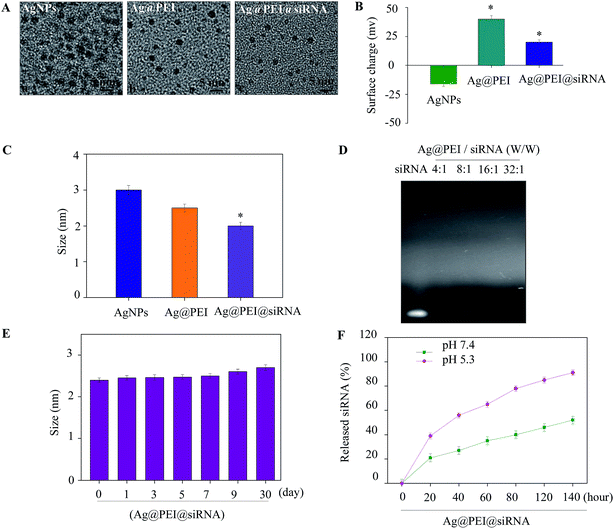
A simple method was used to synthesize functionalized AgNPs through the self-assembly of ligands onto the surface of silver nanoparticles. AgNPs were capped with PEI and VP1 siRNA to form more stable and compact globular nanocomposites. The morphology of the as-prepared Ag@PEI@siRNA was first characterized by TEM. As shown in Fig. 1A, compared with AgNPs, Ag@PEI@siRNA presented a uniformity and monodisperse spherical particles. The formation of silver nanoparticles was achieved as evidenced via UV-vis spectroscopy (Fig. S1†). The UV-vis analysis clearly indicated the formation of silver nanoparticles by displaying strong absorption bands at 420 nm. The observation of absorption bands at this wavelength demonstrates the formation of spherical AgNPs. The zeta potential (Fig. 1B) of Ag@PEI@siRNA has a positive charge and explains how Ag@PEI@siRNA can more easily enter into cells. The size distribution results of different groups of nanoparticles, as shown in Fig. 1C and E, revealed that Ag@PEI@siRNA remained stable for at least 30 days, as observed via dynamic light scattering. The DLS distribution curve is shown in Fig. S2.† The PDI value of Ag@PEI@siRNA was 0.065. The small size of Ag@PEI@siRNA with zeta potential contributed to the highly stable nanostructures and these nanoparticles were able to cross the cell membrane. The binding capacity to VP1 siRNA was further detected. As shown in Fig. 1D, the binding capability of Ag@PEI to VP1 siRNA enhanced with the weight ratio. When the weight ratio was over 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, almost all the VP1 siRNA was retained on the nanocomposites. The siRNA release capacity from Ag@PEI@siRNA was detected at pH 7.4 and pH 5.3 (Fig. 1F). The much more rapid release rate of siRNA from Ag@PEI@siRNA at pH 5.3 should be due to the acidolysis of the nanoparticles. This hydrolysis behavior of siRNA in response to acidic pH conferred the function of pH-controlled release of nanocomposites to this delivery system.
1, almost all the VP1 siRNA was retained on the nanocomposites. The siRNA release capacity from Ag@PEI@siRNA was detected at pH 7.4 and pH 5.3 (Fig. 1F). The much more rapid release rate of siRNA from Ag@PEI@siRNA at pH 5.3 should be due to the acidolysis of the nanoparticles. This hydrolysis behavior of siRNA in response to acidic pH conferred the function of pH-controlled release of nanocomposites to this delivery system.
