Open Access Article
Open Access ArticleUltra-fast and highly sensitive enzyme-free glucose biosensing on a nickel–nickel oxide core–shell electrode†
Halima Begum,
Mohammad Shamsuddin Ahmed and
Seungwon Jeon *
*
Department of Chemistry, Institute of Basic Science, Chonnam National University, Gwangju 500-757, Republic of Korea. E-mail: swjeon3380@naver.com; Fax: +82 62 530 3389; Tel: +82 62 530 0064
First published on 13th January 2017
Abstract
A series of directly electrodeposited nickel–nickel oxide core–shells (NiNiO) on a glassy carbon electrode (GCE) have been prepared by the variation of cycle number through a facile electrochemical deposition method using a cyclic voltammetry (CV) technique in HCl solution and have been used as a highly sensitive enzyme-free glucose biosensor in 0.1 M NaOH. The characterizations of the biosensor probe revealed that the Ni-core forms as a homogeneous nanoparticle structure during the deposition, and then the NiO-shell is formed on the Ni-core. The electrochemical activity of the modified electrode towards the oxidation of glucose was studied using various electrochemical methods. Under optimum conditions, the glucose biosensor exhibited a long linear-range of glucose concentrations from 2 μM to 14 mM and a lower detection limit of 0.4 μM (S/N = 3), associated with excellent stability, high reproducibility, and favorable selectivity against common interferents. Very importantly, it showed an ultra-fast response time (∼1 s) and higher sensitivity (1889.8 μA mM−1 cm−2) than that ever reported for an electrodeposited Ni-based biosensor for glucose oxidation. Moreover, the proposed method was successfully applied to determine glucose in real samples with satisfactory results.
1. Introduction
The determination of the concentration of glucose is an important analytical target due to not only the increasing demands for diagnosing diabetes but also the urgent requirements in food industry, fermentation industry, environmental protection, and biotechnology.1–3 Particularly, diabetes mellitus is one of the most common chronic diseases, which continues to increase and causes various complications.4 In this regard, a simple and easily prepared system to monitor the blood glucose levels could help to reduce the risk of complications associated with diabetes. Moreover, an easily fabricable, cost-effective, highly efficient, and selective detection of glucose has become essential in many different industries including pharmaceuticals, environmental monitoring, textiles, and food industries. Considering these aspects, the development of an electrochemical glucose biosensor has triggered extensive attention because of its high sensitivity, excellent selectivity, rapid response, and good reliability.5–9 However, two types of electrochemical glucose biosensors, such as enzymatic and non-enzymatic or enzyme-free glucose biosensors, have been commonly reported. Enzyme-free glucose biosensors with the application of various materials, based on carbon materials,12 polymers,13 metals or metal oxides,14,15 have received considerable attention owing to the inclination of enzymes utilization, stress-free electrode fabrication, stability, simplicity, reproducibility, and cost-effectiveness.9–11 Among the metals or metal oxides, particularly, nickel (Ni) and its oxide electrodes are a good option due to their high stability, reproducibility of results, low toxicity, natural abundance, and low-cost.16–18 Ni-Based nanomaterials exhibit a remarkably high catalytic activity for glucose oxidation due to the formation of a redox couple Ni(OH)2/NiOOH in an alkaline medium.9,19Recently, a number of Ni-based glucose biosensors have been investigated and their performance is still being improved;8,14,20–24 however, to improve the performance of Ni-based glucose biosensors, many fabrication processes, such as electrodeposition, composition with graphene and/or polymers, alloying with other metals, 3D flower-like approach, etc., have been applied.8,14,20–26 Among various techniques, electrodeposition is particularly interesting due to the direct-electrodeposition on the electrode surface and trouble-free electrode preparation. Moreover, the process can be simply controlled by adjusting the applied current, scanning potential windows, number of cycles and time.27–29 In addition, since the electrochemical deposition has been directly implemented on an electrode surface, the deposited nanoarchitectures do not require any additional step for adding binders. Unfortunately, the low-sensitivity and short linear-range are the major drawbacks for the Ni-electrodeposited electrodes8,23,25 as compared to the other metal electrodes.10 However, one of the strategies can be applied to improve these problems by changing the morphological structure of the electrodeposited nickel–nickel oxide (NiNiO) core–shell on the electrodes.30
Herein, a series of directly electrodeposited NiNiO core–shells have been synthesized by a simple electrodeposition method and successfully applied for the development of an enzyme-free glucose biosensor for the first time. The enzyme-free glucose biosensor performance was evaluated by cyclic voltammetry (CV), differential pulse voltammetry (DPV), and chronoamperometry (CA) methods. The NiNiO core–shell biosensor electrode exhibited a wide concentration range, lower LOD, higher sensitivity, and an ultra-fast response time. Moreover, it was very stable, reproducible, repeatable, and selective towards glucose sensing in the alkaline media. To date, the observed sensitivity and response time of the electrodeposited Ni-based biosensor have not been reported.
