Open Access Article
Open Access ArticleIn silico prediction of serious eye irritation or corrosion potential of chemicals†
Qin Wang,
Xiao Li,
Hongbin Yang,
Yingchun Cai,
Yinyin Wang,
Zhuang Wang,
Weihua Li ,
Yun Tang
,
Yun Tang * and
Guixia Liu
* and
Guixia Liu *
*
Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China. E-mail: gxliu@ecust.edu.cn; ytang234@ecust.edu.cn; Fax: +86-21-64251033; Tel: +86-21-64250811
First published on 20th January 2017
Abstract
Rapidly and correctly identifying eye irritants or corrosive chemicals is an important issue in health hazard assessment. The purpose of this study is to describe the development of in silico methods for the classification of chemicals into irritants/corrosives or non-irritants/non-corrosives. A total of 5220 chemicals for a serious eye irritation (EI) dataset and 2299 chemicals as an eye corrosion (EC) dataset were collected from available databases and literature. Structure–activity relationship (SAR) models were developed to separately predict serious EI or EC via machine learning methods. According to the overall prediction accuracy, the Pub-SVM model gave the best results for both serious EI (overall classification accuracy CA = 0.946) and EC (CA = 0.959). The sensitivity and specificity of serious EI were 97.3% and 86.7% for the training set, and 96.9% and 82.7% for the external validation set, respectively. Similarly, the sensitivity and specificity of EC were 95.5% and 96.2% for the training set, and 94.9% and 96.2% for the external validation set, respectively. The high specificity and sensitivity indicated that our models were reliable and robust, which can be used to predict the potential seriousness of EI/EC of compounds. Moreover, several structural alerts for characterizing serious EI/EC were identified using the combination of information gain and substructure frequency analysis.
1 Introduction
Assessing the eye irritation/corrosion (EI/EC) potential of a chemical is a necessary component of risk assessment. Cornea and conjunctiva tissues comprise the anterior surface of the eye, and hence cornea and conjunctiva tissues are directly exposed to the air and easily suffer injury by chemicals. There are several substances, such as chemicals used in manufacturing, agriculture and warfare, ocular pharmaceuticals, cosmetic products, and household products, that can cause EI or EC.1 To safeguard public health, toxicological assessments to the eye must be conducted prior to the production, transportation, and sale of chemicals and finished products.2Historically, the Draize in vivo rabbit test was used as a standard protocol to assess the EI potential of a chemical.3 However, it has several limitations such as being expensive, time-consuming and has been criticized for its cruelty. Hence, alternative methods to evaluate chemical toxicity of EI/EC are in high demand.
Since the 1990s, enormous efforts have been made to develop alternative in vitro and in silico methods to predict EI/EC responses. Currently, three methods have been adopted by the Organization for Economic Cooperation and Development (OECD) as partial replacement of the Draize test, according to the OECD test guidelines (TG) 437,4 438![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 and 460.6 Although these methods appeared more efficient and cost effective than an in vivo animal test, no single in vitro assay has been fully accepted as a regulatory replacement for the Draize test. In addition, in vitro EI tests have several limitations including being time-consuming and resource-intensive because they require samples of compounds as the test materials and animal eyes as the test tissues.
5 and 460.6 Although these methods appeared more efficient and cost effective than an in vivo animal test, no single in vitro assay has been fully accepted as a regulatory replacement for the Draize test. In addition, in vitro EI tests have several limitations including being time-consuming and resource-intensive because they require samples of compounds as the test materials and animal eyes as the test tissues.
Compared with experimental testing protocols, in silico methods save time and are applicable for virtual molecules before they are synthesized. Previously, several QSAR (quantitative structure–activity relationship) models7–12 have been constructed to predict eye irritants, and those models are mainly local models based on one chemical category or several chemical categories of the European Center for Ecotoxicology and Toxicology of Chemicals (ECETOC). The number of those datasets was very small. For a summary of QSAR studies in ocular toxicity, three previously published reviews13–15 were recommended. Until two years ago,16 a larger database of 1860 chemicals was compiled from in vivo rabbit EI data. However, it is not available publicly.