3.2 In vitro antiviral of Ag@PEI@siRNA
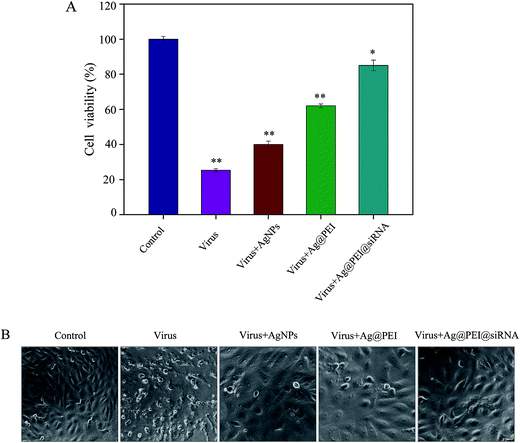
The cytotoxic effects of the EV71 virus on Vero cells and the protective effects of Ag@PEI@siRNA were detected using the MTT assay. Vero cells that were treated with EV71 virus showed a cell viability of 25%; treatment with AgNPs and Ag@PEI increased the cell viability to 40% and 62%, respectively, as shown in Fig. 2A. However, the cell viability was significantly increased to 85% by Ag@PEI@siRNA. These results suggest that the antiviral ability of AgNPs was effectively amplified by the assembly with PEI and siRNA on the surface of the nanoparticles. The protective effects of Ag@PEI@siRNA were further confirmed by phase-contrast observations. As shown in Fig. 2B, Vero cells treated with EV71 virus showed cytoplasmic shrinkage, reduction in cells numbers and loss of cell-to-cell contact. These EV71 virus-induced cell morphological changes were attenuated by co-treatment with Ag@PEI@siRNA and the cells appeared healthy with regularity in shape. Co-delivery platforms for siRNA and chemotherapy agents have been developed to treat various diseases. AgNPs and VP1 siRNA both inhibited the proliferation of EV71 virus. The results suggest that Ag@PEI@siRNA effectively inhibited proliferation of the EV71 virus.3.3 Intracellular localization of Ag@PEI@siRNA
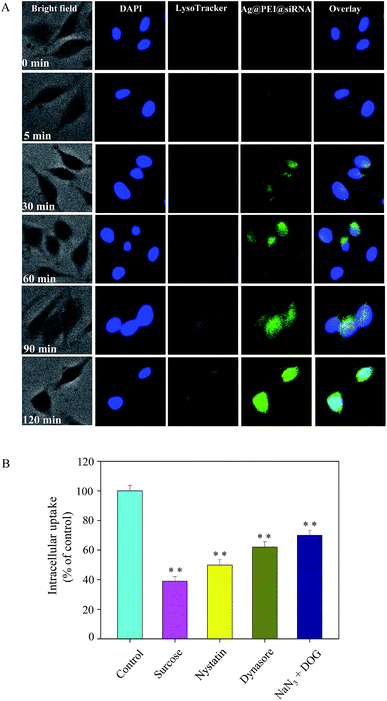
Endocytosis is an important pattern for the cellular uptake of extracellular nanoparticles. As shown in Fig. 3A, the co-localization of lysosomes was found in Vero cells and it increased in a time-dependent manner. Ag@PEI@siRNA escapes from lysosomes after 60 min and are transported into the cytosol and distributed into the cells after 90 min. We used different endocytosis inhibitors to treat the cells before the addition of Ag@PEI@siRNA to further dissect the Ag@PEI@siRNA endocytosis mechanism. As shown in Fig. 3B, treatment with sucrose markedly inhibited the internalization of Ag@PEI@siRNA to 39% of the control, indicating that clathrin-mediated endocytosis was the main pathway. Nystatin and dynasore decreased Ag@PEI@siRNA internalization to 50% and 62% of the control, suggesting that dynamin-mediated pathway was the main pattern of lipid raft-dependent endocytosis. NaN3 and DOG induced the uptake of Ag@PEI@siRNA to 70%, which suggests that Ag@PEI@siRNA is transported into the cells by means of energy-dependent endocytosis. This result demonstrates that the lysosome is the target organelle of Ag@PEI@siRNA.3.4 Morphologic abnormalities in Ag@PEI@siRNA-treated EV71
Negatively stained EV71 virus was observed via TEM to illustrate the interaction between EV71 and Ag@PEI@siRNA. The EV71 virus control, approximately 20 nm, showed typical elliptical or spherically shaped normal EV71 virus containing virus matrix and capsid (Fig. 4A). After interaction of Ag@PEI@siRNA with EV71 virus for 30 minutes (Fig. 4B), the EV71 virus morphology was found to change. Parts of virus edges were lost and the virus morphological structure was almost destroyed. These results show that Ag@PEI@siRNA can interact directly with virus particles, destroy morphological virus structures and inhibit the infectivity of EV71 virus, all of which leads to disruption of virus function in vitro.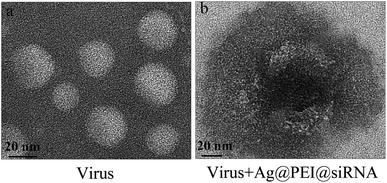 | ||
