Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Sensitive DNA detection using cascade amplification strategy based on conjugated polyelectrolytes and hybridization chain reaction
Biqing Bao,
Jin Zhu,
Lina Gong,
Jia Chen,
Yanrui Pan and
Lianhui Wang*
Key Laboratory for Organic Electronics and Information Displays (KLOEID), Institute of Advanced Materials (IAM), Jiangsu National Synergistic Innovation Center for Advanced Materials (SICAM), Nanjing University of Posts and Telecommunications (NUPT), Nanjing 210023, Jiangsu, China. E-mail: iamlhwang@njupt.edu.cn
First published on 13th January 2017
Abstract
In this paper, a novel homogeneous hybridization chain reaction (HCR) assay is developed by using cationic conjugated polyelectrolytes (CPEs) as an indicator for the detection of DNA. In this assay, two hairpin probes H1 and H2 are labeled with fluorescein at their 5′-end and 3′-end respectively. After the HCR, the two hairpin probes are triggered by target DNA to form a long two-stranded DNA polymer with numerous fluorescein labelling in the strand, which is resistant to the S1 nuclease degradation. When the CPEs are added, strong electrostatic interaction between fluorescein-labeled DNA and CPEs occurred, which allowed fluorescence resonance energy transfer (FRET) from the CPEs to the fluorescein-labeled DNA and an obvious fluorescence colour change of the conjugated polymer solution. With HCR amplification and CPEs to amplify the fluorescence signal, homogeneous HCR for sensitive DNA detection is performed successfully. This proposed cascade amplification strategy can not only extend the application of HCR but also provides an alternative platform for homogeneous detection of DNA.
Introduction
In recent years, sensitive and reliable methods for the detection of DNA have become significantly important in medical diagnosis, mutational analysis, gene therapy, biological studies and specific genomic techniques.1–3 So far, a number of methods such as chemiluminescence,4,5 fluorescence,6–8 electrochemical,9 optical,10–12 electrical13 or microgravimetric14 sensing have been developed for detecting DNA. Since the specific DNA sequences may be present in very low amounts, various signal amplification strategies have been developed for sensitive DNA detection.15,16 Generally, polymerase chain reaction (PCR),17–19 rolling circle amplification (RCA)20–22 and ligase chain reaction (LCR)23–25 have evolved as the frequently used methods for detecting low levels of DNA targets. A limitation associated with the above strategy is the fact that these methods need expensive instruments and may produce false-positive results during template replication due to the cross-contamination from amplicons. The hybridization chain reaction (HCR)26–29 is a process in which the DNA initiator strand triggers a cascade of hybridization events between two alternating H1 and H2 hairpin probes to form a long double-stranded DNA structure. This approach is another innovative method for DNA-amplification techniques which can increase the sensitivity to target molecules at very low concentrations and does not require any expensive equipment.Fluorescent conjugated polyelectrolytes (CPEs), which combine the unique optical amplification properties of π-conjugated polymers and the electrostatic behaviors of polyelectrolytes, are of significant interest due to their potential applications in highly sensitive chemo/biosensors.30–34 Compared to small molecule counterparts, the large number of absorbing units along the CPEs backbone increases the probability of exciton energy transfer to fluorophore reporters. Because of their unique light-harvesting and signal amplification abilities, sensing assay based on fluorescence resonance energy transfer (FRET) from cationic polyelectrolytes to fluorescent molecules labeled in DNA probe can transduce the hybridization events of DNA probe and target DNA (ssDNAc) to fluorescent signal with amplified sensitivity. Therefore, it is possible to combine the two amplification methods based on HCR and CPEs for designing a simple and highly sensitive platform for homogeneous DNA detection.
In this paper, we propose a novel cascade amplification strategy that combines the sensitivity of CPEs molecular wire effects with the signal amplification capability of the HCR to construct a new homogeneous HCR methodology for DNA detection. The method takes advantage of the hybridization chain reaction to discriminate single-base mismatch and the cascade amplification strategy to detect DNA with high sensitivity. In this devised HCR-based sensing system, both probe hairpin pairs are labeled with fluorescein at their sticky ends and the fluorescein-labeled single-stranded region in both probes can be digested to the mono- or oligonucleotides by S1 nuclease. Then the target strand can trigger the chain reaction between alternating hairpin probes to form DNA nanowires, which would prevent the cleavage of fluorescein from DNA strand by S1 nuclease. When the CPEs are added, ratiometric fluorescence response of the solution can be observed due to the strong electrostatic interactions between CPEs and fluorescein-labeled DNA. The combination of CPEs with HCR-based detection method thus allows highly sensitive DNA detection without the need for expensive instruments and sophisticated operations.