The United Nation Globally Harmonized System (GHS) of Classification and Labeling of Chemicals provides a foundation that is used for internationally harmonization of regulations and rules on chemicals.17 Two separate models were built in this investigation to classify the labels H314 (severe eye damage)/H318 (serious eye damage) and H319 as described below, which are referred to as eye corrosion (EC) and serious eye irritation (EI) in this study.
According to the list of GHS hazard statements,17 the definition that is used for modeling in this study is as follows:
(1) H314: causes severe skin burns and eye damage (corrosion).
H318: serious eye damage; the production of tissue damage in the eye, or serious physical decay of vision, following application of a test substance to the anterior surface of the eye, which is not fully reversible within 21 days of application.
(2) H319: causes serious eye irritation; the production of changes in the eye following the application of a test substance to the anterior surface of the eye, which are fully reversible within 21 days of application.
The main goal of this study was to develop in silico methods for estimating the potential of serious EI/EC of substantial various chemicals. In this study, we collected high-quality diverse data of EC and EI from the databases and literature. Then, nine chemical fingerprints were used to represent the chemicals, and six machine learning methods were applied to build binary classification models for the prediction of EI/EC. Five-fold cross validation and external validation were used to determine the predictive ability of those models. Moreover, structural alerts18 of an eye irritant or corrosive were analyzed by information gain and substructure frequency analysis methods, and several important structural alerts were obtained.
2 Materials and methods
2.1 Data collection and preparation
Chemicals were collected with their Chemical Abstracts Service (CAS) registry numbers and Simplified Molecular Input Line Entry System (SMILES) codes from 20 databases of QSAR Toolbox 3.3 (https://www.qsartoolbox.org/home). Those with the same or wrong CAS numbers were deleted by EXCEL. All the SMILES of positive chemicals (corrosives/irritants) were double checked with ChemIDplus Advanced (http://chem.sis.nlm.nih.gov/chemidplus). Then, to search toxicological information of those chemicals, we mapped CAS numbers to X-MOL (http://www.x-mol.com/). Negative chemicals were extracted from one article.19 The SMILES of negative chemicals were collected from ChemIDplus Advanced.All of the data (including positive and negative chemicals) were prepared by removing all false SMILES strings, inorganic substances, organometallic salts, ammonium salts, mixtures and duplicated compounds. When curating the data set, the compounds that existed in both negative chemicals and positive chemicals were removed. Finally, 2299 chemicals (including 887 positive and 1412 negative chemicals) for eye corrosion (EC) models, and 5220 chemicals (including 3874 positive and 1346 negative chemicals) for eye irritation (EI) models were obtained. Then, both the datasets (Table 1) were randomly divided into the training set and the external validation set in the ratio of 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 by Discovery Studio 3.5 Client.
20 by Discovery Studio 3.5 Client.
| Total number | Training set | External validation set | |||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| EC | 2299 | 691 | 1148 | 196 | 264 |
| EI | 5220 | 3107 | 1069 | 767 | 277 |
2.2 Molecular description
In this study, molecular fingerprints were used to represent molecules. Each molecule was described as a binary string of structural keys. Nine molecular fingerprints of all the chemicals were calculated by PaDEL-Descriptor,20 including the Atom Pair 2D fingerprint (AP2D, 780 bits), Estate fingerprint (Estate, 79 bits), CDK extended fingerprint (Extended, 1024 bits), CDK fingerprint (FP, 1024 bits), CDK graph only fingerprint (Graph, 1024 bits), Klekota–Roth fingerprint (KR, 4860 bits), MACCS fingerprint (MACCS, 166 bits), PubChem fingerprint (PubChem, 881 bits) and Substructure fingerprint (SubFP, 307 bits). The detailed description of these fingerprints can be found in the original publication.202.3 Model building
Among a multitude of available binary classification methods, we applied six machine learning methods that are highly effective, robust and extensively successful in the field of drug discovery that include the support vector machine (SVM),21 artificial neural network (ANN),22 C4.5 decision tree (C4.5),23 random forest (RF),24 Naïve Bayes (NB),25 and k-nearest neighbor (kNN).26The SVM algorithm was employed in the open source LIBSVM 3.2 package,27 while the others were implemented in Orange 2.7 (freely available at http://orange.biolab.si/orange2/). The parameters of SVM (C and gamma) were optimized through a python script in the LIBSVM 3.2 package. The number of trees of RF was set to 50, the k parameter of kNN was set to 15, and other parameters were set by default in Orange.