| Fig. 4 Morphologic abnormalities in Ag@PEI@siRNA-treated EV71 virus via HR-TEM negative staining. (A) EV71 virus control (B) EV71 virus were interacted with Ag@PEI@siRNA. | ||
3.5 Depletion of mitochondrial membrane potential (ΔΨm) and translocation of phosphatidylserine induced by Ag@PEI@siRNA
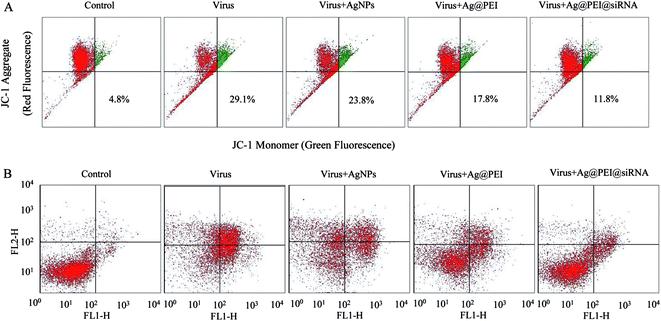
Mitochondria participate in the regulation of apoptosis by integrating the apoptotic signal from intrinsic pathways. Loss of ΔΨm is considered as an initial and irreversible step. Vero cells were treated with the mitochondrial-selective JC-1 dye. As shown in Fig. 5A, treatments of Vero cells with EV71 virus resulted in the elevation of mitochondrial depolarization and the percentage of mitochondrial dysfunction in Vero cells (29.1%). When Vero cells were exposed to Ag@PEI@siRNA, the percentage of mitochondrial membrane potential was significantly decreased (11.8%). These results demonstrate that Ag@PEI@siRNA inhibited virus-triggered apoptotic pathways in Vero cells through induction of mitochondrial dysfunction. The translocation of phosphatidylserine (PS) to the outer leaflet of the cellular membrane is a crucial step in the apoptosis of the Vero cell. An annexin-V-FITC labeling assay was also used to characterize the apoptosis induced by Ag@PEI@siRNA. As shown in Fig. 5B, Vero cells treated with Ag@PEI@siRNA revealed a decrease in Annexin-V positive cells. The mitochondria-mediated pathway may play a more important role in cell apoptosis. These results demonstrate that Ag@PEI@siRNA inhibited EV71 virus infection of Vero cells mainly through inhibiting apoptosis.3.6 Inhibition of EV71 infection of MDCK cells by Ag@PEI@siRNA
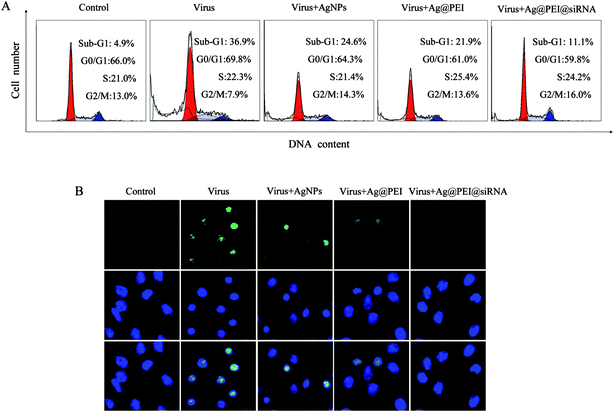
Flow cytometric and TUNEL–DAPI were employed to examine the antiviral mechanism of Ag@PEI@siRNA. As shown in Fig. 6A, the cell population of sub-G1 that represents apoptosis was significantly increased in the DNA histogram with EV71 infection. However, Ag@PEI@siRNA significantly decreased the apoptotic cells population to 11.1%. The induction of apoptosis was further confirmed by DNA fragmentation and nucleus condensation by TUNEL and DAPI co-staining assay. As shown in Fig. 6B, Vero cells exhibited the typical apoptotic features with EV71 virus of DNA fragmentation and nuclear condensation. Ag@PEI@siRNA effectively blocked EV71 virus-induced cell changes in the nuclear morphology. The role of apoptosis in the action of antivirus by Ag@PEI@siRNA has become increasingly clear. These results indicated that Ag@PEI@siRNA rescues Vero cells from EV71 virus-induced apoptosis.3.7 Inhibition of caspase-mediated PARP cleavage by Ag@PEI@siRNA
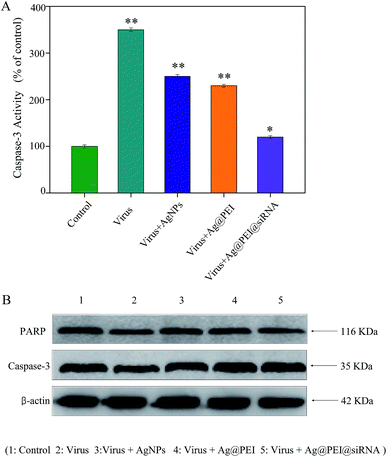