2. Experimental section
2.1 Synthesis of the NiNiO core–shell
The mirror-like surface of a GCE that had been polished with a 0.05 mm alumina suspension and a polishing cloth (Bioanalytical Systems) was used for electrodeposition. The NiNiO core–shell was electrodeposited on the GCE by the CV technique in a 2.6 mg mL−1 NiCl2·6H2O in a 10 mM HCl solution under high purity argon (Ar) for 30 min for 10, 20, and 30 cycles at a 50 mV s−1 scan rate. The NiNiO core–shell deposition was monitored within the potential window of ±1.5 V (Fig. S1†). The potential window with the higher overpotential, which is known as the oxygen evolution region, was selected. The electrochemical measurement and instrumental characterizations were carried out using various instruments (see the ESI†).3. Results and discussions
3.1 Characterization of the NiNiO core–shell
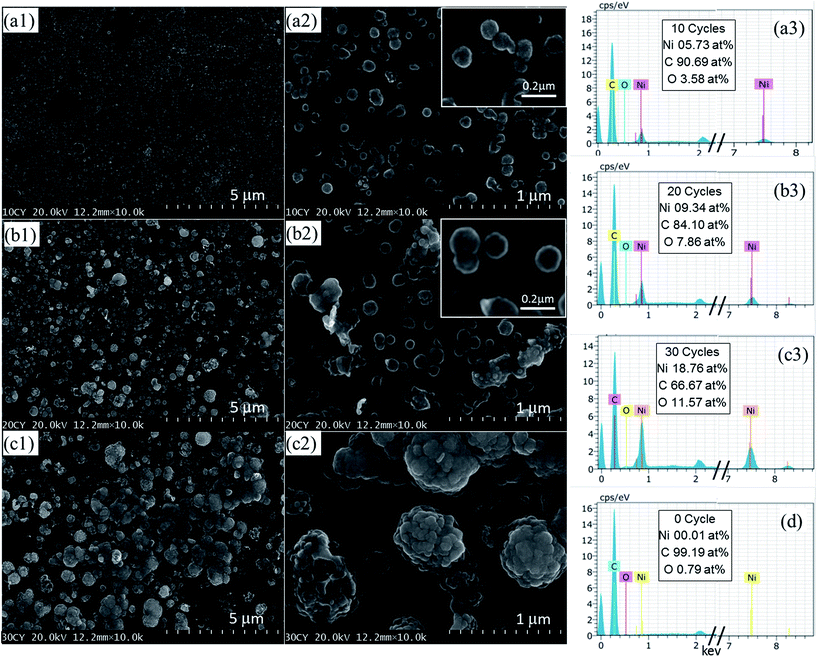
SEM images of the as-prepared NiNiO core–shell on the GC plate are shown in Fig. 1, and the NiNiO was obtained in various CV cycles. The size and number of NiNiO varied with an increase in the cycle number. Over all, all NiNiO were spherical in shape with a nearly homogeneous size. In the case of 10th cycle, a large amount of NiNiO was deposited on the GC plate (Fig. 1a1) and during the 10th cycle, NiNiO was much smaller in size (Fig. 1a2). On increasing the cycle number up to 20th cycle, the size of NiNiO increased (Fig. 1b1), whereas the number decreased due to aggregation (Fig. 1b2). In the case of 30th cycle, NiNiOs were comparatively lower in population (Fig. 1c1) with very high aggregation (Fig. 1c2) because NiNiO had enough time to deposit on the GC plate. Generally, for metal (i.e. Ni)-electrodeposition, an extremely aggregated and thick-film like microstructures can be observed in the whole GC plate, as reported by Sivasakthi et al.25 and Kong et al.31 However, in our study, NiNiO was grown as nanoparticle (NPs)-like core–shells and the size was increased on increasing the CV cycle number, which was similar to that of an another study on the graphene substrate,6 indicating that an advantageous hierarchical structure was directly formed on the GC plate. Note that, as can be seen in the high resolution SEM images (figure insets), all NP surfaces (shell) were comparatively brighter than that of the NPs center (core), indicating a possible unlikely surface. Especially, after 20 cycles, the deposited NiNiO was more sharply visible than that after the 10 cycles of deposited NiNiO NPs, indicating an excellent formation of the core–shell.The EDX analyzer was fitted to the SEM chamber to perform the elemental analysis and determine the composition of all NiNiO core–shells. The EDX spectrum of all NiNiO samples (Fig. 1a3, b3 and c3) showed that all NiNiO contained only Ni with oxygen (O), and the contents (at%) are listed (C comes from the GC plate) in the insets of the corresponding figures. However, Ni and O increased with an increase in the cycle number, signifying the layer-by-layer assembly of NiNiO and probably O originated from the formation of NiO/Ni(OH)2 on the NiNiO surface. The numerical results (Ni and O) of all three samples are plotted in Fig. S2.† For comparison, the EDX spectrum of bare GC is also displayed (Fig. 1d), where Ni is absent.