2.4 Evaluation of model performance
In this study, the robustness of all the models was assessed by five-fold cross validation, and the prediction accuracy of the models was evaluated by the external validation set. In addition, all models were evaluated using the counts of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). The sensitivity (SE), specificity (SP) and the overall classification accuracy (CA) of the models were calculated using the following equations:28| SE = TP/(TP + FN) | (1) |
| SP = TN/(TN + FP) | (2) |
| CA = (TP + TN)/(TP + TN + FP + FN) | (3) |
The binary classification models were also evaluated by the receiver operating characteristic (ROC). The principle is that if the plot has a surface area (AUC) of 1, the classifier is perfect, and if the area equals 0.5, the classifier is a useless random classifier.29
2.5 Analysis of structural alerts
Structural alerts are known as molecular functional groups that make chemicals toxic. They are of importance for predicting toxicity because they are directly derived from mechanistic knowledge.30 In this study, structural alerts were analyzed using the methods of information gain31 and substructure frequency analysis.32If a substructure is more frequently presented in the corrosive or irritant chemical class, this substructure is called a structural alert involved in chemical corrosion or irritation. The frequency of a fragment in toxic chemicals was determined by the frequency of a fragment enrichment factor, which is defined as follows:33
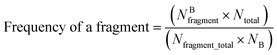 | (4) |
3 Results
3.1 The datasets of EC and EI
After processing original databases, we obtained 887 positive chemicals for EC and 3874 positive chemicals for the EI model. We also extracted 1412 negative chemicals for EC and 1346 negative chemicals for EI from the literature. A total of 5220 chemicals for eye irritation models and 2299 chemicals for eye corrosion models were collected from available databases and the literature. Then, the data set was randomly divided into a training set and an external validation set in the ratio of 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 by Discovery Studio 3.5 Client. The detailed statistical descriptions of the EC and EI datasets are listed in Table 1.
20 by Discovery Studio 3.5 Client. The detailed statistical descriptions of the EC and EI datasets are listed in Table 1.
For the EC model, chemicals of EC were represented as 1 and negative chemicals as 0. For the EI model, chemicals of EI were represented as 1 and negative chemicals as 0.
The SMILES strings and classification of all chemicals for EC and EI models can be found in Tables S1 and S2 of ESI,† respectively.
3.2 Chemical diversity analysis
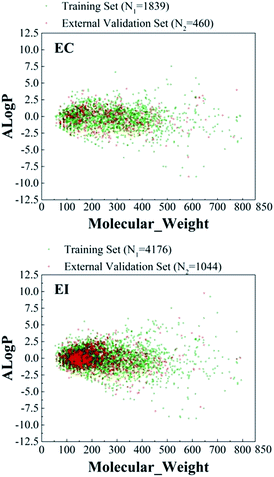
The sums of the chemicals in the EC and serious EI datasets were 2299 and 5220, respectively, as shown in Table 1. To build a robust prediction model, chemical diversity of a data set is a key issue. Therefore, the chemical space and Tanimoto similarity were used to investigate the chemical diversity.We used the molecule weight (MW) and Ghose–Crippen log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kow (A
Kow (A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) of each class in the database to investigate the chemical space distribution. The chemical space distribution scatter diagrams of the two datasets are presented in Fig. 1, which illustrated that the external validation set shared similar chemical space with the training set.
P) of each class in the database to investigate the chemical space distribution. The chemical space distribution scatter diagrams of the two datasets are presented in Fig. 1, which illustrated that the external validation set shared similar chemical space with the training set.