Caspase-3 was confirmed as an important mediator of cell apoptosis. Caspase-3 and PARP were examined via Western blotting to determine cell apoptosis. As shown in Fig. 7A, the treatment of Vero cells with EV71 virus remarkably increased the activity of caspase-3, but Ag@PEI@siRNA decreased the activity of caspase-3. As shown in Fig. 7B, compared to the virus, Ag@PEI@siRNA significantly reduced the activation of caspase-3 and PARP. Caspases act as important mediators of apoptosis and contribute to the overall apoptotic morphology by cleavage of various cellular substrates, such as PARP. The results show that the synthetic nanosystem inhibits EV71 virus activity through reducing cell apoptosis.3.8 Suppression of VP1 mRNA and protein expression by Ag@PEI@siRNA
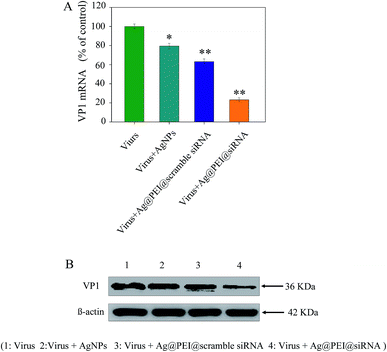
The efficacy of silencing VP1 mRNA with Ag@PEI@siRNA in Vero cells was examined via Q-PCR and Western blotting. As shown in Fig. 8A, the level of VP1 mRNA expression was reduced in the cells exposed to Ag@PEI@siRNA. Compared with Ag@PEI@scramble siRNA, the higher silencing efficiency of Ag@PEI@siRNA could be due to its positive charge. The expression of VP1 in protein levels was detected by Western blotting. As shown in Fig. 8B, Ag@PEI@siRNA effectively restrained the expression of VP1 in Vero cells. Both Q-PCR and Western blotting proved that Ag@PEI@siRNA exhibits the ability to silence VP1 at the mRNA and protein levels, which may control the Vero cell fate by regulation of downstream effectors. RT-PCR and Western blotting data showed that the Ag@PEI had the ability to deliver siRNA and down-regulate VP1 in Vero cell.3.9 TEM image of thin section characterizations of Ag@PEI@siRNA
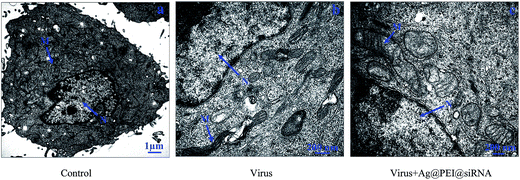
The morphology change of Vero cells treated with Ag@PEI@siRNA was observed via TEM. As shown in Fig. 9, untreated cells exhibited stretched, polygonal and abundant microvilli on the surface, plenty of mitochondria were observed and no morphological changes were observed. When incubated with the EV71 virus, the TEM image shows fewer cells with a reduced volume, the disappearance of microvilli, a shrinking cytoplasm, distorted or lysed organelles and condensed chromatin located along nuclear envelope, indicating apoptosis of the Vero cells. After treatment with Ag@PEI@siRNA, the percentage of Vero cells that lost adhesion and shrunk was decreased and there was a significant decrease in apoptotic cells. The results revealed that Ag@PEI@siRNA can inhibit the infectivity of EV71 virus in vitro.3.10 Inhibition of ROS generation by Ag@PEI@siRNA
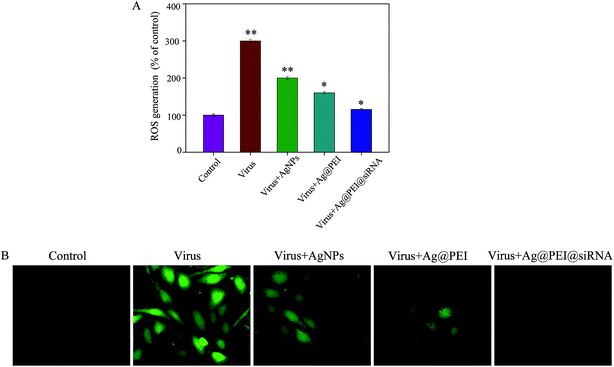
The ROS generation was monitored through DCF assay to indicate the action mechanisms of Ag@PEI@siRNA. As shown in Fig. 10A, the EV71 virus remarkably increased the generation of ROS in Vero cells. Ag@PEI@siRNA significantly decreased the intracellular ROS generation. A stronger fluorescence intensity of DCF was found in Vero cells infected with the EV71 virus, as shown in Fig. 10B. Compared with Ag@PEI@siRNA, the fluorescence intensity of DCF treated with EV71 virus was much stronger. The mitochondrial respiratory chain is a potential source of ROS. Overproduction of ROS impairs mitochondrial ATP synthesis and leads to mitochondrial dysfunction. The results indicate the involvement of ROS in the antiviral action of Ag@PEI@siRNA.3.11 Activation of ROS-mediated signaling pathways by Ag@PEI@siRNA
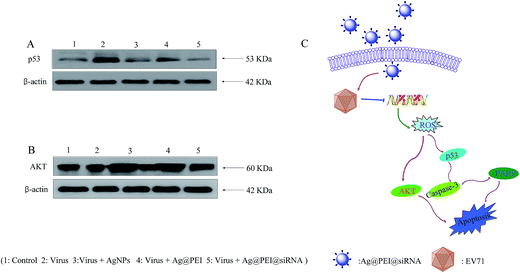