Experimental section
Materials and apparatus
S1 nuclease was purchased from TaKaRa Biotechnology Co., Ltd (Dalian, China). The cationic poly[(9,9-bis{6′-[(N,N-diethyl)-N-methylammonium]hexyl}-2,7-fluorenyleneethynylene)-alt-co-(2,5-bis{3′-[(N,N-diethyl)-N-methylammonium]-1′-oxapropyl}-1,4-phenylene)]tetraiodide (PFEPN+) (see Scheme 1) used as the CPEs donor in the FRET experiments was synthesized according to the previously reported method.35 HPLC-purified DNA were obtained from Sangon Biotechnology Co., Ltd (Shanghai, China). The sequences of the probes and targets are listed in Table 1. All solutions were prepared in deionized and sterilized water. The other reagents were of analytical reagent grade and used as purchased without further purification. A Shimadzu RF-5301PC spectrophotometer equipped with a xenon lamp was used to obtain the fluorescence spectra.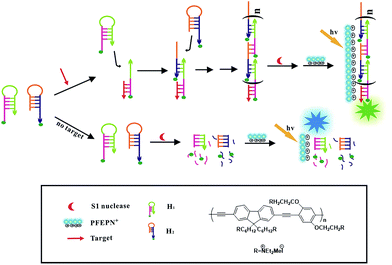 | ||
| Scheme 1 Schematic illustration of the cascade signal amplification strategy for DNA detection based on HCR and CPEs. | ||
| Name | Sequence (from 5′ to 3′) |
|---|---|
| a The design of the hairpin probes was adapted from the literature.36,37 In the hairpin sequences, loops are italicized, sticky ends are underlined and mismatch bases are bolded. | |
| H1 | 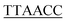 CACGCCGAATCCTAGACTCAAAGTAGTCTAGGATTCGGCGTG CACGCCGAATCCTAGACTCAAAGTAGTCTAGGATTCGGCGTG |
| H2 | AGTCTAGGATTCGGCGTGGGTTAACACGCCGAATCCTAGACT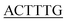 |
| T (ssDNAc) | AGTCTAGGATTCGGCGTGGGTTAA |
| T (1-mis) | AGTCTAGGATTGGGCGTGGGTTAA |
| T (3-mis) | AGTCTAGTATTCGGCATGGGTTAC |
| T (random) | TCAGATCCTAAGCCGCACCCAATT |
Detection of target DNA by fluorescence measurements
Hairpin probes H1 and H2 were respectively heated at 95 °C for 2 min and cooled down slowly at room temperature for at least 2 h to form the hairpin structure before use. Afterwards, target DNA was incubated with a mixture of 1.2 μL H1 and 1.2 μL H2 in HCR reaction solution (50 mM CH3COONa, 1 M NaCl, pH 7.5) for 2 h to form double-stranded DNA. After incubation with 0.75 U μL−1 S1 nuclease at 37 °C for 25 min, 10 μL PFEPN+ was added and the resulting mixture was incubated for another 5 min. Fluorescence measurements (excited at 400 nm) were carried out at room temperature in a 50 μL quartz cuvette.Gel electrophoresis
Hairpin probes H1 (10 μM) and H2 (10 μM) were respectively heated at 95 °C for 2 min and cooled down slowly at room temperature for at least 2 h to form the hairpin structure before use. Target DNA (1 μM) and non-complementary DNA (1 μM) were annealed with hairpin probes H1 (2 μM) and H2 (2 μM) to form double-stranded DNA. The GelRed was used as DNA stain and mixed with the DNA samples. A 12.5% native polyacrylamide gel was prepared with 1× TBE buffer (pH 8.0). 6 μL of each DNA sample was loaded into the lane and performed at a constant potential of 60 V for 150 min with 1× TBE (pH 8.0) as the running buffer. Gel electrophoresis was performed on a Bio-Rad Mini-PROTEAN Tetra electrophoresis system.Results and discussion
Sensing mechanism
The principle of fabricated cascade amplified strategy for target DNA detection based on HCR and conjugated polyelectrolytes PFEPN+ is shown in Scheme 1. Two complementary DNA hairpin probes H1 and H2, which have 18-base-pair stems and 6-base-pair loops, are both labeled with fluorescein at their 6-base-pair sticky ends. In the absence of ssDNAc, both probes (H1 and H2) are stable and adopt hairpin-shaped structure with the protruding DNA fragments. In this case, S1 nuclease can preferentially bind to and cleave the single strand DNA fragment of the H1 and H2 hairpin probes, which cause the release of fluorescein from the stem region of H1 and H2. After addition of the cationic conjugated polyelectrolytes PFEPN+, blue fluorescent color of PFEPN+ can be observed under UV