The Tanimoto coefficient was used to evaluate the diversity of chemicals in the two datasets. The heat maps of the Tanimoto similarity index of EC and EI datasets are shown in Fig. 2. Colors close to red in the heat map (with high Tanimoto similarity index) indicate that the compounds are more similar. On the contrary, colors close to dark blue (with low Tanimoto similarity index) indicate that the compounds have higher diversity. The average Tanimoto similarity indexes were 0.24 for the EC training set and 0.23 for the external validation set. The indexes were 0.21 for the EI training set and 0.21 for the external validation set. For the entire EC and EI datasets, the indexes were 0.24 and 0.21, respectively. From these values, we know that the datasets were chemically diverse.
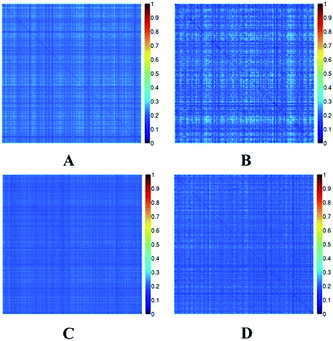 | ||
| Fig. 2 Tanimoto similarity index for the EC-training set (A), EC-external validation set (B), EI-training set (C) and EI-external validation set (D). | ||
3.3 Performance of five-fold cross validation
In this study, the EC and EI binary classification models were built using nine chemical fingerprints combined with six machine learning methods, including NB, ANN, kNN, C4.5 DT, SVM and RF. The models were validated using five-fold cross validation and external set validation. The detailed values of CA, AUC, SE and SP for the five-fold cross validation of EC and EI models are summarized in Tables S3 and S4 of ESI,† respectively.As shown in Table S3,† for the models of EC, the CA values in all models ranged from 81.9% to 95.9%, the AUC values in all models ranged from 88.1% to 99.2%, the SE values in all models ranged from 80.6% to 99.0%, and the SP values in all models ranged from 72.0% to 96.9%. It was encouraging that the SP values in only two models were less than 80%. Among these models, Pub-SVM gave the best result (CA = 0.959, AUC = 0.992, SE = 0.955, SP = 0.962). According to the overall prediction accuracy, the top five models were Pub-SVM, MACCS-ANN, MACCS-SVM, FP-SVM and FP-ANN.
As shown in Table S4,† for the models of EI, the CA values in all models ranged from 83.8% to 94.6%, the AUC values in all models ranged from 77.9% to 97.6%, the SE values in all models ranged from 84.0% to 99.2%, and the SP values in all models ranged from 59.3% to 91.7%. Although the SP values were less than the SE values, according to the SP values ranging from maximum to minimum, the CA values in the top 10 models ranged from 83.8% to 94.6%, the AUC values in the top 10 models ranged from 88.1% to 97.6%, the SE values ranged from 84.0% to 97.5%, and the SP values in the top 10 models ranged from 82.8% to 91.7%. It was encouraging that there were numerous models with SP values more than 80%. Among these 54 models, Pub-SVM gave the best result (CA = 0.946, AUC = 0.976, SE = 0.973, SP = 0.867). According to the overall prediction accuracy, the top five models were Pub-SVM, MACCS-SVM, Pub-ANN, FP-SVM and KR-SVM.
According to the results of the five-fold cross validation of EC and EI, three conclusions could be obtained. The first one was that most of the models exhibited good overall predictive performance for the training set. The second one was that six machine learning methods greatly differed in prediction ability. The third one was that the models with good performance were mainly developed via the SVM machine learning method combined with PubChem and MACCS as attributes.