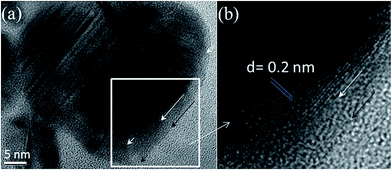
The resolution of SEM is not sufficient to precisely and clearly observe the change in the NiNiO surface. Thus, high-resolution TEM (HRTEM) images of the NiNiO core–shell were obtained (Fig. 2), which showed a highly ordered crystalline structure of the core and an amorphous shell (between arrow signs in Fig. 2a). The lattice lines of the core are visibly clear with a lattice d-spacing of 0.2 nm, which corresponds to the (111) plane of Ni0.32 On the other hand, the outer layer was amorphous in nature and the amorphous nature does not represent any lattice spacing,33 confirming the formation of NiO on the Ni0 surface (Fig. 2b). Typically, Ni(OH)2 also shows the lattice d-spacing, which is slightly higher than 0.2 nm.19 Although, the core–shell formation mechanism under this condition was not intensively studied, it is rational that the metallic Ni was deposited during the continuous CV cycles at a lower overpotential (Fig. S1a†). Then, the NiO shell was formed at higher overpotential, which is known as the oxygen evolution region (Fig. S1b†). Recently, the oxygen evolution mechanism on the Ni surface was nicely explained by Kauffman et al.34 Moreover, Ni can be easily oxidized by water and air.18,33 Therefore, the metallic core of Ni0 and the NiO shell were confirmed by the HRTEM analysis.
Fig. 3a shows the XRD patterns of three different NiNiO core–shells, which were prepared during the various CV cycles. Each pattern of Fig. 3a represents three major characteristic typical peaks. The peaks at 2θ = 44.5°, 52.3°, and 77.2° are allocated to the Ni (111), (200), and (220) planes, respectively. Moreover, the comparatively smaller peaks at 36.8° and 63.6° result from the NiO (111) and (200) planes,35,36 respectively. This is due to presence of a NiO layer as a shell on the Ni0 core.36 The Ni (111) XRD peak from the 20 cycles of the deposited NiNiO shifted towards a lower 2θ value as compared to that of the other NiNiO samples. These kinds of anomalies have been attributed to the prevalence of point defects such as oxide formation.37 We suspect that among three different samples, the ration of shell/cores (O/Ni in Fig. S2†) at 20 cycles of the deposited NiNiO were higher due to the comparatively higher NiO shell formation. Fig. 3b shows a core level XPS spectrum of Ni2p for only 20 cycles of a deposited NiNiO core–shell sample. The two strong peaks near 855.1 eV and 872.6 eV in Fig. 3b can be attributed to Ni2p3/2 and Ni2p1/2, respectively. The spectrum also shows two other additional peaks at ∼862.1 eV and 881 eV, which reveal the presence of NiO species.18,38
3.2 EIS measurement
The interfacial properties of the modified electrode, which are vitally significant for the electrical conductivity and the electrocatalytic features, were first analyzed by EIS measurement and the EIS experiment was carried out in a 5 mM [Fe(CN)6]3−/4− redox probe containing 0.1 M KCl in the frequency range from 105 to 0.01 Hz in Fig. 3c. The electron transfer characteristics were interpreted using the Randles circuit. The Randles circuit consisted of electrolyte resistance (Rs), electron transfer resistance (Rct), double layer capacitance (Cdl), and Warburg impedance (W) (Fig. 3c inset). As can be observed in Fig. 3c, the Nyquist plot of the bare GCE displayed a well-defined, enlarged semicircle with an Rct of about 2.1 kΩ at a high frequency. Compared with the bare GCE, the diameter of the semicircle in the Nyquist plot of the NiNiO/GCE (electrodeposited up to the 20th CV cycle) dramatically decreased, featuring a lower Rct of 290 Ω. This result indicates that NiNiO is an excellent conducting core–shell and could act as tiny conductive centers to promote the electron transfer.3.3 Electrochemical glucose oxidation
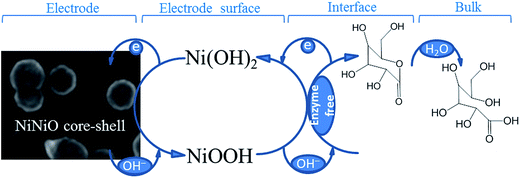
The electrochemical properties for the 20th cycle number of the deposited NiNiO core–shell at 50 mV s−1 scan rate were evaluated by the CV technique, as shown in Fig. 4a. The CVs were obtained in an Ar-saturated 0.1 M NaOH at a scan rate of 50 mV s−1 and agreed well with those in the reports.6,7,39 On comparison, these redox peaks were invisible in the case of bare GCE [curve (i) in Fig. 4a]. In the absence of glucose, the neat NiNiO/GCE exhibited well-defined redox peak potentials (anodic, Epa and cathodic, Epc) of 0.5 V and 0.42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) V [curve (ii) in Fig. 4a], respectively, resembling that of the neat NiNiO/GCE.39,40 In this context, the presence of NiNiO should play a key role for the observed redox behavior, which may be explained by the mechanism proposed by Ding et al.1 and Wang et al.,41 as mentioned below and in Scheme 1.
V [curve (ii) in Fig. 4a], respectively, resembling that of the neat NiNiO/GCE.39,40 In this context, the presence of NiNiO should play a key role for the observed redox behavior, which may be explained by the mechanism proposed by Ding et al.1 and Wang et al.,41 as mentioned below and in Scheme 1.| NiO + OH− → NiOOH + e− | (1) |
The electrochemical activity based on eqn (1) for all CV cycles (10, 20, and 30 with 5 and 15 for a better comparison) of the deposited NiNiO core–shell was evaluated by the CVs in an Ar-saturated 0.1 M NaOH at a 50 mV s−1 scan rate (see Fig. S3†). At a constant scan rate (50 mV s−1), among all the CV cycles of the deposited NiNiO core–shell-modified electrodes, a superior Ipa and better negatively shifted Epa were observed at 20 cycles of the deposited NiNiO/GCE. Therefore, we will consider only 20 cycles of the deposited NiNiO/GCE for the subsequent experiments.
To investigate the applicability of the NiNiO core–shell in an enzyme-free glucose detection, a noticeable increase in the Ipa along with a decrease in the Ipc was observed [curve (iii) in Fig. 4a] on the addition of 1 mM glucose at the NiNiO/GCE, which may be ascribed to the formation of gluconolactone through the oxidation of glucose by NiOOH (as can be seen in Scheme 1 and eqn (2)).9,39,41
| NiOOH + C6H12O6 → Ni(OH)2 + C6H10O6 | (2) |
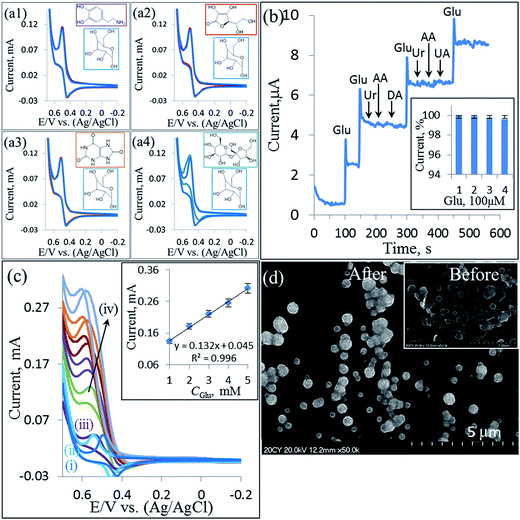
Moreover, the CVs of the NiNiO/GCE in an Ar-saturated 0.1 M NaOH with the sequential addition of glucose are shown in Fig. 4b. The oxidation Ipa corresponding to the transformation of Ni(OH)2 to NiOOH significantly increased (according to eqn (1)) with an increase in the glucose concentration (Cglu), and the Ipc accordingly decreased. This is due to the more consumption of NiOOH on increasing the Cglu (according to eqn (2)). A linear correlation was also found between the Ipa and Cglu up to 14 mM glucose in an Ar-saturated 0.1 M NaOH solution (Fig. 4b inset) with the linear regression equation of Ipa (mA) = 0.1332 × Cglu (mM) + 0.045 and correlation coefficient R2 = 0.9998, which confirms that the NiNiO/GCE have an excellent electrocatalytic activity for the enzyme-free glucose oxidation within 14 mM. Also, to further examine the enzyme-free glucose oxidation, we employed DPV and a similar result was observed (see Fig. S4†). Therefore, a wide range up to 14 mM with a high correlation coefficient of 0.9998 was confirmed once again for the NiNiO core–shell-modified GCE.
Fig. 4c displays the CV curves of the NiNiO/GCE obtained in an Ar-saturated 0.1 M NaOH solution at different scan rates (10 to 200 mV s−1) in the absence of glucose. Both Ipa and Ipc peak currents as well as their peak-to-peak separation were found to linearly increase with an increasing scan rate (Fig. 4c inset), indicating the occurrence of a surface-controlled electrochemical process.31 Moreover, in the presence of 1 mM glucose, the CV curves exhibited a similar behavior, as shown in Fig. 4d. A linear correlation was also observed for both Ipa and Ipc vs. scan rate (Fig. 4d inset), revealing again a surface-controlled process.
3.4 Amperometric determination of glucose
Amperometry is a sensitive and reliable technique to evaluate the electroactivities of the catalysts applicable for the electrochemical glucose biosensors. The amperometric responses obtained for the NiNiO/GCE in an Ar-saturated 0.1 M NaOH solution for the successive addition of different concentrations of glucose at an optimized applied potential of 0.5 V (vs. Ag/AgCl) (see Fig. S5†) are shown in Fig. 5. The NiNiO/GCE exhibited a sensitive response towards the different concentrations of glucose and an obvious increase in the oxidation current was observed for the increased glucose concentration. From Fig. 5, it is clear that the electrochemical response of the NiNiO/GCE reached the dynamic equilibrium state and generated a steady state current within ∼1 s (see Fig. S6†). It indicates good electrocatalytic oxidation and fast electron transfer properties of the NiNiO core–shell on the GCE. The calibration curve for the enzyme-free electrochemical responses of the NiNiO/GCE towards glucose is shown in the inset (left). A similar CV slope value was observed in the linear regression equation of Ipa (μA) = 0.1336 × Cglu (μM) + 1.17 with a similar R2 value (0.9997). Moreover, the enlarged CA curve shows that the NiNiO/GCE responded at 2 μM glucose with a distinguished increase in the current (inset, right). Accordingly, a lower limit of detection limit (LOD) at 0.4 μM was determined according to the formula LOD = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Stb/B (where Stb is the standard deviation of the blank signal, and B is the slope of the calibration curve).42,43 The obtained sensitivity of the glucose biosensor was derived to be 1889.8 μA mM−1 cm−2, surpassing most of the reported Ni-based composite electrodes (Table 1).
Stb/B (where Stb is the standard deviation of the blank signal, and B is the slope of the calibration curve).42,43 The obtained sensitivity of the glucose biosensor was derived to be 1889.8 μA mM−1 cm−2, surpassing most of the reported Ni-based composite electrodes (Table 1).
| Electrode materials | Media | LOD (μM) | Linear range (μM) | Sensitivity (μA mM−1 cm−2) | Response time (s) | References |
|---|---|---|---|---|---|---|
| a Electrodeposition method for electrode preparation. | ||||||
| NiNiO core–shell | 0.1 M NaOH | 0.4 | 2–14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1889.8 | ∼1 | This worka |
| NiO–Ag nanofibers | 0.1 M NaOH | 1.37 | ∼590 | 19.3 | 1 | |
| NiNPs/PEDOT/RGO | 0.1 M NaOH | 0.8 | 1–5100 | 36.15 | 6 | |
| Ni3S2/NF | 0.1 M NaOH | 0.82 | 0.5–3000 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 460 460 |
7 | |
| NiO/Pt/ERGO | 0.05 M NaOH | 0.2 | 2–5660 | 668.2 | 8a | |
| PVdF–HFP/Ni/Co | 0.1 M NaOH | 0.26 | 1–7000 | 7.56 | <10 | 9 |
| Porous NiO | 0.1 M NaOH | 0.34 | 0–1000 | 1680 | <4 | 14 |
| PVP–GNs–NiNPs–CS | 0.1 M NaOH | 0.03 | 0.1–500 | 103.8 | 20 | |
| ITO/Ni/Cu | 0.1 M NaOH | 0.23 | 1–2000 | 2530 | 2 | 21 |
| Ni7S6 flower | 0.1 M NaOH | 0.15 | 5–3700 | 271.8 | 22 | |
| EAuNi(OH)2 | 0.1 M KOH | 370 | ∼2000 | 23a | ||
| Ni–ZnO | 0.1 M KOH | 0.28 | 1–8100 | 824.34 | 4 | 24 |
| Ni–ITO nanocomposite | 0.1 M NaOH | 3.74 | 20–3000 | 610 | 25a | |
| Pt3Ni7/MWCNTs–NF | 0.1 M PBS | 0.3 | ∼15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
940 | ∼3 | 26a |
| NiO nanoflowers | 0.1 M NaOH | 1 | 10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
120 μA mM−1 | <10 | 30 |
| NiHCF/PANI/graphene | 0.1 M NaOH | 0.5 | 1–765 | 487.33 | 31a | |
| Ni paste with carbon | 0.1 M NaOH | 400 | 1000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1.3 mA cm−2 | 38 | |
| NiO–HSs@RGO–NF | 0.1 M NaOH | 0.03 | 258–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
2721 | ∼3 | 39 |
| HAC/NiO | 0.1 M NaOH | 0.055 | 5–4793 | 1721.5 | 40 | |
| NiO nanoflake | 0.1 M NaOH | 1.2 | 10–800 | 41 | ||
| Ni–MWCNT | 0.1 M NaOH | 0.89 | 3.2–17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
67.2 | <2 | 44a |
| RGO–Ni(OH)2 | 0.1 M NaOH | 0.6 | 2–3100 | 11.43 | 45 | |
| NiNPs/SMWNTs | 0.1 M NaOH | 0.5 | 1–1000 | 1438 | 3 | 46 |
| Nafion/NiO–rGO | 0.1 M NaOH | 0.77 | 2–600 | 1100 | <5 | 47 |
| Ni/ITO | 0.1 M NaOH | 0.5 | 1–350 | 189.5 | 48 | |
| Graphene/NiO | 0.1 M NaOH | 1 | 5–2800 | 1571 | 3 | 49 |
| Ni(OH)2@oPPyNW | 0.1 M NaOH | 0.3 | 1–4863 | 1049.2 | <10 | 50 |
| Ni(OH)2/Au | 0.1 M NaOH | 0.92 | 5–2200 | 371.2 | 51 | |
| NiO/CVD–graphene | 1 M KOH | 0.13 | 2–40 | 40.6 | 5 | 52a |
| NiNPs/ATP/RGO | 0.1 M NaOH | 0.37 | 1–710 | 1414.4 | 2 | 53a |
| GCE–P–Ni | 0.1 M NaOH | 0.690 | 2.5–1115 | 6.64 | 54a | |
| CS–RGO–NiNPs | 0.1 M NaOH | 4.1 | ∼9000 | 318.4 | 55 | |
| Ni–MG–BDD | 0.5 M NaOH | 0.24 | 0.5–15.5 | 660.8 | 56 | |
3.5 Selective determination of glucose
To assess the specificity of the enzyme-free glucose biosensor for real-time applications, the CVs were employed first. As can be observed in Fig. 6a, no significant interference was observed in the determination of 1 mM glucose in the presence of the following coexisting molecules: 0.5 mM dopamine (DA, a1), 1 mM ascorbic acid (AA, a2), 0.5 mM uric acid (UA, a3), and 0.2 mM sucrose (a4). It was concluded that the effects of these bioorganic molecules were negligible in the glucose oxidation, suggesting that the prepared NiNiO/GCE had a high selectivity towards the enzyme-free sensing of glucose. Moreover, an amperometric study was performed on the NiNiO/GCE under sequential addition of 100 μM glucose (Glu) and other electroactive interferences, such as DA, AA, and UA, in the presence of 20 μL of the pretreated (ESI†) urine (Ur, equivalent to 6.7 mL L−1), as shown in Fig. 6b. Obviously, the reported NiNiO core–shell-modified GCE was highly selective for glucose sensing and insensitive towards other interfering biomolecules. Moreover, the reported enzyme-free glucose biosensor is also insensitive to urine, which contains a variety of proteins and other common interfering molecules and ions. By comparing the amperometric responses of the biosensor in the presence of glucose with and without increasing the urine content (Fig. 6b inset), it is clear that the NiNiO/GCE was indeed sensitive and selective for the detection of glucose in a real clinical application. In addition, to explore the industrial use of the NiNiO core–shell-modified GCE, we employed this electrode for the determination of glucose in the presence of 200 μL (equivalent to 40 mL L−1) tap water (TW) and 200 μL Ur as an industrial wastewater sample, as shown in Fig. 6c. There was no significant increase in the Ipa in the presence of the abovementioned amounts of the real samples (see Fig. S7†). After sequential addition of glucose, the Ipa accordingly increased with the increasing glucose concentration. A linear regression equation of Ipa (mA) = 0.132 × Cglu (mM) + 0.045 with a R2 value of 0.996 was obtained, which is similar to the linear regression equation in the CVs investigation, as shown in the inset of Fig. 4b. Therefore, the NiNiO/GCE was also sensitive and selective for the detection of glucose in an industrial application.3.6 Stability, reproducibility, and repeatability
The operational stability of the NiNiO core–shell-modified GCE was also evaluated through the electrooxidation of glucose every 3 days by obtaining the CV curves in an Ar-saturated 0.1 M NaOH electrolyte solution in the presence of 1 mM glucose, and the coated electrode was stored under normal atmospheric conditions when not in use (Fig. S8a†). It was found that the NiNiO/GCE retained about 92% of its initial response for glucose oxidation after 30 days of storage. The NiNiO/GCE surface was evaluated by SEM after long term stability evaluation, as shown in Fig. 6d, and no significant difference in the shape of the NiNiO NPs was observed, however, the size and population may be reduced due to the decay nature for 33 days catalysis, indicating a homogeneous catalysis at the surface of every single NiNiO NP. The reproducibility of this enzyme-free biosensor was evaluated by assaying the CV response towards 1 mM glucose for five separately prepared NiNiO/GCEs under the same conditions (Fig. S8a inset†). A relative standard deviation (RSD) of 2.2% was observed for a series of NiNiO/GCEs, signifying the good reproducibility of the prepared biosensors. The repeatability of the reported glucose biosensor was also tested at a single NiNiO/GCE with the CV in the presence of 1 mM glucose for 10 consecutive CV responses. It was found that the glucose biosensor retained 0.64% RSD (Fig. S8b†), revealing an excellent repeatability. Moreover, only 0.68% RSD was found when the repeatability was evaluated in five different glucose solutions (Fig. S8b inset†). Also, we found that this biosensor was sensing well up to 60 °C, which is an another advantage over the enzyme-based glucose biosensors15 (Fig. S9†).3.7 Real sample analysis
The practical application of the NiNiO/GCE was tested by determining the glucose concentration in a human urine sample, which was obtained from a healthy volunteer. Informed consents were obtained from human participants for this study. The glucose concentration was detected at the NiNiO/GCE in the presence of 200 μL of urine. The analytical recoveries were determined for the addition of 1 mM, 3 mM, and 5 mM of the standard glucose to the urine sample. The recoveries of glucose at the NiNiO/GCE were in the range of 99.4–102% with a good RSD value of 3.6% (n = 3), suggesting that the glucose determination in the real samples had good accuracy (Table S1†).4. Conclusion
In summary, a NiNiO core–shell was prepared using a simple and easy electrodeposition method, which was then used to investigate the enzyme-free glucose sensing. The fabricated biosensor was a single component system combining the GCE with a NiNiO core–shell catalyst including the advantages of being binder free, time and cost-effective along with a stable attachment of the catalyst. The NiNiO–GCEs not only exhibited a high sensitivity and selectivity for the detection of glucose in the presence of biointerferences, such as AA, DA, and UA, but also detected glucose in the real samples such as human urine and tap water. Importantly, the NiNiO-electrode detected a wide range of glucose concentrations from 2 μM to 14 mM with a high sensitivity (1889.8 μA mM−1 cm−2). Additionally, a lower LOD and an ultra-fast response time of 0.4 μM and ∼1 s were observed, respectively. Thus, the NiNiO-based electrode, which showed superior electrochemical performances surpassing the other Ni-based electrodes, as shown in Table 1, should have perspective applications as a high-performance glucose biosensor even in real samples.Acknowledgements
This research study was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (NRF-2015R1D1A1A09059344).References
- Y. Ding, Y. Wang, L. Su, H. Zhang and Y. Lei, J. Mater. Chem., 2010, 20, 9918 RSC.
- X. Cao, N. Wang, S. Jia and Y. Shao, Anal. Chem., 2013, 85, 5040 CrossRef CAS PubMed.
- P. Yang, S. Y. Jin, Q. Z. Xu and S. H. Yu, Small, 2013, 9, 199 CrossRef CAS PubMed.
- P. Zimmet, K. G. M. M. Alberti and J. Shaw, Nature, 2001, 414, 782 CrossRef CAS PubMed.
- J. E. Choe, M. S. Ahmed and S. Jeon, J. Electrochem. Soc., 2016, 163, B113 CrossRef CAS.
- N. Hui, S. Wang, H. Xie, S. Xu, S. Niu and X. Luo, Sens. Actuators, B, 2015, 221, 606 CrossRef CAS.
- S. Kim, S. H. Lee, M. Cho and Y. Lee, Biosens. Bioelectron., 2016, 85, 587 CrossRef CAS PubMed.
- M. Li, X. Bo, Z. Mu, Y. Zhang and L. Guo, Sens. Actuators, B, 2014, 192, 261 CrossRef CAS.
- N. Senthilkumar, K. J. Babu, G. G. Kumar, A. R. Kim and D. J. Yoo, Ind. Eng. Chem. Res., 2014, 53, 10347 CrossRef CAS.
- H. S. Han, M. S. Ahmed, H. Jeong and S. Jeon, J. Electrochem. Soc., 2015, 162, B75 CrossRef CAS.
- Y. Zhang, L. Luo, Z. Zhang, Y. Ding, S. Liu, D. Deng, H. Zhaoa and Y. Chen, J. Mater. Chem. B, 2014, 2, 529 RSC.
- J.-M. You, Y. N. Jeong, M. S. Ahmed, S. K. Kim, H. C. Choi and S. Jeon, Biosens. Bioelectron., 2011, 26, 2287 CrossRef CAS PubMed.
- H. S. Han, H. Seol, D. H. Kang, M. S. Ahmed, H. Jeong and S. Jeon, Sens. Actuators, B, 2014, 204, 289 CrossRef CAS.
- F. J. Garcia-Garcia, P. Salazar, F. Yubero and A. R. González-Elipe, Electrochim. Acta, 2016, 201, 38 CrossRef CAS.
- D.-M. Kim, S. J. Cho, C.-H. Cho, K. B. Kim, M.-Y. Kim and Y.-B. Shim, Biosens. Bioelectron., 2016, 79, 165 CrossRef CAS PubMed.
- M. Fleischmann, K. Korinek and D. Pletcher, J. Electroanal. Chem. Interfacial Electrochem., 1971, 31, 39 CrossRef CAS.
- R. E. Reim and R. M. Van Effen, Anal. Chem., 1986, 58, 3203 CrossRef CAS.
- M. S. Ahmed and S. Jeon, ACS Catal., 2014, 4, 1830 CrossRef CAS.
- M. S. Ahmed and S. Jeon, J. Electrochem. Soc., 2014, 161, F1300 CrossRef.
- Z. Liu, Y. Guo and C. Dong, Talanta, 2015, 137, 87 CrossRef CAS PubMed.
- P. Salazar, V. Rico and A. R. González-Elipe, Sens. Actuators, B, 2016, 226, 436 CrossRef CAS.
- W. Wu, Y. Li, J. Jin, H. Wu, S. Wang and Q. Xia, Sens. Actuators, B, 2016, 232, 633 CrossRef CAS.
- A. L. Rinaldi and R. Carballo, Sens. Actuators, B, 2016, 228, 43 CrossRef CAS.
- Y. Yang, Y. Wang, X. Bao and H. Li, J. Electroanal. Chem., 2016, 775, 163 CrossRef CAS.
- P. Sivasakthi, G. N. K. R. Bapu and M. Chandrasekaran, Mater. Sci. Eng., C, 2016, 58, 782 CrossRef CAS PubMed.
- Y. Zhao, L. Fan, B. Hong, J. Ren, M. Zhang, Q. Que and J. Ji, Sens. Actuators, B, 2016, 231, 800 CrossRef CAS.
- K. Lee, M. S. Ahmed and S. Jeon, J. Power Sources, 2015, 288, 261 CrossRef CAS.
- Y.-F. Li, J.-J. Lv, M. Zhang, J.-J. Feng, F.-F. Li and A.-J. Wang, J. Electroanal. Chem., 2015, 738, 1 CrossRef CAS.
- H. Zhang, W. Zhou, Y. Du, P. Yang and C. Wang, Electrochem. Commun., 2010, 12, 882 CrossRef CAS.
- Z. H. Ibupoto, K. Khun, V. Beni and M. Willander, Soft Nanosci. Lett., 2013, 3, 46 CrossRef CAS.
- Y. Kong, Y. Sha, Y. Tao, Y. Qin, H. Xue and M. Lu, J. Electrochem. Soc., 2014, 161, B269 CrossRef.
- S. Helveg, C. Lopez-Cartes, J. Sehested, P. L. Hansen, B. S. Clausen, J. R. Rostrup-Nielsen, F. Abild-Pedersen and J. K. Nørskov, Nature, 2004, 427, 426 CrossRef CAS PubMed.
- A. Dutta and J. Datta, J. Mater. Chem. A, 2014, 2, 3237 CAS.
- D. R. Kauffman, D. Alfonso, D. N. Tafen, J. Lekse, C. Wang, X. Deng, J. Lee, H. Jang, J. Lee, S. Kumar and C. Matranga, ACS Catal., 2016, 6, 1225 CrossRef CAS.
- H. Pang, Q. Lu, Y. Zhang, Y. Li and F. Gao, Nanoscale, 2010, 2, 920 RSC.
- J.-H. Kim, S. H. Kang, K. Zhu, J. Y. Kim, N. R. Neale and A. J. Frank, Chem. Commun., 2011, 47, 5214 RSC.
- V. Grover, R. Shukla, R. Kumari, B. P. Mandal, P. K. Kulriya, S. K. Srivastava, S. Ghosh, A. K. Tyagi and D. K. Avasthi, Phys. Chem. Chem. Phys., 2014, 16, 27065 RSC.
- W.-Y. Jeon, Y.-B. Choi and H.-H. Kim, Sensors, 2015, 15, 31083 CrossRef CAS PubMed.
- P. Lu, J. Yu, Y. Lei, S. Lu, C. Wang, D. Liu and Q. Guo, Sens. Actuators, B, 2015, 208, 90 CrossRef CAS.
- V. Veeramani, R. Madhu, S.-M. Chen, P. Veerakumar, C.-T. Hung and S.-B. Liu, Sens. Actuators, B, 2015, 221, 1384 CrossRef CAS.
- G. Wang, X. Lu, T. Zhai, Y. Ling, H. Wang, Y. Tong and Y. Li, Nanoscale, 2012, 4, 3123 RSC.
- H. Begum, M. S. Ahmed and S. Jeon, RSC Adv., 2016, 6, 50572 RSC.
- M. Yun, J. E. Choe, J.-M. You, M. S. Ahmed, K. Lee, Z. Ustundag and S. Jeon, Food Chem., 2015, 169, 114 CrossRef CAS PubMed.
- A. Sun, J. Zheng and Q. Sheng, Electrochim. Acta, 2012, 65, 64 CrossRef CAS.
- Y. Zhang, F. G. Xu, Y. J. Sun, Y. Shi, Z. W. Wen and Z. Li, J. Mater. Chem., 2011, 21, 16949 RSC.
- H. Nie, Z. Yao, X. Zhou, Z. Yang and S. Huang, Biosens. Bioelectron., 2011, 30, 28 CrossRef CAS PubMed.
- Y. Zhang, Y. Wang, J. Jia and J. Wang, Sens. Actuators, B, 2012, 171–172, 580 CrossRef CAS.
- H. Tian, M. Jia, M. Zhang and J. Hu, Electrochim. Acta, 2013, 96, 285 CrossRef CAS.
- S. J. Li, N. Xia, X. L. Lv, M. M. Zhao, B. Q. Yuan and H. Pang, Sens. Actuators, B, 2014, 190, 809 CrossRef CAS.
- J. Yang, M. Cho, C. Pang and Y. Lee, Sens. Actuators, B, 2015, 211, 93 CrossRef CAS.
- J. Chen and J. Zheng, J. Electroanal. Chem., 2015, 749, 83 CrossRef CAS.
- A. Rengaraj, Y. Haldorai, C. H. Kwak, S. Ahn, K.-J. Jeon, S. H. Park, Y.-K. Han and Y. S. Huh, J. Mater. Chem. B, 2015, 3, 6301 RSC.
- Z. Shen, W. Gao, P. Li, X. Wang, Q. Zheng, H. Wu, Y. Ma, W. Guan, S. Wu, Y. Yu and K. Ding, Talanta, 2016, 159, 194 CrossRef CAS PubMed.
- M. Mazloum-Ardakani, E. Amin-Sadrabadi and A. Khoshroo, J. Electroanal. Chem., 2016, 775, 116 CrossRef CAS.
- J. Yang, J.-H. Yu, J. R. Strickler, W.-J. Chang and S. Gunasekaran, Biosens. Bioelectron., 2013, 47, 530 CrossRef CAS PubMed.
- Z. Deng, H. Long, Q. Wei, Z. Yu, B. Zhou, Y. Wang, L. Zhang, S. Li, L. Ma, Y. Xie and J. Min, Sens. Actuators, B, 2016 DOI:10.1016/j.snb.2016.09.176.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra25459d |
| This journal is © The Royal Society of Chemistry 2017 |