The overexpression of ROS could lead to DNA damage and result in cell apoptosis through regulating p53 and AKT signaling pathways. As a transcriptional factor, p53 affects the DNA repair system, cell cycle arrest and cell apoptosis. AKT kinases regulate several cellular processes that comprise cell proliferation and survival. Western blotting was used to examine the effects on the ROS-mediated signaling pathways for the detection of ROS overproduction in Vero cells exposed to EV71 virus. Western blot analysis showed that virus treatment significantly up-regulated the p53, but treatments with Ag@PEI@siRNA evidently restrained the expression levels of p53 in Vero cells, as shown in Fig. 11A. Similarly, for the AKT signaling pathway, virus treatment significantly inhibited the AKT, but Ag@PEI@siRNA increased the level of AKT protein (Fig. 11B). These results reveal that Ag@PEI@siRNA inhibited EV71 virus-induced Vero cells apoptosis by ROS-mediated p53 and AKT signaling pathways (Fig. 11C).4 Conclusion
In conclusion, this study described the preparation of PEI and siRNA surface decorated silver nanoparticles. Ag@PEI@siRNA exhibits superior abilities for enhanced cellular uptake and block EV71 virus infection. Ag@PEI@siRNA significantly decreased the apoptotic cell population, which prevented the spread of the EV71 virus. The underlying molecular mechanism of the study revealed that the main pattern of cell death was reduced by Ag@PEI@siRNA through reducing cell apoptosis. The mechanisms indicated that Ag@PEI@siRNA inhibited caspase-3 mediated apoptosis through ROS generation. More importantly, AKT and p53 are apoptotic signaling pathway triggered by the Ag@PEI@siRNA in Vero cells. Therefore, the designed surface decorated AgNPs with PEI and siRNA might be a prospective silver species with antiviral properties for treatment of viral diseases.Acknowledgements
This work was supported by the China Postdoctoral Science Foundation (2015M582366), the Technology Planning Project of Guangdong Province (No. 2014A020212697), the Technology Planning Project of Guangdong (No. 201607010120).Notes and references
- R. Li, L. Liu, Z. Mo, X. Wang, J. Xia, Z. Liang, Y. Zhang, Y. Li, Q. Mao, J. Wang, L. Jiang, C. Dong, Y. Che, T. Huang, Z. Jiang, Z. Xie, L. Wang, Y. Liao, Y. Liang, Y. Nong, J. Liu, H. Zhao, R. Na, L. Guo, J. Pu, E. Yang, L. Sun, P. Cui, H. Shi, J. Wang and Q. Li, N. Engl. J. Med., 2014, 370, 829–837 CrossRef CAS PubMed.
- F. Zhu, W. Xu, J. Xia, Z. Liang, Y. Liu, X. Zhang, X. Tan, L. Wang, Q. Mao, J. Wu, Y. Hu, T. Ji, L. Song, Q. Liang, B. Zhang, Q. Gao, J. Li, S. Wang, Y. Hu, S. Gu, J. Zhang, G. Yao, J. Gu, X. Wang, Y. Zhou, C. Chen, M. Zhang, M. Cao, J. Wang, H. Wang and N. Wang, N. Engl. J. Med., 2014, 370, 818–828 CrossRef CAS PubMed.
- F. C. Zhu, F. Y. Meng, J. X. Li, X. L. Li, Q. Y. Mao, H. Tao, Y. T. Zhang, X. Yao, K. Chu, Q. H. Chen, Y. M. Hu, X. Wu, P. Liu, L. Y. Zhu, F. Gao, H. Jin, Y. J. Chen, Y. Y. Dong, Y. C. Liang, N. M. Shi, H. M. Ge, L. Liu, S. G. Chen, X. Ai, Z. Y. Zhang, Y. G. Ji, F. J. Luo, X. Q. Chen, Y. Zhang, L. W. Zhu, Z. L. Liang and X. L. Shen, Lancet, 2013, 381, 2024–2032 CrossRef CAS.