3.4 Performance of external validation
The external validation set was used to evaluate the performance of the best five binary classification models, and the detailed results are given in Table 2. As shown in Table 2, for the external validation set of the EC models, the best result with the highest accuracy of 95.9% was the FP fingerprint combined with the SVM algorithm, and the other four models also exhibited excellent overall predictive performance for the external validation set. The model performing the best (Pub-SVM) in the five-fold cross validation was the second best in the external validation set. As shown in Table 2, for the external validation set of EI models, the best result was with the highest accuracy of 93.8% using the MACCS fingerprint combined with the SVM algorithm. The model (MACCS-SVM) was the second best in the five-fold cross validation. Considering the values of SP and SE, although SP values were lower than SE values for EI models, SP values were higher than 82% for all the external validation results of the five models (Table 2). Therefore, we can conclude that the prediction results demonstrated the stable robustness and precise prediction accuracy of the models.| Type | Model | CA | AUC | SE | SP |
|---|---|---|---|---|---|
| EC | Pub-SVM | 0.957 | 0.994 | 0.949 | 0.962 |
| MACCS-ANN | 0.950 | 0.991 | 0.939 | 0.958 | |
| MACCS-SVM | 0.957 | 0.991 | 0.939 | 0.970 | |
| FP-SVM | 0.959 | 0.991 | 0.964 | 0.955 | |
| FP-ANN | 0.954 | 0.989 | 0.949 | 0.958 | |
| EI | Pub-SVM | 0.931 | 0.968 | 0.969 | 0.827 |
| MACCS-SVM | 0.938 | 0.972 | 0.967 | 0.856 | |
| Pub-ANN | 0.929 | 0.962 | 0.960 | 0.845 | |
| FP-SVM | 0.933 | 0.969 | 0.971 | 0.827 | |
| KR-SVM | 0.936 | 0.958 | 0.967 | 0.848 |
3.5 Identification of structural alerts
To investigate structural features between ocular toxic and nontoxic compounds, substructure frequency analysis and information gain methods were applied on the entire datasets of EC and EI (containing the training set and the external validation set) using the PubChem fingerprint. Four and five important structural alerts (Table 3) were identified to appear more frequently in corrosive/irritant chemicals than in non-corrosive/non-irritant chemicals, respectively.| Type | Structural alert | Positive frequencya | Negative frequencya | Limits |
|---|---|---|---|---|
| a Positive frequency and negative frequency represent the “frequency of a fragment” in positive chemicals and negative chemicals respectively, which were used to pick structural alerts.b The number in the parenthesis means the number of the fragment-containing chemicals in the positive class.c The number in the parenthesis means the number of the fragment-containing chemicals in the negative class. | ||||
| EC | 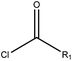 |
2.51 (90)b | 0 (0)c | R1 = –OR/–aryl/–alkyl |
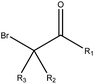 |
2.45 (37) | 0.04 (1) | R1 = –H/–OH/–NH2/–OR/–aryl/–X, R2–3 = any | |
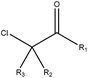 |
2.14 (29) | 0.23 (5) | R1 = –H/–OH/–NH2/–OR/–aryl/–X, R2–3 = any | |
 |
2.51 (31) | 0 (0) | R1–3 = any (e.g., further halogen) | |
| EI | 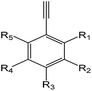 |
1.24 (189)b | 0.32 (17)c | R1–5 = –OH/–NH2/–X/–alkyl/–aryl |
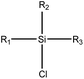
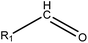 |
1.23 (182) | 0.35 (18) | R1 = –OR/–aryl/–alkyl | |
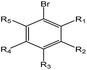 |
1.24 (138) | 0.31 (12) | R1–5 = –H/–X/–OH/–COOH/–alkyl/–aryl | |
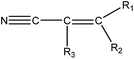 |
1.21 (90) | 0.38 (10) | R1–2 = –H/–alkyl/–aryl, R3 = –H/–alkyl/–X (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C double bond may be –aryl) C double bond may be –aryl) |
|
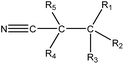 |
1.23 (172) | 0.33 (16) | R1–3 = any, R4–5 = –H/–alkyl | |
As shown in Table 3, the first structural alert is an acylating agent. Recently, acylation was one reaction mechanism of electrophilic reactivity-based profiling which was used to confirm serious eye irritants or corrosives.34 The second structural alert has a bromine methyl that can abstract an electron and consequently accelerate acylation. The last structural alert is in agreement with a previously published study.35 Therefore, the former three structural alerts were proven as new structural alerts.
