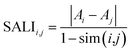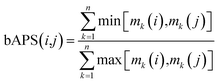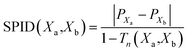Open Access Article
Open Access ArticleGetting SMARt in drug discovery: chemoinformatics approaches for mining structure–multiple activity relationships†
Fernanda I. Saldívar-González a,
J. Jesús Naveja
a,
J. Jesús Naveja abc,
Oscar Palomino-Hernández
abc,
Oscar Palomino-Hernández a and
José L. Medina-Franco
a and
José L. Medina-Franco *a
*a
aFacultad de Química, Departamento de Farmacia, Universidad Nacional Autónoma de México, Avenida Universidad 3000, Mexico City 04510, Mexico. E-mail: medinajl@unam.mx; jose.medina.franco@gmail.com; Tel: +52-55-5622-3899 ext. 44458
bFacultad de Medicina, PECEM, Universidad Nacional Autónoma de México, Avenida Universidad 3000, Mexico City, 04510, Mexico
cInstitute of Bioinformatics and Systems Biology, Helmholtz Zentrum München, 85764, Neuherberg, Germany
First published on 3rd January 2017
Abstract
In light of the high relevance of polypharmacology, multi-target screening is a major trend in drug discovery. As such, the increasing amount of available structure–activity data requires the application of chemoinformatic approaches to mine structure–multiple activity relationships. To this end, activity landscape methods, initially developed to explore the structure–activity relationships for compounds screened against one target, have been adapted to mine Structure–Multiple Activity Relationships (SMARt). Herein, we survey advances in the chemoinformatic approaches to retrieve SMARt from screening data sets. Case studies relevant to modern drug discovery are discussed. The methods covered in this survey are general and can be implemented to explore the SMARt of other data sets screened across multiple biologically endpoints.
Introduction
Analysis of structure–activity relationships (SAR) is a common practice in many areas of chemistry. Most medicinal and computational chemists working on drug discovery obtain SAR of compound data sets on a routine basis. This is true not only in academic settings but also in the pharmaceutical industry and research institutes. In several current drug discovery projects, compound data sets are screened across more than one biological endpoint. Depending on the project, it is desirable to identify selective compounds or identify molecules with activity across multiple endpoints. Moreover, in light of the increasing awareness of polypharmacology1 and multi-target drug discovery,2 screening small compound data sets or large chemical libraries across more than one biological endpoint is a fundamental task. Therefore, getting Structure–Multiple Activity Relationships (SMARt) is a common need in drug discovery.Methods to get SMARt can be broadly classified into qualitative and quantitative. Qualitative approaches can be applied without the need of computational tools and depend on the experience of the chemist analyzing the data. Thus, qualitative methods are suitable to handle small-to-medium size data sets. In contrast, large screening data sets, in particular those tested across several endpoints, usually require the application of computational procedures in addition to the experience of the chemist.3 In these cases, in silico methods can be performed for either predictive or descriptive purposes. As discussed previously, understanding the SAR of compound data sets should be performed before developing predictive models4 such as QSAR and QSPR in order to predict novel, potent, and selective compounds.5,6 In this regard, new computational models that combine multi-target QSAR with machine learning such as artificial neural network algorithms have been developed with the aim of predict the interactions of multiple molecules to targets involved in many diseases and processes of neuroprotection.5,7
Activity landscape modeling (ALM) is a chemoinformatic strategy to mine the SAR of compound data sets and it is actively used in academia, industry and other research settings. ALM can be regarded as part of computer-aided drug design and it is an important component in medicinal chemistry.8 For more than ten years several groups have worked on the development of ALM. These approaches relay on the quantitative comparison of structure similarity with activity similarity (or potency difference) for all pairs of compounds in a screening data set. Over the years a large number of quantitative and visual methods have been developed. Most of these methods started with the main goal of identifying ‘activity cliffs’: pairs of compounds with very similar structure but unexpected high activity difference.9 Activity cliffs have a ‘dual face’ with a large impact in medicinal and computational chemistry.10 It has been largely advised that after identifying activity cliffs, molecular modeling studies should be conducted that help to explain, at the molecular level, the reason associated with the large change in activity due to a small modification the chemical structure. Such studies are highly valuable because add three-dimensional information to the system. To this end, mechanistic studies towards the structural interpretation of activity cliffs in three dimensions have been published.11,12 Overall, the specific reasons that are associated with the formation of the activity cliffs depend on the system. An alternative approach to add three-dimensional information to the system and consider additional effects of functional groups, conformations and configurations, molecular descriptors that take into account the coordinates space of the compounds or even using several different conformations of the molecules in the data set have been reported.13,14
ALM seeks not only to identify activity cliffs but other significant areas of the activity landscape such as ‘similarity cliffs’ (which are related to scaffold hops)15 and other continuous regions of the activity landscape. The broad applicability of ALM in medicinal chemistry has been reviewed.16 Initially developed to describe SARs, ALM has been tested for predictive purposes.17,18 Similarly, ALM was originally applied to describe the SAR of compound data sets screened for one biological endpoint, for instance, for a single target. However, several methods used in ALM have been adapted to mine SMARts.
The goal of this work is to survey the progress of ALM to get SMARt in drug discovery. We put special emphasis on the development and application of Structure–Activity Similarity (SAS) maps which were one of the first approaches used in ALM.19 Four years ago the authors reviewed the development of SAS maps to explore SARs.20 In contrast, this review covers the most recent developments and applications aimed to explore SMARts. As part of the recent developments the concept of ‘pro-activity cliffs’ is introduced. The manuscript is organized in five main sections: after this introduction a brief overview of the SAS maps is presented with special emphasis on the development of density SAS maps and activity landscape sweeping strategies. The section after that describes the adaptation of ALM from single to multi-target activity analysis. This section is followed by a discussion of future trends in SAR and SMARt analysis using ALM. Concluding remarks are presented at the end.
Structure–activity similarity (SAS) maps
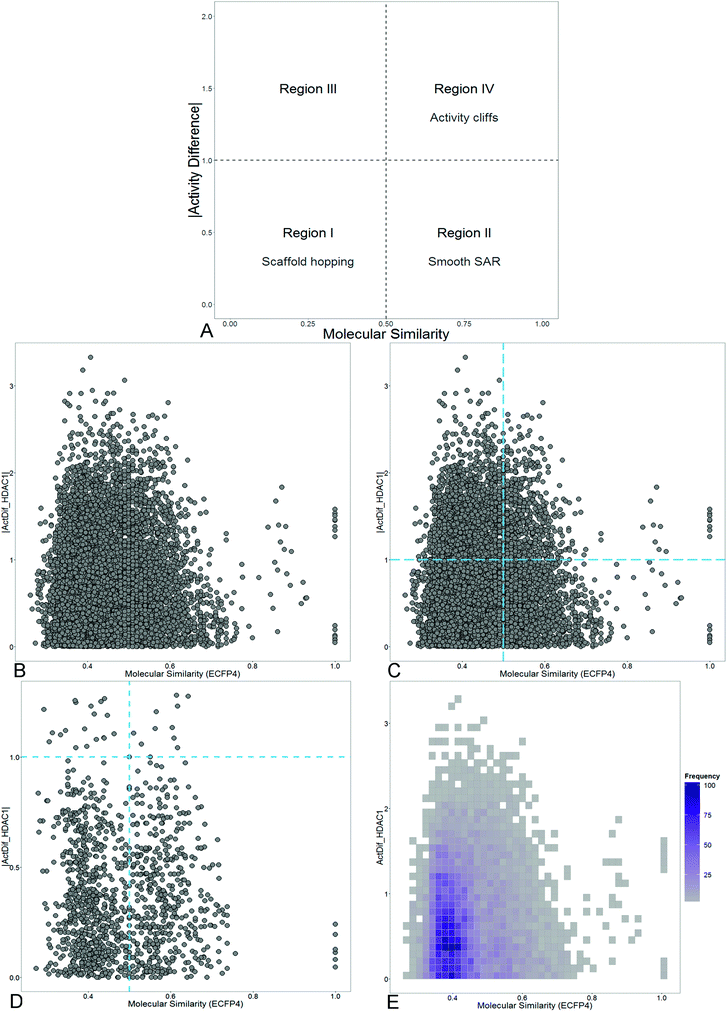
SAS maps were proposed in 2001.19 The basic idea of a SAS map is to plot in two-dimensions (2D) the pairwise structure similarity (usually plotted on the X-axis) and activity difference (plotted on the Y-axis) for all pairs of compounds in a data set. A general form of a SAS map is shown in Fig. 1A. To aid in the interpretation, a SAS map can be roughly divided in four major quadrants each one distinguishing pairs of compounds with high/low activity difference and high/low structure similarity. Activity cliffs are located in the quadrant that identifies pairs of molecules with high structure similarity and high activity difference (region IV). Compound pairs with a smooth SAR have high structure similarity and low activity difference (region II). Scaffold hops (or similarity cliffs) are located in the opposite quadrant of the activity cliffs (region I). Noteworthy, even in the absence of the thresholds with formally defined quadrants, SAS maps are helpful to differentiate major regions in the landscape.One of the known limitations of the SAS maps is the quantitative criteria to define the thresholds along the X- and Y-axis. A number of approaches to address this issue are discussed elsewhere.20 Briefly, the thresholds that define high/low activity difference depend on the goal of the project. Usual cutoffs are one, two or more potency units. The thresholds to define high/low structure similarity can be set up based on the distribution of the similarity values of the data set. In some instances, heuristic values of similarity are considered based on author's experience.
Another limitation of the SAS maps is the large amount of data points that could be generated. Therefore, for large data sets it is challenging the visual interpretation of the SAS maps. To address this issue several strategies have been proposed which are discussed below.