- F. C. Zhu, Z. L. Liang, X. L. Li, H. M. Ge, F. Y. Meng, Q. Y. Mao, Y. T. Zhang, Y. M. Hu, Z. Y. Zhang, J. X. Li, F. Gao, Q. H. Chen, Q. Y. Zhu, K. Chu, X. Wu, X. Yao, H. J. Guo, X. Q. Chen, P. Liu, Y. Y. Dong, F. X. Li, X. L. Shen and J. Z. Wang, Lancet, 2013, 381, 1037–1045 CrossRef CAS.
- Y. P. Li, Z. L. Liang, J. L. Xia, J. Y. Wu, L. Wang, L. F. Song, Q. Y. Mao, S. Q. Wen, R. G. Huang, Y. S. Hu, X. Yao, X. Miao, X. Wu, R. C. Li, J. Z. Wang and W. D. Yin, J. Infect. Dis., 2014, 209, 46–55 CrossRef CAS PubMed.
- C. C. Liao, A. T. Liou, Y. S. Chang, S. Y. Wu, C. S. Chang, C. K. Lee, J. T. Kung, P. H. Tu, Y. Y. Yu, C. Y. Lin, J. S. Lin and C. Shih, J. Virol., 2014, 88, 12485–12499 CrossRef PubMed.
- H. Zhang, L. Song, H. Cong and P. Tien, J. Virol., 2015, 89, 10031–10043 CrossRef CAS PubMed.
- M. H. Ooi, S. C. Wong, P. Lewthwaite, M. J. Cardosa and T. Solomon, Lancet Neurol., 2010, 9, 1097–1105 CrossRef PubMed.
- Y. Zhang, Z. Zhu, W. Yang, J. Ren, X. Tan, Y. Wang, N. Mao, S. Xu, S. Zhu, A. Cui, Y. Zhang, D. Yan, Q. Li, X. Dong, J. Zhang, Y. Zhao, J. Wan, Z. Feng, J. Sun, S. Wang, D. Li and W. Xu, J. Virol., 2010, 7, 94 CrossRef PubMed.
- J. Qing, Y. Wang, Y. Sun, J. Huang, W. Yan, J. Wang, D. Su, C. Ni, J. Li, Z. Rao, L. Liu and Z. Lou, PLoS Pathog., 2014, 10, e1004422 Search PubMed.
- Z. Reed and M. J. Cardosa, Vaccine, 2016, 34, 2967–2970 CrossRef CAS PubMed.
- S. Y. Leong, B. K. Ong and J. J. Chu, PLoS Pathog., 2014, 11, e1004686 Search PubMed.
- L. C. Wang, S. O. Chen, S. P. Chang, Y. P. Lee, C. K. Yu, C. L. Chen, P. C. Tseng, C. Y. Hsieh, S. H. Chen and C. F. Lin, J. Virol., 2015, 89, 7028–7037 CrossRef CAS PubMed.
- P. Chong, C. C. Liu, Y. H. Chow, A. H. Chou and M. Klein, Clin. Infect. Dis., 2014, 60, 797–803 CrossRef PubMed.
- K. L. Shingler, J. O. Cifuente, R. E. Ashley, A. M. Makhov, J. F. Conway and S. Hafenstein, J. Virol., 2014, 89, 1900–1908 CrossRef PubMed.
- X. Ye, C. Fan, Z. Ku, T. Zuo, L. Kong, C. Zhang, J. Shi, Q. Liu, T. Chen, Y. Zhang, W. Jiang, L. Zhang, Z. Huang and Y. Cong, PLoS Pathog., 2016, 12, e1005454 Search PubMed.
- P. Plevka, R. Perera, J. Cardosa, R. J. Kuhn and M. G. Rossmann, Science, 2012, 336, 1274 CrossRef CAS PubMed.
- C. Kataoka, T. Suzuki, O. Kotani, N. Iwata-Yoshikawa, N. Nagata, Y. Ami, T. Wakita, Y. Nishimura and H. Shimizu, PLoS Pathog., 2015, 11, e1005033 Search PubMed.
- Z. H. Chen, T. Yu, B. F. Zhou, J. H. Wei, Y. Fang, J. Lu, L. Guo, W. Chen, Z. P. Liu and J. H. Luo, Biomaterials, 2016, 81, 125–134 CrossRef CAS PubMed.
- X. Huang, A. Pallaoro, G. B. Braun, D. P. Morales, M. O. Ogunyankin, J. Zagadzinski and N. O. Reich, Nano Lett., 2014, 14, 2046–2051 CrossRef CAS PubMed.
- H. J. Kim and C. Leal, ACS Nano, 2015, 9, 10214–10226 CrossRef CAS PubMed.
- H. He, N. Zheng, Z. Y. Song, K. H. Kim, C. Yao, R. J. Zhang, C. L. Zhang, Y. H. Huang, F. M. Uckun, J. J. Cheng, Y. F. Zhang and L. C. Yin, ACS Nano, 2016, 10, 1859–1870 CrossRef CAS PubMed.
- F. Perche, Y. Yi, L. Hespel, P. Mi, A. Dirisala, H. Cabral, K. Miyata and K. Kataoka, Biomaterials, 2016, 90, 62–71 CrossRef CAS PubMed.
- L. Li, M. Munoz-Culla, U. Carmona, M. P. Lopez, F. Yang, C. Trigueros, D. Otaegui, L. Zhang and M. Knez, Biomaterials, 2016, 98, 143–151 CrossRef CAS PubMed.
- C. F. Xu, H. B. Zhang, C. Y. Sun, Y. Liu, S. Shen, X. Z. Yang, Y. H. Zhu and J. Wang, Biomaterials, 2016, 88, 48–59 CrossRef CAS PubMed.
- P. Zhang, C. Wang, J. Zhao, A. Xiao, Q. Shen, L. Li, J. Li, J. Zhang, Q. Min, J. Chen, H. Y. Chen and J. J. Zhu, ACS Nano, 2016, 10, 3637–3647 CrossRef CAS PubMed.
- K. Moller, K. Muller, H. Engelke, C. Brauchle, E. Wagner and T. Bein, Nanoscale, 2016, 8, 4007–4019 RSC.
- C. F. Xu, Y. Liu, S. Shen, Y. H. Zhu and J. Wang, Biomaterials, 2015, 51, 1–11 CrossRef CAS PubMed.
- T. E. Park, B. Singh, H. Li, J. Y. Lee, S. K. Kang, Y. J. Choi and C. S. Cho, Biomaterials, 2015, 38, 61–71 CrossRef CAS PubMed.
- B. Zhu, Y. H. Li, Z. F. Lin, M. Q. Zhao, T. T. Xu, C. B. Wang and N. Deng, Nanoscale. Res. Lett., 2016, 11, 1–8 CrossRef CAS PubMed.
- H. Wu, J. Lin, P. D. Liu, Z. H. Huang, P. Zhao, H. Z. Jin, C. L. Wang, L. P. Wen and N. Gu, Biomaterials, 2015, 62, 47–57 CrossRef CAS PubMed.
- H. Qin, H. L. Cao, Y. C. Zhao, C. Zhu, T. Cheng, Q. J. Wang, X. C. Peng, M. Q. Cheng, J. X. Wang, G. D. Jin, Y. Jiang, X. L. Zhang, X. Y. Liu and P. K. Chu, Biomaterials, 2014, 35, 9114–9125 CrossRef CAS PubMed.
- J. P. Shi, X. Sun, Y. Lin, X. Y. Zou, Z. J. Li, Y. Y. Liao, M. M. Du and H. W. Zhang, Biomaterials, 2014, 35, 6657–6666 CrossRef CAS PubMed.
- L. L. Huo, R. Chen, L. Zhao, X. F. Shi, R. Bai, D. X. Long, F. Chen, Y. L. Zhao, Y. Z. Chang and C. Y. Chen, Biomaterials, 2015, 61, 307–315 CrossRef CAS PubMed.
- D. W. Guo, L. Y. Zhu, Z. H. Huang, H. X. Zhou, Y. Ge, W. J. Ma, J. Wu, X. Y. Zhang, X. F. Zhou, Y. Zhang, Y. Zhao and N. Gu, Biomaterials, 2013, 34, 7884–7894 CrossRef CAS PubMed.
- A. Z. Bekele, K. Gokulan, K. M. Williams and S. Khare, Foodborne Pathog. Dis., 2016, 13, 239–244 CrossRef CAS PubMed.
- Y. N. Chen, Y. H. Hsueh, C. T. Hsieh, D. Y. Tzou and P. L. Chang, Int. J. Environ. Res. Public Health, 2016, 13, 430 CrossRef PubMed.
- V. D. Badwaik, E. Aicart, Y. A. Mondjinou, M. A. Johnson, V. D. Bowman and D. H. Thompson, Biomaterials, 2016, 84, 86–98 CrossRef CAS PubMed.