For the structural alerts for ocular irritant chemicals, the basic skeleton of the first structural alert was the irritant, phenylacetylene. When R1–5 of phenylacetylene was replaced by other substituents excluding hydrogen, the new chemicals may produce EI via multiple mechanisms. For instance, if R is a hydroxyl, this chemical is a phenol. Thus, it refers to alcohols that may cause serious EI.36 It is easy for the second structural alert to undergo a Michael addition reaction, which has been mentioned in a previously published study.34 After checking our entire data set, we found 138 positive chemicals that have the bromine atom together with an aromatic ring. Therefore, the third structural alert could be proven as a new structural alert.
4 Discussion
4.1 Diversity of dataset
The diversity of the dataset plays an important role for the results of classification prediction models. Previously, several QSAR models have been constructed to predict eye irritants, and those models were mainly local models based on one chemical category or several chemical categories, including organic acids, salicylates, and alcohols.7–12 The number of congeneric sets of chemicals or diverse chemical structures of those data sets ranged from 7 to 54. We could say that those models only provided reliable predictions within a limited chemical space. Only most recently, Verma and Matthews developed c-QSAR (classification QSAR) models using an artificial neural network for the prediction of EI based on 2928 substances.19 However, they only had 917 positive chemicals.In our study, we collected diverse chemicals from databases and the literature (1839 compounds for EC and 4176 ones for EI) to build global models. The training sets shared a similar chemical space with the external validation sets, as shown in Fig. 1. In addition, the average Tanimoto similarity indexes of the entire datasets were about 0.2, indicating that our datasets were very diverse. Therefore, our models would have a wide range of applicability domain.
4.2 Performance of the binary classification models
Six modeling methods combined with nine fingerprints were used to develop binary classification models for predicting eye corrosives and eye irritants. Despite the imbalance of corrosives and non-corrosives (corrosives are less than non-corrosives), it was clear that the values of SE were nearly the same with SP in almost 54 binary classification models of EC (Table S3†). In addition, the lowest AUC value was 0.88, and hence we can conclude that the quality of the datasets of EC was very high. The results of 54 binary classification models of EI showed that the values of SE were higher than SP. We deduced that this phenomenon might be caused due to the imbalance of the eye irritants and non-irritants since the number of eye irritants (N = 3107) was more than non-irritants (N = 1069) in the training sets. Thus, the values of SE and SP may be improved by adjusting the ratio of eye irritants and non-irritants in the datasets. In addition, we found that the overall performance of SVM was almost better than that of five other algorithms, which was consistent with the conclusion in our previous study that SVM is a better method for chemical toxicity prediction.33 As mentioned above, Verma and Matthews developed c-QSAR models for the prediction of EI based on 2928 substances along with sensitivities in the 80–90% range, which were lower than our results ranging from 90–97% for the top five models. Among the EC models, Pub-SVM gave the best result (CA = 0.959, AUC = 0.992, SE = 0.955, SP = 0.962), which was comparable to the result of c-QSAR models for corrosion potential based on 504 substances.37In this study, nine chemical fingerprints were used to build binary classification prediction models unlike the traditional models that were always built with molecular descriptors. The method of fingerprints made a direct connection between the chemical 2D structure and the toxicity endpoint of chemicals. From the results of Table S4,† it was found that different chemical fingerprints, along with the same algorithm or different algorithms along with the same fingerprint, can produce different predictions. Therefore, it is necessary to pick a suitable algorithm and suitable fingerprint to characterize an entire dataset.
4.3 Advantages and limitations of our models
To the best of our knowledge, we have collected the biggest database of EC or EI for the construction of classification prediction models. The average Tanimoto similarity indexes of the entire datasets were about 0.2, indicating that our datasets were very diverse. The predictive ability of EC or EI classification prediction models, in this study, was comparable to those in any of the previous studies. The results of classification prediction models suggested that the constructed classification models could be reliably used for industry and regulatory agencies in initial prediction of EC or EI potential for diverse chemicals. Moreover, several structural alerts for characterizing serious eye irritation/corrosion were identified using the combination of information gain and substructure frequency analysis. These structural alerts can be used for structural optimization in future drug design and ocular toxicity safety assessment of chemicals.In this study, inorganic salts, organometallic salts and ammonium salts were removed. Clearly, pH values play important roles in EC or EI potential of chemicals. In general, salts and their relevant chemicals have different pH values. Therefore, we cannot directly transfer the 2D structures of salts into their corresponding acids or bases, and the models could not predict ocular toxicity of salts. Although small part of salts could not be predicted, in the future we will try to combine other in silico methods to solve this problem.