irradiation. When the target is present in the reaction solution, probes H1 and H2 will open and the hybridization chain reaction is triggered to obtain long double-stranded DNA bearing a lot of fluorescein in the strand. Such designs enable the hybridization products to keep with the fluorescein labeled at their double-strand DNA, which prevent the HCR products from being digested by S1 nuclease because S1 nuclease is not active on the double-stranded DNA. After addition of PFEPN+, the emission intensity of PFEPN+ decreased and the fluorescent color changed from blue to green due to the much stronger electrostatic interaction between PFEPN+ and HCR product than that of PFEPN+ and separated fluorescein-labeled nucleotide. Thus, the visual detection of target DNA can be successfully realized in a homogeneous manner based on this excellent cascade amplification technique. Nevertheless, the addition of single-strand-specific S1 nuclease manifests high specificity of this developed sensing system for the detection of target DNA.Amplification performance of CPEs and HCR
To investigate whether the employment of CPEs and HCR strategy amplified the signal, fluorescence responses of PFEPN+ were recorded. The normalized PL spectra of PFEPN+ with or without the ssDNAc are shown in Fig. 1. The solution of PFEPN+ exhibits a strong emission at 450 nm, which correspond to a bright blue fluorescence. After addition of H1, H2 hairpin probes and S1 nuclease, the control solution in the absence of ssDNAc gives blue emission at 450 nm and some fluorescein emission signal at 530 nm since the released fluorescein also interacts with PFEPN+. However, the blue emission band of PFEPN+ at 450 nm displays fluorescence quenching along with the obvious fluorescence color change to green in the presence of ssDNAc, which suggests the strong electrostatic interaction of PFEPN+ and HCR product and efficient FRET from PFEPN+ to fluorescein. This target-mediated FRET process indicates that the target-triggered HCR products could effectively protect the hairpin probes from the S1 nuclease digestion and lead to the formation of stable polyelectrolytes complexes between PFEPN+ and the long fluorescein-labeled DNA polymer by electrostatic interactions. The close proximity of PFEPN+ to fluorescein thus results in a cascade amplified fluorescence signal. 4.5-fold higher emission ratio of the fluorescein to PFEPN+ in the presence of ssDNAc to that without ssDNAc is observed, indicating that the target DNA detection could be realized by monitoring the PL intensity and fluorescent color difference of PFEPN+ solution. Furthermore, it can be observed that the emission at 450 nm exhibits blue-shift in the presence of ssDNAc, which may be attributed to the torsion of polymer backbone and the decrease conjugation length of polymer caused by increased interaction between the polymer and the HCR products.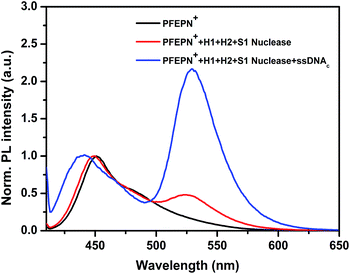 | ||
| Fig. 1 Normalized fluorescence spectra of PFEPN+ for the detection of ssDNAc using the cascade signal amplification strategy. [S1 nuclease] = 0.75 U μL−1. | ||
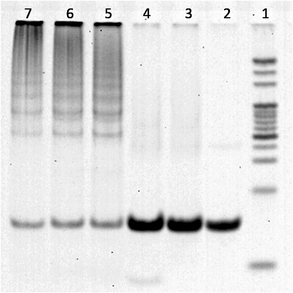
To confirm the occurrence of hybridization chain reaction, native gel electrophoresis was performed in the presence and absence of ssDNAc. As shown in Fig. 2, there are only low molecular weight bands in lane 2 (only H1), lane 3 (H1/H2) and lane 4 (H1/H2/random ssDNA). Lane 3 in Fig. 2 indicates that the mixture of H1 and H2 do not open and self-assembled to form HCR products, and lane 4 shows that the hairpin probes with a random sequence could not result in the HCR events either. Only in the presence of the ssDNAc, high molecular weight bands are observed, which indicates that ssDNAc can trigger hybridization chain reaction between H1 and H2 to form a long dsDNA polymer, with amplification of the fluorescence signal.