- K. Sarkar, S. L. Banerjee, P. P. Kundu, G. Madras and K. Chatterjee, J. Mater. Chem. B, 2015, 3, 5266–5276 RSC.
- J. Ramos, T. Potta, O. Scheideler and K. Rege, ACS Appl. Mater. Interface, 2014, 6, 14861–14873 CrossRef CAS PubMed.
- D. D. Sun, W. W. Zhang, N. Li, Z. W. Zhao, Z. P. Mou, E. D. Yang and W. Y. Wang, Mater. Sci. Eng., C, 2016, 63, 522–534 CrossRef CAS PubMed.
- N. Kasyanenko, V. Bakulev, I. Perevyazko, T. Nekrasova, O. Nazarova, A. Slita, Y. Zolotova and E. Panarin, J. Biotechnol., 2016, 236, 78–87 CrossRef CAS PubMed.
- Y. H. Li, X. L. Li, Y. S. Wong, T. F. Chen, H. B. Zhang, C. R. Liu and W. J. Zheng, Biomaterials, 2011, 32, 9068–9076 CrossRef CAS PubMed.
- Y. H. Li, X. L. Li, W. J. Zheng, C. D. Fan, Y. B. Zhang and T. F. Chen, J. Mater. Chem. B, 2013, 1, 6365–6372 RSC.
- W. Y. Li, Y. J. Liu, J. W. Du, K. F. Ren and Y. X. Wang, Nanoscale, 2015, 7, 8476–8484 RSC.
- K. D. Wegner and N. Hildebrandt, Chem. Soc. Rev., 2015, 44, 4792–4834 RSC.
- Y. H. Li, Z. F. Lin, M. Q. Zhao, T. T. Xu, C. B. Wang, H. M. Xia, H. Z. Wang and B. Zhu, Int. J. Nanomed., 2016, 11, 3065–3076 CrossRef CAS PubMed.
- T. Senthilkumar, N. Parekh, S. B. Nikama and S. K. Asha, J. Mater. Chem. B, 2016, 4, 299–308 RSC.
- H. L. Wu, X. L. Li, W. Liu, T. F. Chen, Y. H. Li, W. J. Zheng, C. W. Y. Man, M. K. Wong and K. H. Wong, J. Mater. Chem., 2012, 22, 9602–9610 RSC.
- W. Q. Cao, W. J. Zheng and T. F. Chen, Sci. Rep., 2015, 5, 9157 CrossRef CAS PubMed.
- Y. B. Zhang, X. L. Li, Z. Huang, W. J. Zheng, C. D. Fan and T. F. Chen, J. Nanomed. Nanotechnol., 2013, 9, 74–84 CrossRef CAS PubMed.
- C. P. Zheng, J. S. Wang, Y. N. Liu, Q. Q. Yu, Y. Liu, N. Deng and J. Liu, Adv. Funct. Mater., 2014, 24, 6872–6883 CrossRef CAS.
- F. Yang, Q. M. Tang, X. Y. Zhong, Y. Bai, T. F. Chen, Y. B. Zhang, Y. H. Li and W. J. Zheng, Int. J. Nanomed., 2012, 7, 835–844 CrossRef CAS PubMed.
- T. Liu, L. L. Zeng, W. T. Jiang, Y. T. Fu, W. J. Zheng and T. F. Chen, J. Nanomed. Nanotechnol., 2015, 11, 947–958 CrossRef CAS PubMed.
- W. Liu, X. L. Li, Y. S. Wong, W. J. Zheng, Y. B. Zhang, W. Q. Cao and T. F. Chen, ACS Nano, 2012, 6, 6578–6591 CrossRef CAS PubMed.
- W. J. Zheng, T. T. Yin, Q. C. Chen, X. Y. Qin, X. Q. Huang, S. Zhao, T. Y. Xu, L. M. Chen and J. Liu, Acta Biomater., 2016, 31, 197–210 CrossRef CAS PubMed.
- J. M. Li, Y. Y. Wang, M. X. Zhao, C. P. Tan, Y. Q. Li, X. Y. Le, L. N. Ji and Z. W. Mao, Biomaterials, 2012, 33, 2780–2790 CrossRef CAS PubMed.
- B. Yu, X. L. Li, W. J. Zheng, Y. X. Feng, Y. S. Wong and T. F. Chen, J. Mater. Chem. B, 2014, 2, 5409–5418 RSC.
- L. Zhou, X. M. Duan, S. Zeng, K. Men, X. Y. Zhang, L. Yang and X. Li, Int. J. Nanomed., 2015, 10, 5205–5218 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra26472g |
| This journal is © The Royal Society of Chemistry 2017 |