5 Conclusions
In this study, nine chemical fingerprints combined with six machine learning methods were used to build binary classification models based on datasets of 2299/5220 organic chemicals for predicting the potential EC/EI of compounds. Based on the values of CA and AUC, Pub-SVM models were the best for both EC and EI, which could provide robust and reliable predictions for EC and EI potentials of chemicals. Moreover, the structural alerts were identified, which could be used to distinguish eye corrosives/irritants and non-corrosives/non-irritants, by means of information gain and substructure frequency analysis. These structural alerts appear more frequently in compounds with EC/EI, and thus they should be responsible for acute eye toxicity, which would be helpful for understanding the reaction mechanism. In summary, this study provided a series of predictive models and toxic substructures for EC/EI, which might be helpful for drug screening in early drug discovery.Acknowledgements
We gratefully acknowledge the financial support from the National Natural Science Foundation of China (Grants 81273438 and 81373329) and the 111 Project (Grant B07023).Notes and references
- K. R. Wilhelmus, Surv. Ophthalmol., 2001, 45, 493–515 CrossRef CAS PubMed.
- M. K. Robinson, C. Cohen, A. D. B. de Fraissinette, M. Ponec, E. Whittle and J. H. Fentem, Food Chem. Toxicol., 2002, 40, 573–592 CrossRef CAS PubMed.
- J. H. Draize, G. Woodard and H. O. Calvery, J. Pharmacol. Exp. Ther., 1944, 82, 377–390 CAS.
- OECD, Test No. 437: Bovine Corneal Opacity and Permeability Test Method for Identifying (i) Chemicals Inducing Serious Eye Damage and (ii) Chemicals Not Requiring Classification for Eye Irritation or Serious Eye Damage, OECD Publishing.
- OECD, Test No. 438: Isolated Chicken Eye Test Method for Identifying (i) Chemicals Inducing Serious Eye Damage and (ii) Chemicals Not Requiring Classification for Eye Irritation or Serious Eye Damage, OECD Publishing.
- OECD, Test No. 460: Fluorescein Leakage Test Method for Identifying Ocular Corrosives and Severe Irritants, OECD Publishing.
- S. Sugai, K. Murata, T. Kitagaki and I. Tomita, J. Toxicol. Sci., 1991, 16, 111–130 CrossRef CAS PubMed.
- M. T. D. Cronin, D. A. Basketter and M. York, Toxicol. in Vitro, 1994, 8, 21–28 CrossRef CAS PubMed.
- M. H. Abraham, Chem. Soc. Rev., 1993, 22, 73–83 RSC.
- M. H. Abraham, R. Kumarsingh, J. E. Cometto-Muniz and W. S. Cain, Toxicol. in Vitro, 1998, 12, 201–207 CrossRef CAS PubMed.
- M. H. Abraham, R. Kumarsingh, J. E. Cometto-MuÑIz and W. S. Cain, Ann. N. Y. Acad. Sci., 1998, 855, 652–656 CrossRef CAS PubMed.
- A. Kulkarni, A. J. Hopfinger, R. Osborne, L. H. Bruner and E. D. Thompson, Toxicol. Sci., 2001, 59, 335–345 CrossRef CAS PubMed.
- A. G. Saliner, G. Patlewicz and A. P. Worth, QSAR Comb. Sci., 2008, 27, 49–59 CAS.
- C. J. Somps, N. Greene, J. A. Render, M. D. Aleo, J. H. Fortner, J. A. Dykens and G. Phillips, Cutaneous Ocul. Toxicol., 2009, 28, 1–18 CrossRef CAS PubMed.
- G. Patlewicz, R. Rodford and J. D. Walker, Environ. Toxicol. Chem., 2003, 22, 1862–1869 CrossRef CAS PubMed.