Optimization of experimental conditions for target DNA detection
In order to establish optimum conditions for the ssDNAc detection, the experimental conditions were systematically investigated. First, the concentration of S1 nuclease plays an important role in the sensing process. To investigate the optimal concentration of S1 nuclease, the probe H1, H2, and ssDNAc were mixed with different concentrations of S1 nuclease, ranging from 0 U μL−1 to 1.33 U μL−1. The fluorescence emission ratio of the fluorescein to PFEPN+ (I530/I450) are plotted as a function of the S1 nuclease concentration. As shown in the inset in Fig. 3a, the emission ratio (I530/I450) decreases with the increase of the concentration of S1 nuclease from 0 U μL−1 to 0.75 U μL−1 in the absence of ssDNAc, implying enhanced digestion of fluorescein-labeled single strand DNA by S1 nuclease. Meanwhile, in the presence of ssDNAc, it can be observed that the emission ration (I530/I450) also decreases slightly as the concentration of S1 nuclease increases due to the non-specific S1 nuclease cleavage of duplex DNA (Fig. 3b). The results indicate that the optimum concentration of S1 nuclease is 0.75 U μL−1 by considering the signal-to-noise level.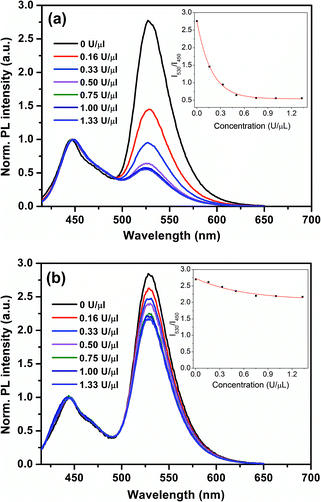 | ||
| Fig. 3 Normalized fluorescence response of PFEPN+ to different S1 nuclease concentrations (a) without and (b) with ssDNAc for 20 min at 37 °C. [PFEPN+] = 1.6 × 10−6 M. | ||
The incubation time of S1 nuclease with hairpin probes and HCR products also has significant effect on the sensitivity of the detection process. To investigate the effect of incubation time, the changes of the FRET efficiency at different cleavage time intervals with and without the addition of ssDNAc into the system were recorded. As shown in Fig. 4a, in the absence of ssDNAc, the emission ratio (I530/I450) decreases at a fast rate with the increase of incubation time and reaches a plateau at 25 min. After addition of ssDNAc, increase of the incubation time could lead to a slow decrease of the emission ratio (I530/I450) in the initial stage due to the non-specific S1 nuclease digestion. However, the emission ratio (I530/I450) keep a plateau after 25 min since the HCR process forms long double-stranded DNA, which could protect the sticky ends at the hairpin probes from S1 nuclease-mediated digestion. Therefore, for best signal-to-noise level, 25 min is selected for the cleavage reaction.
 | ||
| Fig. 4 Normalized fluorescence response of PFEPN+ to different digestion time of S1 nuclease (a) without and (b) with ssDNAc at 37 °C. [PFEPN+] = 1.6 × 10−6 M. | ||
Specificity and sensitivity of the assay
To test the sensitivity of the cascade amplified sensing system for ssDNAc detection, a series concentration of ssDNAc (0–100 nM) were incubated with hairpin probes H1 and H2 under the optimized conditions. After the cleave reaction steps by S1 nuclease at 37 °C for 25 min, PFEPN+ was subsequently added to each sample, and the normalized PL spectra of PFEPN+ solution upon excitation at 400 nm are shown in Fig. 5. It can be seen that the emission ratio (I530/I450) increases obviously with an increase of the concentration of ssDNAc in the range from 10 to 40 nM ssDNAc. When the concentration of ssDNAc reaches 60 nM, the PL spectra of the polymer then keep essentially unchanged with further increasing the ssDNAc concentration, indicating there exists a saturation point. Color change of the emission fluorescence from the solution with and without ssDNAc can also be monitored by naked-eye under UV irradiation. As shown in the insets of Fig. 5, addition of ssDNAc induce effective HCR and generate a noticeable blue-to-green fluorescence color change of the solution.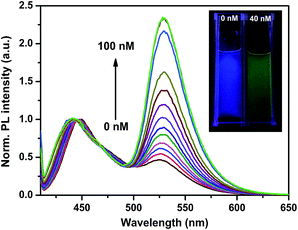 | ||
| Fig. 5 The fluorescence response of PFEPN+ in the presence of ssDNAc with [ssDNAc] ranging from 0 to 100 nm in CH3COONa solution. Insets: fluorescence photographs of PFEPN+ with and without ssDNAc. | ||
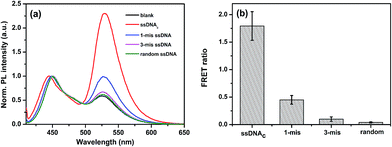
The specificity of the proposed HCR-based PFEPN+ sensor was evaluated by interference experiment. Herein, a series of control experiments by using single-mismatched, 3-mismatched and random ssDNA were performed under the same condition, in which all concentrations of them were 40 nM. As shown in Fig. 6, the highest FRET ratio is observed for the solution containing the target sequence (red curve in Fig. 6a). This is due to the fact that the fluorescein-labeled HCR products, which are triggered by the ssDNAc and have a large amount of negative charges, are able to effectively interact with cationic conjugated polyelectrolytes PFEPN+ and generate effective FRET from donor PFEPN+ to the acceptor fluorescein. Furthermore, the FRET ratio decreases with an increasing number of mismatched base pairs in DNA strands. The results show that single base pair mismatch could be easily detected and the cascade strategy based on HCR and CPEs could be used to improve both the sensitivity and selectivity.