Density SAS maps
To aid in the visualization of the SAS and related maps three major strategies have been developed: (1) categorical SAS maps;13 (2) filtered SAS maps showing only the most relevant data points (for instance, the ‘active pairs’ of compounds defined as pair of molecules containing at least one active compound in the pair) and, more recently (3) density SAS maps that display the amount of data points using a continuous color scale.21 Fig. 1 shows examples of ‘simplified’ SAS maps: categorical, filtered and density SAS map for a data set of 140 pyrimidine hydroxyl amide compounds tested with histone deacetylase 1 (HDAC1). These compounds were synthesized and tested as part of a program of optimization to find potent and selective inhibitors of HDAC6, enzyme required for the formation of the aggresome and survival of cancer cells.22 HDAC is a major epigenetic target and the computational analysis of the SAR can be regarded as part of the emerging research field of Epi-Informatics.23 SAR analysis of HDAC inhibitors is particularly useful for the treatment of proliferative diseases and disorders by protein deposition, likewise, it is useful for probing biological pathways. A full discussion of the SAR of HDAC inhibitors is out of the scope of this Short Review that is focused on ALM. Fig. S1 in the ESI† shows additional examples of simplified SAS maps for a data set of 91 compounds tested against the parasite Giardia intestinalis. Note that density SAS maps provide better information regarding the general distribution of the data points, though sacrificing the chance of including information regarding the individual activity of any of the compounds in the pair.Several analyses have shown that the similarity cliff region is one of the most populated for several data sets.13 Results of Maggiora et al. further confirmed these observations analyzing many data sets.15 This is also the case in the activity landscape depicted in Fig. 1 and S1.† Density SAS maps have been employed to analyze the ALM of 5α-alpha reductase inhibitors24 and inhibitors of DNA metiltransferases (DNMTs), other major epigenetic target.21
Activity landscape sweeping
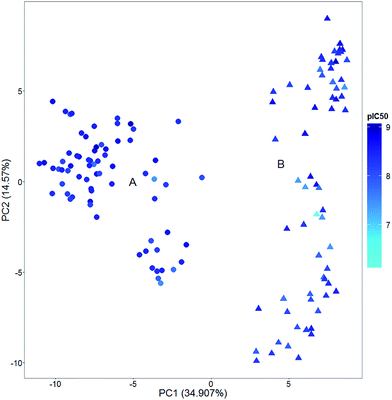
Activity landscape sweeping is a strategy recently developed to ‘clean’ the SAR/SMARt of a data set by filtering first the compounds that are considered to analyze the landscape. An approach is to classify the compounds by the types of molecular scaffold25 or the relative position in chemical space, to name two criteria. Then, the ALM would be centered on the local SAR of the filtered molecules. In a broad sense, activity landscape sweeping is an approach to analyze local models of SAR/SMARts. Despite the fact that such models are not general, activity landscape sweeping gives rise to focused analysis of the most interpretable areas of the activity landscape.In order to illustrate the filtering of compounds before ALM, i.e., activity landscape sweeping, Fig. 2 shows a visual representation of the chemical space of a series of 140 pyrimidine hydroxyl amide compounds synthesized and evaluated as HDAC6 inhibitors. Two main clusters (A: circles, B: triangles) are readily distinguished: compounds of cluster A correspond to formulas IV–VIII described by Van Duzer et al. while compounds of cluster B correspond to formulas I–III described in the same work.22 The main difference between these two groups is the carbon attached to the nitrogen of 2-amino-N-hydroxypyrimidine-5-carboxamide. In group A, this carbon is tertiary, while in group B is primary or secondary. Representative chemical structures are shown in Fig. S2 of the ESI.†
Activity landscape sweeping has been recently applied to DNMT inhibitors21 and 5α-reductase inhibitors.24 In both instances activity landscape sweeping was used in conjunction with SAS maps. This approach helped to ‘clean’ the landscape and facilitated the visual analysis of the SAS maps. Activity landscape sweeping has been used in conjunction with SAS maps but could be implemented in combination with any other ALM strategy such as Structure–Activity Landscape Index (SALI)26 or other methods.
SAS maps and PLIFS
Protein–ligand interaction fingerprints (PLIFS) are convenient representations to capture protein–ligands contacts in a systematic manner. PLIFS are at the interface of chemoinformatics and molecular modeling27 and have been designed to ‘capture a 1D representation of the interactions between ligand and protein either in complexes of known structure or in docked poses’.28 Recently SAS maps have been adapted to analyze structure–protein ligand interactions giving rise to the protein–ligand interaction cliffs.27 These are defined as pairs of compounds with high structure similarity, high protein–ligand contact similarity but very different activity profile. That study was conducted for a series of kinase inhibitors. In that work, Méndez-Lucio et al. integrated PLIFS to a multi-target kinase activity landscape analysis. Three data sets, containing the crystallographic structure of the ligand bound to a kinase were used. The authors employed three data sets, containing the crystallographic structure of the ligand bound to a kinase. Pairwise interaction similarity was assessed using PLIFs and the Tanimoto coefficient, whereas twelve 2D and 3D molecular descriptors were used to compute pairwise molecular similarity. Pairwise structure-similarity analysis revealed no correlation with interaction similarity in none of the data sets despite the fact that the kinase ATP binding site is highly conserved. On average, only 33% of the molecular pairs categorized as highly similar showed similar interactions. This approach not only provided structural information of activity cliffs but it also was useful to identify hot spots in the target protein associated with selectivity.27,29Tuning ALM to get SMARt
In addition to SAS maps several other methods have been developed for ALM analysis.16,20,26,30,31 For instance SALI, the first index developed to rapidly identify activity cliffs, is calculated with the expression:26where Ai and Aj are the activities of the ith and jth molecules, and sim(i,j) is the similarity coefficient between the two molecules. Also, the research group of Bajorath has developed a large number of approaches for ALM.16
Several of ALM methods have been adapted to handle SMARt. For instance, a straightforward extension of SALI to measure SMARt is replacing the numerator of the SALI with the biological profile similarity of the compound pair computed with the Tanimoto coefficient (giving rise to a Structure–Multiple Activity Landscape Index).32 Representative case studies of the adaptation of ALM to get SMARt are summarized in Table 1 and discussed in the next sections.
| Study | Major outcome (method) | Major outcome (interpretation) | Ref. |
|---|---|---|---|
| SMART of >50 benzimidazoles tested with T. vaginalis and G. intestinalis | Dual activity difference maps with fingerprint and sub-structure representation | ‘Activity switches’ are introduced: pairs of compounds where one small change in the structure is associated with a different and opposite change in the activity of two biological endpoints | 34 and 35 |
| SMART of a series of purine analogs screened against the cysteine protease cathepsins | Triple activity difference maps | The concept of structure–property–activity (SPA) similarity in SAR studies are introduced. SPA maps are analyzed to determine the extent to which property similarities could be applied to characterize SARs | 14 |
| SMART of compounds in PubChem | Structure multiple Activity Similarity (SmAS) maps | Bioassay activity landscape is introduced to study the relationship between the structure and bioactivity profiles | 37 |
| ADMET analysis of 166 compounds screened for kappa-opioid receptor activity | ADMET property–activity pairwise similarity maps with ADMET descriptors and dimensional ‘violation bit vector’ representing | Study of the range of ADMET property violations that arise from structural changes, subtle and significant | 41 |
SMART of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 252 compounds screened across 100 diverse proteins reported by Clemons et al.38 252 compounds screened across 100 diverse proteins reported by Clemons et al.38 |
SPID measure (Structure–Promiscuity Index Difference) | Structure promiscuity index is introduced to identify the pairs of compounds with high structure–similarity but large activity difference | 39 |
SMARt with few biological endpoints
One of the first applications of SAS maps applied to analyze data sets across more than one biological endpoint was the SMARt exploration of more than 50 benzimidazole analogues tested for their ability to inhibit the growth of the protozoa Trichomonas vaginalis and Giardia intestinalis.33 A tool to analyze simultaneously the difference in activity data for both parasites was the Dual Activity Difference (DAD) maps. DAD maps represent in 2D changes in potency difference for two targets.34 One of the major outcomes of the DAD maps are ‘activity switches’ defined as pairs of compounds where one small change in the structure is associated with a very different but opposite change in the activity for both biological endpoints. Activity switches have been reviewed in detail.20 Triple-Activity Difference (TAD) maps where developed later as a natural extension of the DAD maps to analyze SMARts.14More recently, DAD maps were used to analyze systematically the activity landscape of a series of 91 benzimidazoles tested with the parasites T. vaginalis and G. intestinalis.35 In that work the chemical structure of the 91 benzimidazoles was encoded using a fragment-based approach that indicated the presence or absence of six substituents around a common benzimidazole nucleus. Using DAD maps, single and dual substitutions around the benzimidazole scaffold were identified that were associated with large changes in potency for each of the two parasites. Furthermore, single and dual substitutions associated with large and opposite changes in activity for the two parasites were found.35
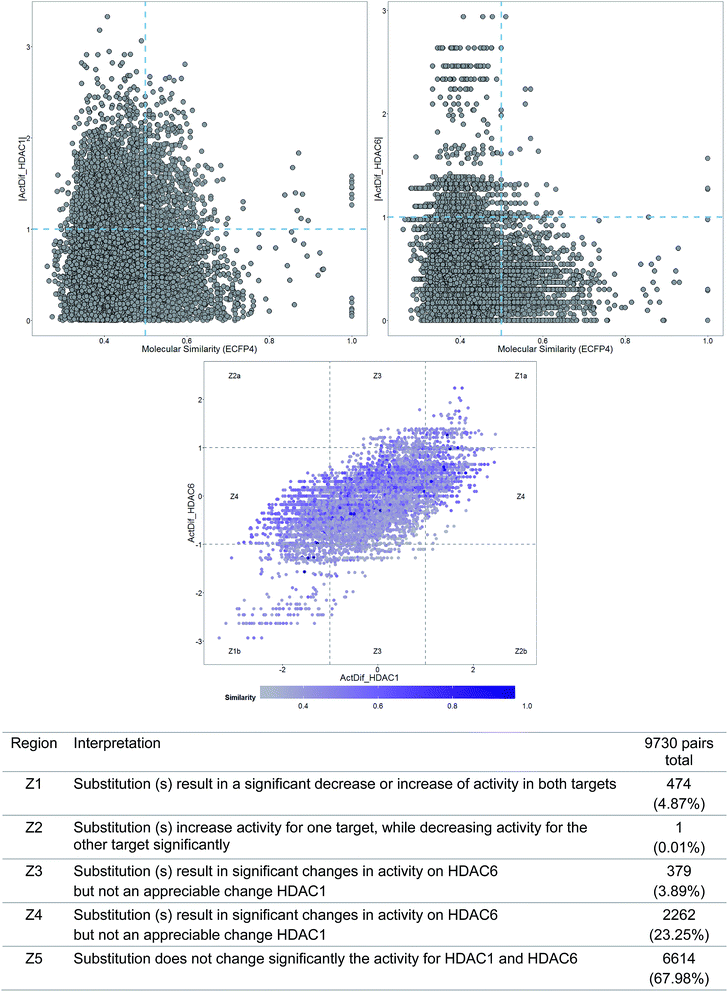
To illustrate a DAD map, Fig. 3 shows a plot of a data set of 140 molecules tested as HDAC1 and HDAC6 inhibitors.22 As reference, Fig. 3 also shows the corresponding SAS maps for HDAC1 and HDAC6. In general, the DAD map in Fig. 3 shows that the larger amount of pairs of compounds are located in the region Z5 of the plot (close to 68%), indicating that most of the compounds show activity values very similar for both enzymes. The pairs identified in the Z3 and Z4 regions (simple activity cliffs) suggest that changes in the scaffold are more susceptible to present changes in activity against HDAC1 compared with HDAC6. The increased presence of pairs of compounds in the Z1 region compared to Z2 region indicates that there is a greater likelihood that the modifications affect the activity of both enzymes in the same magnitude and direction.