- E. Adriaens, J. Barroso, C. Eskes, S. Hoffmann, P. McNamee, N. Alépée, S. Bessou-Touya, A. De Smedt, B. De Wever, U. Pfannenbecker, M. Tailhardat and V. Zuang, Arch. Toxicol., 2014, 88, 701–723 CrossRef CAS PubMed.
- About the GHS – UNECE, http://www.unece.org/trans/danger/publi/ghs/ghs_welcome_e.html.
- N. L. Kruhlak, J. F. Contrera, R. D. Benz and E. J. Matthews, Adv. Drug Delivery Rev., 2007, 59, 43–55 CrossRef CAS PubMed.
- R. P. Verma and E. J. Matthews, Regul. Toxicol. Pharmacol., 2015, 71, 318–330 CrossRef CAS PubMed.
- C. W. Yap, J. Comput. Chem., 2011, 32, 1466–1474 CrossRef CAS PubMed.
- S. Suthaharan, in Machine Learning Models and Algorithms for Big Data Classification: Thinking with Examples for Effective Learning, Springer US, Boston, MA, 2016, pp. 207–235, DOI:10.1007/978-1-4899-7641-3_9.
- H. Parhizgar, M. R. Dehghani, A. Khazaei and M. Dalirian, Ind. Eng. Chem. Res., 2012, 51, 2775–2781 CrossRef CAS.
- J. R. Quinlan, C4.5: programs for machine learning, Morgan Kaufmann Publishers Inc., 1993 Search PubMed.
- L. Breiman, Mach. Learn., 2001, 45, 5–32 CrossRef.
- H. Sun, J. Med. Chem., 2005, 48, 4031–4039 CrossRef CAS PubMed.
- G. W. Kauffman and P. C. Jurs, J. Chem. Inf. Comput. Sci., 2001, 41, 1553–1560 CrossRef CAS PubMed.
- C.-C. Chang and C.-J. Lin, ACM Trans. Intell. Syst. Technol., 2011, 2, 1–27 CrossRef.
- Y. Chen, F. Cheng, L. Sun, W. Li, G. Liu and Y. Tang, Ecotoxicol. Environ. Saf., 2014, 110, 280–287 CrossRef CAS PubMed.
- J. Li and P. Gramatica, SAR QSAR Environ. Res., 2010, 21, 657–669 CrossRef CAS PubMed.
- R. Benigni and C. Bossa, Mutat. Res., Rev. Mutat. Res., 2008, 659, 248–261 CrossRef CAS PubMed.
- J. Shen, F. Cheng, Y. Xu, W. Li and Y. Tang, J. Chem. Inf. Model., 2010, 50, 1034–1041 CrossRef CAS PubMed.
- B. F. Jensen, C. Vind, P. B. Brockhoff and H. H. F. Refsgaard, J. Med. Chem., 2007, 50, 501–511 CrossRef CAS PubMed.
- F. Cheng, Y. Ikenaga, Y. Zhou, Y. Yu, W. Li, J. Shen, Z. Du, L. Chen, C. Xu, G. Liu, P. W. Lee and Y. Tang, J. Chem. Inf. Model., 2012, 52, 655–669 CrossRef CAS PubMed.
- B. Bhhatarai, D. M. Wilson, A. K. Parks, E. W. Carney and P. J. Spencer, Chem. Res. Toxicol., 2016, 29, 810–822 CrossRef CAS PubMed.
- E. Hulzebos, J. D. Walker, I. Gerner and K. Schlegel, QSAR Comb. Sci., 2005, 24, 332–342 CAS.
- P. Zhang, M. Liu and R. Liao, Mol. Med. Rep., 2012, 6, 33–38 CAS.
- R. P. Verma and E. J. Matthews, Regul. Toxicol. Pharmacol., 2015, 71, 331–336 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The name, SMILES strings and classification of all chemicals for EC models and EI models were listed in Tables S1 and S2. The values of CA, AUC, SE and SP for the five-fold cross validation of EC and EI models were summarized in Tables S3 and S4, respectively. See DOI: 10.1039/c6ra25267b |
| This journal is © The Royal Society of Chemistry 2017 |