Conclusions
In summary, we have proposed a novel DNA-amplified detection method that combined the amplifying capacity of HCR and the molecular wire effect property of CPEs. The target DNA not only act as a trigger to initiate HCR for first signal amplification but also protect the fluorescein-labeled probes from S1 nuclease degradation. Strong electrostatic interaction between the CPEs and fluorescein-labeled HCR products effective FRET can be observed by subsequently addition of CPEs, which leads to cascade amplification fluorescence response for the improved sensitivity. Therefore, the homogeneous HCR for DNA detection is carried out successfully through monitoring the fluorescence color changes of the solution by naked eyes. This method is simple, and does not require expensive instruments or sophisticated operations. Moreover, this proposed assay strategy extends the application of HCR and provides an alternative platform for homogeneous detection of DNA as well as mutational analysis and clinical diagnosis.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (21574069, 21475064), the Natural Science Foundation of Jiangsu Province (BK20151503), the Jiangsu National Synergistic Innovation Center for Advanced Materials (SICAM), the Program for Changjiang Scholars and Innovative Research Team in University (IRT_15R37), the Sci-tech Support Plan of Jiangsu Province (BE2014719), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, YX03001).Notes and references
- H. Aoki, Chem.–Asian J., 2015, 10, 2560–2573 CrossRef CAS PubMed.
- K. Chang, S. Deng and M. Chen, Biosens. Bioelectron., 2015, 66, 297–307 CrossRef CAS PubMed.
- M. L. Ermini, S. Mariani, S. Scarano and M. Minunni, Biosens. Bioelectron., 2014, 61, 28–37 CrossRef CAS PubMed.
- S. Bi, Z. Zhang, Y. Dong and Z. Wang, Biosens. Bioelectron., 2015, 65, 139–144 CrossRef CAS PubMed.
- S. M. Khoshfetrat, M. Ranjbari, M. Shayan, M. A. Mehrgardi and A. Kiani, Anal. Chem., 2015, 87, 8123–8131 CrossRef CAS PubMed.
- Q. Guo, Z. Bai, Y. Liu and Q. Sun, Biosens. Bioelectron., 2016, 77, 107–110 CrossRef CAS PubMed.
- Y. Zhou, Z. Xu and J. Yoon, Chem. Soc. Rev., 2011, 40, 2222–2235 RSC.
- Y. Liu, M. Ye, Q. Ge, X. Qu, Q. Guo, X. Hu and Q. Sun, Anal. Chem., 2016, 88, 1768–1774 CrossRef CAS PubMed.
- A. M. Debela, S. Thorimbert, B. Hasenknopf, C. K. O'Sullivan and M. Ortiz, Chem. Commun., 2016, 52, 757–759 RSC.
- B. L. Li, H. L. Zou, L. Lu, Y. Yang, J. L. Lei, H. Q. Luo and N. B. Li, Adv. Funct. Mater., 2015, 25, 3541–3550 CrossRef CAS.
- P. Toren, E. Ozgur and M. Bayindir, Anal. Chem., 2015, 87, 10920–10926 CrossRef CAS PubMed.