SMARt with many biological endpoints
ALM have also been applied to analyze the SMARt of screening collections tested across a large number of biological endpoints. Different ALM methods have been used including SAS maps. For instance, SAS maps were employed to analyze the SMARt obtained from Pubchem.32 In a proof-of-concept study, Medina-Franco and Wadell analyzed the bioassay activity landscape of 618 molecules tested across 244 confirmatory bioassays. One of the particular challenges in that work was that each bioassay in PubChem has its own specific definition of active, inactive, or inconclusive. A second major challenge was that not all 618 compounds were tested in all 244 bioassays. A distinctive feature of the SAS-like maps proposed to address those two challenges was the calculation of a pairwise bioassay activity profile similarity (bAPS): for each of the 618 compounds tested in any of the 244 confirmatory assays the bioassay activity profile was represented as a multiset fingerprint encoding of the activity data as follows: ‘active’ was set to ‘2’; ‘inactive’ as ‘1’; inconclusive or not tested as ‘0’; the pairwise bAPS was calculated using the Tanimoto coefficient:36where bAPS(i,j) is the bioassay activity profile similarity of the ith and jth molecules, mk(i) and mk(j) are the activity encodings of the ith and jth molecules, respectively, and n is the total number of assays that the molecules were screened across. This encoding of the activity data enabled the systematic structure- and bioprofile activity similarity and identified bioassay activity profile cliffs i.e., pairs of compounds with high structure similarity but very different bioassay activity profiles.37
In a separate work Yongye et al. analyzed the ALM of a chemogenomics data set released by Clemons et al. The data set contained more than 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds from different sources (commercial compounds, natural products and synthetic molecules) that where screened across 100 sequence-unrelated proteins.38 SMARt analysis using SAS maps led to the identification of structural changes that differentiated highly specific from promiscuous compounds. It was also concluded that, in general, similar synthetic structures from academic groups showed greater promiscuity differences than do commercial compounds and natural products.39 A characteristic metric employed in that work was the Structure-Promiscuity Index Difference (SPID); for each pair of compounds, the relationship between structure similarity and the different number of proteins to which each compound in the pair binds was computed using the expression:
000 compounds from different sources (commercial compounds, natural products and synthetic molecules) that where screened across 100 sequence-unrelated proteins.38 SMARt analysis using SAS maps led to the identification of structural changes that differentiated highly specific from promiscuous compounds. It was also concluded that, in general, similar synthetic structures from academic groups showed greater promiscuity differences than do commercial compounds and natural products.39 A characteristic metric employed in that work was the Structure-Promiscuity Index Difference (SPID); for each pair of compounds, the relationship between structure similarity and the different number of proteins to which each compound in the pair binds was computed using the expression:
Similar to activity landscape analysis with one biological endpoint, the structural interpretation of SMARt with many biological endpoints would require further molecular modeling studies with the three dimensional structures of the targets, if available. An alternative is to incorporate three dimensional molecular descriptors to describe the chemical structures. It is particularly interesting to provide a further rationale of the source of selectivity or promiscuity of the compounds.
Future directions
In principle, methods employed in ALM can be implemented to explore the SAR or SMART of any screening data evaluated across multiple biological endpoints. Moreover, several methods can be extended to mine biological fingerprints. SMART studies can be further extended to analyze properties such as toxicity. In this regard, Austin et al. introduced ADMET property–activity pairwise similarity maps to analyze the relationships between activity, structure and ADMET violations/compliance with particular emphasis on determining structural changes that have a large impact on the ADMET compliance.41In drug discovery, big data is typically obtained from high-throughput screening (HTS). HTS usually is conducted in two general steps: assays at a single dose followed by confirmatory assays at multiple doses. Despite the fact that biological assays at single-dose concentrations have not been considered for activity landscape analysis,42 such assays do provide valuable information that could be considered in preliminary activity landscape studies. We propose that this is a relevant future direction not only in ALM but in SMARt studies in general.
Pro-activity cliffs
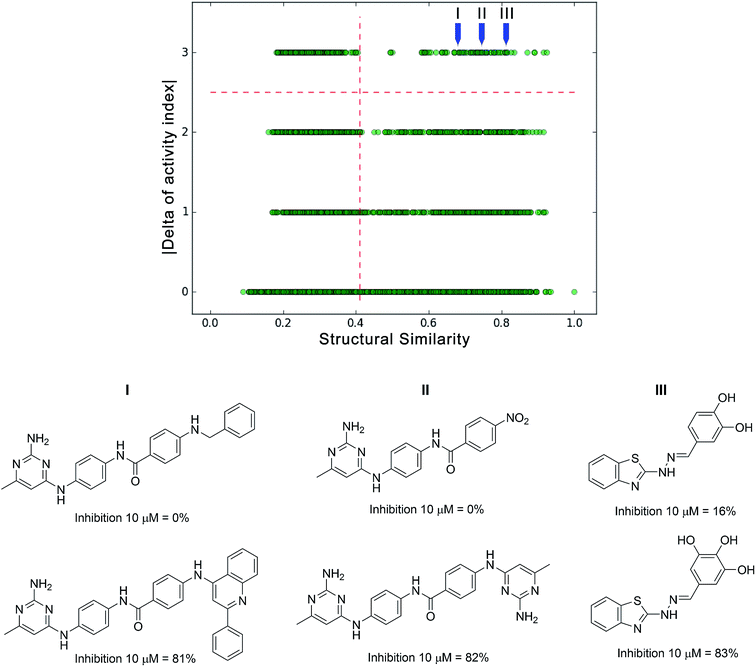
Relevant regions in the activity landscape are analyzed using high quality biological activity data that are obtained after multiple-dose inhibition assays. Due to its rigorous determination, it has been proposed that only those values can be used within the realm of ALM methods in order to minimize errors.42,43 While the latter remains as the ideal case, there are several cases where only single-dose biological activity data for a given target is available. Herein is proposed that this data can be used for a preliminary activity landscape analysis in order to identify potential areas of interest and guide the next steps towards the acquisition of high quality information. Thus, identification of potential activity cliffs i.e., pro-activity cliffs is valuable, as they can be prioritized for additional experimental evaluation.To temporarily address both the lack of multiple-dose/high quality data and the error involved in the biological activity measurement, the percentage of inhibition frequently obtained at single dose evaluations can be distributed in different categories; for instance, potentially very active, active, inactive and potentially very inactive. Integer indices can be assigned to the different classes: e.g., an integer index of 1 for the least active compounds and 4 for the most active ones. The limits of the inhibitory activity can be fitted to the distribution of the data set; e.g., those compounds with less than 25% of inhibition can be regarded as potentially inactive (e.g., activity index of 1), while those with more than 75% inhibition can be considered as potentially active compounds (e.g., activity index of 4). After classifying activity data and generating a categorical structure–activity similarity map, four horizontal zones can be defined as the result of comparing the activity index: 0 (as the result of comparing members of the same group), 1 (by comparing members of one unit of difference in the activity index), and so on. Thus, pro-activity cliffs can be defined as pairs of compounds with high structure similarity where one is highly probable to be active and the other is highly probable to be inactive. To illustrate this point, an actual set of single-dose activity data is exemplified for a group of inhibitors of DNA methyltransferase 3A (DNMT3A); for a large number of compounds, only percentages of inhibition obtained at single dose are available (10 μM). Fig. 4 shows a categorical SAS map for 106 compounds tested as potential modulators of DNMT3A. The SAS map in this figure has 5565 data points; the x-axis represents the pair-wise structure similarity computed as the mean of the Tanimoto similarity values computed with Extended Connectivity Fingerprints (radius 2) and MACCS keys (166 bits). The y-axis represents the four regions defined by the difference of the activity indices. The vertical dashed line is marked in the 3rd quartile of the pair-wise mean similarity values of the data set (mean similarity of 0.41). In Fig. 4 upper right quadrant identifies the pro-activity cliffs. The same figure illustrates three specific examples of pro-activity cliffs. As shown in Fig. 4, the three pairs of compounds show a remarkable resemblance, and a high difference in their inhibition activities. For instance, the only structural difference in pro-activity cliff “III” is a hydroxyl group. Further multiple-dose testing would confirm or not the status of the potential activity cliffs.
Concluding remarks
ALM is a quantitative approach to analyze systematically SAR of compound data sets. In many drug discovery programs compound data sets are screened against two, three or many more biological endpoints. To rapidly mine the usually large data generated, ALM have been adapted to analyze the associated SMARt. Among ALM approaches, SAS maps have evolved rapidly to address the increasing need of analyzing SMARt. To date, several successful applications have been reported including the analysis of SAR, SMARt and protein–ligand interaction cliffs. As part of the development of the SAS maps, a number of metrics and visualization approaches have been developed. Since SMARt analysis usually involves analysis of large amount of data, getting smart in drug discovery may require using information available in large screening campaigns that include incomplete chemogenomics data sets or activity data obtained at single concentrations. Bioactivity-profile similarity, activity landscape sweeping and pro-activity cliffs are examples of recently proposed concepts to advance the SMARt analysis in drug discovery. One of the major perspectives in the field is to incorporate the principles of quantum mechanics to refine the SMARt models and further improve their applicability in drug discovery projects.Conflict of interest
The authors declare that they do not have any conflict of interest related to this manuscript.Acknowledgements
This work was supported by the Universidad Nacional Autónoma de México (UNAM) grant ‘Programa de Apoyo a Proyectos para la Innovación y Mejoramiento de la Enseñanza’ (PAPIME) PE200116 and grant ‘Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica’ (PAPIIT) IA204016. We also thank funding from the ‘Programa de Apoyo a la Investigación y el Posgrado’ (PAIP) 50009163, Facultad de Química, UNAM. OP-H is grateful to CONACyT for the fellowships granted No. 765897/606003. JJ-N is grateful with PECEM MD PhD program for the organization of the fellowship. Rich discussions with Oscar Méndez-Lucio, Eli Fernández-de-Gortari and Mario Omar García-Sánchez are highly acknowledged.References
- O. Mendez-Lucio, J. J. Naveja, H. Vite-Caritino, F. D. Prieto-Martinez and J. L. Medina-Franco, J. Mex. Chem. Soc., 2016, 60, 168–181 Search PubMed.
- J. L. Medina-Franco, M. A. Giulianotti, G. S. Welmaker and R. A. Houghten, Drug Discovery Today, 2013, 18, 495–501 CrossRef PubMed.
- X. Hu, Y. Hu, M. Vogt, D. Stumpfe and J. Bajorath, J. Chem. Inf. Model., 2012, 52, 1138–1145 CrossRef CAS PubMed.
- J. L. Medina-Franco, G. Navarrete-Vázquez and O. Méndez-Lucio, Future Med. Chem., 2015, 7, 1197–1211 CrossRef CAS PubMed.
- H. González-Díaz, D. M. Herrera-Ibatá, A. Duardo-Sánchez, C. R. Munteanu, R. A. Orbegozo-Medina and A. Pazos, J. Chem. Inf. Model., 2014, 54, 744–755 CrossRef PubMed.
- G. M. Casañola-Martin, H. Le-Thi-Thu, F. Pérez-Giménez, Y. Marrero-Ponce, M. Merino-Sanjuán, C. Abad and H. González-Díaz, Mol. Diversity, 2015, 19, 347–356 CrossRef PubMed.
- F. Durán, N. Alonso, O. Caamaño, X. García-Mera, M. Yañez, F. Prado-Prado and H. González-Díaz, Int. J. Mol. Sci., 2014, 15, 17035 CrossRef PubMed.
- F. Saldívar-González, F. D. Prieto-Martínez and J. L. Medina-Franco, Educ. Quim., 2017 DOI:10.1016/j.eq.2016.06.002.
- G. M. Maggiora, J. Chem. Inf. Model., 2006, 46, 1535 CrossRef CAS PubMed.
- M. Cruz-Monteagudo, J. L. Medina-Franco, Y. Pérez-Castillo, O. Nicolotti, M. N. D. S. Cordeiro and F. Borges, Drug Discovery Today, 2014, 19, 1069–1080 CrossRef CAS PubMed.
- O. Méndez-Lucio, J. Pérez-Villanueva, R. Castillo and J. L. Medina-Franco, Mol. Inf., 2012, 31, 837–846 CrossRef PubMed.
- J. J. Naveja and J. L. Medina-Franco, RSC Adv., 2015, 5, 63882–63895 RSC.
- J. L. Medina-Franco, K. Martinez-Mayorga, A. Bender, R. M. Marin, M. A. Giulianotti, C. Pinilla and R. A. Houghten, J. Chem. Inf. Model., 2009, 49, 477–491 CrossRef CAS PubMed.
- A. Yongye, K. Byler, R. Santos, K. Martínez-Mayorga, G. M. Maggiora and J. L. Medina-Franco, J. Chem. Inf. Model., 2011, 51, 1259–1270 CrossRef CAS PubMed.
- P. Iyer, D. Stumpfe, M. Vogt, J. Bajorath and G. M. Maggiora, Mol. Inf., 2013, 32, 421–430 CrossRef CAS PubMed.
- D. Dimova and J. Bajorath, Mol. Inf., 2016, 35, 181–191 CrossRef CAS PubMed.
- R. Guha, J. Chem. Inf. Model., 2012, 52, 2181–2191 CrossRef CAS PubMed.
- J. Husby, G. Bottegoni, I. Kufareva, R. Abagyan and A. Cavalli, J. Chem. Inf. Model., 2015, 55, 1062–1076 CrossRef CAS PubMed.
- V. Shanmugasundaram and G. M. Maggiora, presented in part at the 222nd ACS National Meeting, Chicago, IL, United States, August 26–30, 2001 Search PubMed.
- J. L. Medina-Franco, J. Chem. Inf. Model., 2012, 52, 2485–2493 CrossRef CAS PubMed.
- J. J. Naveja and J. L. Medina-Franco, Expert Opin. Drug Discovery, 2015, 10, 1059–1070 CrossRef PubMed.
- J. H. Van duzer, R. Mazitschek, Y. Ding, N. Yu, Y. Cao and Y. Liu, Pyrimidine Hydroxy Amide Compounds as Protein Deacetylase Inhibitors and Methods of Use Thereof, Acetylon Pharmaceuticals, Inc., 2014, EP2640709 Search PubMed.
- A. Dueñas-González, J. Jesús Naveja and J. L. Medina-Franco, in Epi-Informatics, Academic Press, Boston, 2016, pp. 1–20 Search PubMed.
- J. J. Naveja, F. Cortés-Benítez, E. Bratoeff and J. L. Medina-Franco, Mol. Diversity, 2016, 20, 771–780 CrossRef CAS PubMed.
- J. Pérez-Villanueva, O. Méndez-Lucio, O. Soria-Arteche and J. Medina-Franco, Mol. Diversity, 2015, 19, 1021–1035 CrossRef PubMed.
- R. Guha and J. H. VanDrie, J. Chem. Inf. Model., 2008, 48, 646–658 CrossRef CAS PubMed.
- O. Méndez-Lucio, A. J. Kooistra, C. d. Graaf, A. Bender and J. L. Medina-Franco, J. Chem. Inf. Model., 2015, 55, 251–262 CrossRef PubMed.
- S. C. Brewerton, Curr. Opin. Drug Discovery Dev., 2008, 11, 356–364 CAS.
- J. L. Medina-Franco, O. Méndez-Lucio and K. Martinez-Mayorga, Adv. Protein Chem. Struct. Biol., 2014, 96, 1–37 CrossRef CAS PubMed.
- R. Guha, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2012, 2, 829–841 CrossRef PubMed.
- V. F. Kuyoc-Carrillo and J. L. Medina-Franco, Drug Dev. Res., 2014, 75, 313–323 CrossRef CAS PubMed.
- J. Waddell and J. L. Medina-Franco, Bioorg. Med. Chem., 2012, 20, 5443–5452 CrossRef CAS PubMed.
- J. Perez-Villanueva, R. Santos, A. Hernandez-Campos, M. A. Giulianotti, R. Castillo and J. L. Medina-Franco, Bioorg. Med. Chem., 2010, 18, 7380–7391 CrossRef CAS PubMed.
- J. Pérez-Villanueva, R. Santos, A. Hernández-Campos, M. A. Giulianotti, R. Castillo and J. L. Medina-Franco, MedChemComm, 2011, 2, 44–49 RSC.
- R. Aguayo-Ortiz, J. Perez-Villanueva, A. Hernandez-Campos, R. Castillo, N. Meurice and J. L. Medina-Franco, Future Med. Chem., 2014, 6, 281–294 CrossRef CAS PubMed.
- G. M. Maggiora and V. Shanmugasundaram, in Chemoinformatics and Computational Chemical Biology, Methods in Molecular Biology, ed. J. Bajorath, Springer, New York, 2011, vol. 672, pp. 39–100 Search PubMed.
- J. L. Medina-Franco and J. Waddell, J. Mex. Chem. Soc., 2012, 56, 163–168 CAS.
- P. A. Clemons, N. E. Bodycombe, H. A. Carrinski, J. A. Wilson, A. F. Shamji, B. K. Wagner, A. N. Koehler and S. L. Schreiber, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 18787–18792 CrossRef CAS PubMed.
- A. B. Yongye and J. L. Medina-Franco, J. Chem. Inf. Model., 2012, 52, 2454–2461 CrossRef CAS PubMed.
- A. Steffen, T. Kogej, C. Tyrchan and O. Engkvist, J. Chem. Inf. Model., 2009, 49, 338–347 CrossRef CAS PubMed.
- A. B. Yongye and J. L. Medina-Franco, Drug Discovery Today, 2013, 18, 732–739 CrossRef CAS PubMed.
- D. Stumpfe and J. Bajorath, J. Med. Chem., 2012, 55, 2932–2942 CrossRef CAS PubMed.
- J. L. Medina-Franco, G. M. Maggiora, M. A. Giulianotti, C. Pinilla and R. A. Houghten, Chem. Biol. Drug Des., 2007, 70, 393–412 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra26230a |
| This journal is © The Royal Society of Chemistry 2017 |