- L. Gao, C. Lian, Y. Zhou, L. Yan, Q. Li, C. Zhang, L. Chen and K. Chen, Biosens. Bioelectron., 2014, 60, 22–29 CrossRef CAS PubMed.
- J. Hihath, B. Q. Xu, P. M. Zhang and N. J. Tao, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 16979–16983 CrossRef CAS PubMed.
- T. Liu, J. Tang, M. M. Han and L. Jiang, Biochem. Biophys. Res. Commun., 2003, 304, 98–100 CrossRef CAS PubMed.
- Y. Gao and B. X. Li, Anal. Chem., 2014, 86(17), 8881–8887 CrossRef CAS PubMed.
- H. Z. Zhao, J. J. Dong, F. L. Zhou and B. X. Li, Microchim. Acta, 2015, 182, 2495–2502 CrossRef CAS.
- L. Miotke, B. T. Lau, R. T. Rumma and H. P. Ji, Anal. Chem., 2015, 87, 3114 CrossRef CAS PubMed.
- D. Ogunremi, H. Kelly, A. A. Dupras, S. Belanger and J. Devenish, J. Clin. Microbiol., 2014, 52, 4275–4285 CrossRef PubMed.
- Y. Suyama and Y. Matsuki, Sci. Rep., 2015, 5, 16963 CrossRef CAS PubMed.
- S. Bi, L. Li and S. Zhang, Anal. Chem., 2010, 82, 9447–9454 CrossRef CAS PubMed.
- J. Li, T. Deng, X. Chu, R. Yang, J. Jiang, G. Shen and R. Yu, Anal. Chem., 2010, 82, 2811–2816 CrossRef CAS PubMed.
- Y. F. Wang, R. Y. Zhan, T. H. Li, K. Y. Pu, Y. Y. Wang, Y. K. C. Tan and B. Liu, Langmuir, 2012, 28, 889–895 CrossRef CAS PubMed.
- L. Zhou, F. Du, Y. Zhao, A. Yameen, H. Chen and Z. Tang, Biosens. Bioelectron., 2013, 45, 141–147 CrossRef CAS PubMed.
- W. G. Cao, Trends Biotechnol., 2004, 22, 38–44 CrossRef CAS PubMed.
- Y. Cheng, Q. Du, L. Wang, H. Jia and Z. Li, Anal. Chem., 2012, 84, 3739–3744 CrossRef CAS PubMed.
- L. Jia, S. Shi, R. Ma, W. Jia and H. Wang, Biosens. Bioelectron., 2016, 80, 392–397 CrossRef CAS PubMed.
- J. Ikbal, G. S. Lim and Z. Gao, TrAC, Trends Anal. Chem., 2015, 64, 86–99 CrossRef CAS.
- H.-L. Shuai, K.-J. Huang and Y.-X. Chen, J. Mater. Chem. B, 2016, 4, 1186–1196 RSC.
- M. Rana, M. Balcioglu, M. Kovach, M. S. Hizir, N. M. Robertson, I. Khan and M. V. Yigit, Chem. Commun., 2016, 52, 3524–3527 RSC.
- F. Qiu, Y. Huang and X. Zhu, Macromol. Chem. Phys., 2016, 217, 266–283 CrossRef CAS.
- F. Lv, T. Qiu, L. Liu, J. Ying and S. Wang, Small, 2016, 12, 696–705 CrossRef CAS PubMed.
- J. F. Fennell Jr, S. F. Liu, J. M. Azzarelli, J. G. Weis, S. Rochat, K. A. Mirica, J. B. Ravnsbaek and T. M. Swager, Angew. Chem., Int. Ed., 2016, 55, 1266–1281 CrossRef PubMed.
- L.-L. Zhou, M. Li, H.-Y. Lu and C.-F. Chen, Polym. Chem., 2016, 7, 310–318 RSC.
- Z. Chen, P. Wu, R. Cong, N. Xu, Y. Tan, C. Tan and Y. Jiang, ACS. Appl. Mater. Interfaces, 2016, 8, 3567–3574 CAS.
- Y. Q. Huang, Q. L. Fan, X. M. Lu, C. Fang, S. J. Liu, L. H. Yu-Wen, L. H. Wang and W. Huang, J. Polym. Sci., Part A: Polym. Chem., 2006, 44, 5778–5794 CrossRef CAS.
- J. Huang, Y. Wu, Y. Chen, Z. Zhu, X. Yang, C. J. Yang, K. Wang and W. Tan, Angew. Chem., Int. Ed., 2011, 50, 401–404 CrossRef CAS PubMed.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |