Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Carbon-dot–hydrogel for enzyme-mediated bacterial detection†
Sagarika Bhattacharyaa,
Sukhendu Nandi‡
a and
Raz Jelinek *ab
*ab
aDepartment of Chemistry, Ben-Gurion University of the Negev, Beer Sheva 84105, Israel. E-mail: razj@bgu.ac.il; Fax: +972-8-6472943
bIlse Katz Institute for Nanotechnology, Ben-Gurion University of the Negev, Beer Sheva 84105, Israel
First published on 3rd January 2017
Abstract
A hybrid carbon-dot (C-dot)–hydrogel matrix was constructed and employed for detection of bacteria. The transduction mechanism is novel, based upon cleavage of ester bonds within the hydrogel scaffold by bacterially-secreted esterases; the ensuing fluidization of the hydrogel resulted in aggregation of the embedded C-dots and consequent quenching of their fluorescence. We show that the C-dot–hydrogel exhibits high sensitivity and can distinguish among bacterial species through modulation of the emitted fluorescence, depending upon their esterase secretions.
Introduction
Hydrogels, three dimensional flexible networks of water-soluble polymers, are useful host materials enabling encapsulation of high concentrations of varied guest molecules and nanoparticles.1–7 Hydrogels are generally classified according to two categories: (i) thermally-irreversible hydrogels in which the gelator molecules are crosslinked by covalent bonds (defined as chemical gels),5 and (ii) thermally-reversible physical gels in which the gelator molecules exhibit weak physical interactions.5 Hydrogels have been used in varied applications including tissue engineering, drug delivery and controlled-release, and medical implants.5Hydrogel composites have been used as useful biosensing platforms. Redox-based sensing schemes utilizing hydrogel-encapsulated enzymes were reported.8,9 A peptide hydrogel consisting of a Fmoc-diphenylalanine-encapsulating enzyme receptor (e.g., glucose oxidase or horseradish peroxidase) and semiconductor quantum dots (QDs) was reported for detection of glucose and toxic phenolic compounds through quenching of the QDs' fluorescence.9 A hydrogel cross-linked with aptamers was reported to detect very low amount of cocaine through visual detection.10 Porous silica polyacrylamide hydrogel system served as an optical bacterial sensor.11
The hydrogel-based sensors discussed above largely rely upon molecular recognition and/or redox reactions involving embedded enzymes as the components in the transduction mechanism. In such systems, the hydrogel serves only as a host matrix for immobilization of guest biomolecules. Exploiting changes in the physical state of the hydrogel as a sensing strategy could be, in fact, a promising sensing avenue. Varied externally-added molecules were shown to affect the structural features and mechanical properties of hydrogel matrixes.12–14 Hydrogel framework modulation has been employed as a sensing platform.15,16
Carbon dots (C-dots) are newly discovered fluorescent carbonaceous nanoparticles (<10 nm), and have attracted broad interest in light of their intriguing luminescence properties, particularly a broad range of excitation-dependent emission range.17–24 C-dots are biocompatible and less cytotoxic than semiconductor quantum dots, thus constituting promising vehicles for biological studies.19,25,26 Modulation of C-dots' fluorescence has been used as a powerful transduction mechanism in varied sensing applications, for example targeting metal ions,27,28 organic molecules,29,30 and radical species.31,32 C-dots have been recently used as a platform for bacterial sensing and imaging.33 Hybrid carbon-dot hydrogel systems were also used for detection of heavy metal ions.34,35
Herein, we report construction of a new C-dot–hydrogel hybrid material which was used for bacterial detection through modulation of the fluorescence emission of the hydrogel-embedded C-dots. In particular, we found that cleavage of hydrogel scaffolds' ester bonds by bacterially-secreted esterases and lipases resulted in fluidization of the hydrogel, consequent aggregation and concomitant fluorescence quenching of the embedded C-dots. The C-dot–hydrogel constitutes a sensitive bacterial detection platform since the transduction mechanism is based upon secreted enzymes rather than the bacterial cells themselves. Furthermore, the new assay can be employed for distinguishing bacterial species based upon the extent of their esterase/lipase secretion.
Experimental
Materials
D-Glucose, sodium sulfate, pyridine, lauric acid and sodium chloride were purchased from Sigma Aldrich, USA. L-(+)-Tartaric acid was purchased from Alfa-Aesar, England. Lauroyl chloride was purchased from TCI, Japan. Chloroform and n-hexane were bought from Daejung chemicals, Korea. Dimethylformamide (DMF) and acetone were purchased from Frutarom (Haifa, Israel). Ethyl acetate and concentrated hydrochloric acid were purchased from Bio-Lab Ltd (Jerusalem, Israel). Luria–Bertani (LB) agar was purchased from Pronadisa (Spain). All chemicals were used without further purification.C-dot–hydrogel synthesis
The C-dots and hydrogel were prepared from 6-O-(O-O′-dilauroyltartaryl)-D-glucose (compound 1, Fig. 1A) according to a published procedure.36,37 Briefly, we prepared 6-O-acylated fatty acid ester of D-glucose by reacting it with O-O′-dilauroyltartaric acid anhydride. In a Teflon film tightened, septum-capped test tube 100 mg of 1 and 330 μL DI water were placed and then heated in an oven to 125 °C for 2.5 h. Upon completion of the carbonization, the reaction mixture was cooled to room temperature yielding a brown precipitate indicating the formation of carbon dots and purified for further work. For preparation of the C-dot–hydrogel, 5 mg of C-dots were mixed with 50 mg of compound 1 and dissolved in 0.5 mL DI water in a sealed vial by sonicating for 30 min at 50 °C and further heated for 2–3 min until the gelator compound was dissolved completely. Upon cooling to room temperature for about 24 hours, the aqueous solution completely gelified, tested by the “stable to inversion” method.Bacterial growth
The bacteria used in the studies were Bacillus cereus, Bacillus subtilis (stain Bacillus 3610), Pseudomonas aeruginosa PAO1 wild type, and Staphylococcus aureus wild type.38 The bacteria were grown aerobically at 37 °C in sterilized solid LB medium composed of 13.5% yeast extract, 27% peptone, 27% NaCl, and 32.5% agar at pH 7.4. After overnight growth, a colony from each bacterial strain was taken and added to 10 mL sterilized LB medium and incubated at 37 °C. Bacterial growth was monitored in the specific time points through measuring the concentration of the bacteria by visible spectroscopy (108 cells per mL when optical density at 600 nm was 1.0). Bacterial cells were separated from the growth medium through centrifugation followed by washing with sterilized water several times and resuspending in distilled water.Confocal fluorescence microscopy
Confocal microscopy images of the C-dot–hydrogel were acquired on an UltraVIEW system (PerkinElmer Life Sciences, Waltham, MA) equipped with an Axiovert-200M microscope (Zeiss, Oberkochen, Germany) and a Plan-Neofluar 63×/1.4 oil objective. Excitation wavelengths of 405, 488 and 514 nm were produced by an argon/krypton laser. For the experiment the C-dot–hydrogel was added on a glass slide.Scanning electron microscopy (SEM)
Scanning electron microscopy (SEM) images were recorded on a Quanta 200 SEM. For the SEM study, the dilute solution of gel materials was dried on a glass cover slip and gold coating was carried out.Fluorescence spectroscopy and detection of bacteria
A Varioskan 96-well plate reader was used for the fluorescence analysis. 50 μL of C-dot–hydrogel was placed in each well and gelation was carried out. After 24 hours, fluorescence measurement and titration with different bacterial stains at 108 cells per mL bacterial concentrations were carried out using λex = 375 nm at 37 °C by addition of 15 μL aqueous bacterial suspension. Quenching efficiencies (Q. E.) of the C-dot–hydrogel hybrid were calculated by the Stern–Volmer equation:in which I0 and I refer to the fluorescence intensities of the system (λem = 430 nm) in the absence and presence of bacterial suspensions, respectively.
Thermal and chemical treatments on B. cereus
The aqueous suspensions of 108 cells per mL bacterial cells were heated to 100 °C and then cooled to room temperature. From this suspension, 15 μL bacteria were added to the C-dot–hydrogel to observe the change in fluorescence intensity (430 nm emission wavelength at 37 °C, where λex: 375 nm).To determine the contribution of divalent metal ions to the enzymatic activity of B. cereus, 15 μL of 5 mM EDTA solution was added together with 15 μL of aqueous bacterial suspension to the C-dot–hydrogel.
Rheology measurements
Rheology experiments were carried out using an Advanced Rheometer AR 2000 (TA Instruments) by cone and plate geometry in a Peltier plate. The cone diameter was 40 mm, cone angle 4°, and truncation 104 μm. The rheology analysis was performed by changing the shear rate (at 37 °C).NMR experiments
NMR studies were carried out on a Brüker DPX 400 MHz spectrometer at 300 K. Compound concentrations were 10 mm in methanol-d4. Chemical changes were monitored through incubation of suspensions of Bacillus cereus with hydrogel precursor 1 for 30 minutes. The reaction product was lyophilized for 2 h, and 1H and 13C NMR experiments were carried out without further purification.FT-IR analyses
All the FT-IR measurements were performed on a Thermo Scientific Nicolet 6700 spectrometer. The organic compound before and after addition of bacteria were identified by using this technique. The extraction of the organic compound was similar like the NMR experiments.Results and discussion
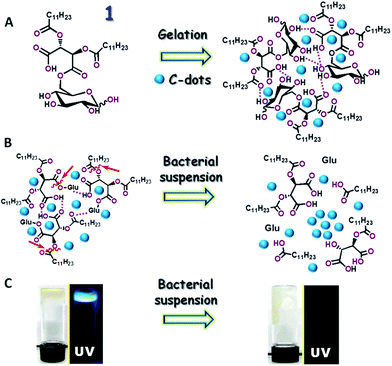
Fig. 1 depicts the preparation scheme of the C-dot–hydrogel hybrid and the enzyme-mediated bacterial detection concept. The hybrid hydrogel was assembled through mixing the precursor molecule 6-O-(O-O′-dilauroyltartaryl)-D-glucose and amphiphilic C-dots in water (Fig. 1A). Electron microscopy analysis confirmed the crystalline structure of the graphitic cores of the C-dots (Fig. S1, ESI†). The C-dots/precursor solution was gently heated until a homogeneous mixture was formed, followed by gelation which occurred at room temperature. Importantly, in the initial gel state prior to addition of bacterial suspensions (Fig. 1B, left), the embedded amphiphilic C-dots were dispersed within the hydrogel host matrix due to affinity to amphiphilic and hydrophobic domains within the internal porous surface. The hydrogel-embedded C-dots exhibited enhanced fluorescence as they were not aggregated, and thus fluorescence quenching was blocked.34,35However, the hydrogel network was disrupted upon addition of bacterial suspensions through ester bond cleavage by bacterially-secreted esterases (enzymatic cleavage sites are indicated by the arrows in Fig. 1B). The collapse of the hydrogel network gave rise to aggregation of the embedded amphiphilic C-dots and concomitant quenching of their fluorescence emission (Fig. 1B, right). The visual and fluorescence photographs in Fig. 1C depict the bacterially-induced transformation of the hybrid C-dot–hydrogel. As shown in Fig. 1C, left, the as-prepared dense gelatinous substance was fluorescent (λex: 375 nm) due to the embedded dispersed C-dots. Following incubation with a bacterial suspension, the gel material became more fluid and non-fluorescent (Fig. 1C, right).
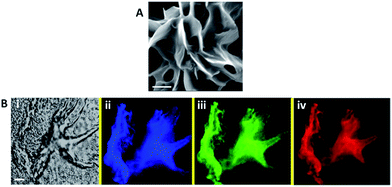
Fig. 2 presents microscopic characterization of the C-dot–hydrogel composite. The representative scanning electron microscopy (SEM) image in Fig. 2A reveals a highly porous sheet morphology of the hydrogel displaying considerable internal surface area. The fluorescence microscopy data in Fig. 2B confirm encapsulation of abundant C-dots within the hydrogel matrix. Specifically, the confocal fluorescence microscopy images in Fig. 2B reveal a highly fluorescent hydrogel scaffold due to the C-dots bound to the hydrogel surface. In particular, the different colors of the hydrogel in Fig. 2B reflect the excitation-dependent fluorescence emissions of the embedded C-dots.21,22 The excitation-dependent emission spectra of the hydrogel-embedded C-dots was distinctive from soluble C-dots (see ESI, Fig. S2†), confirming their encapsulation within the hydrogel scaffold.
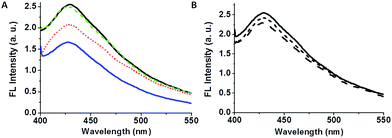
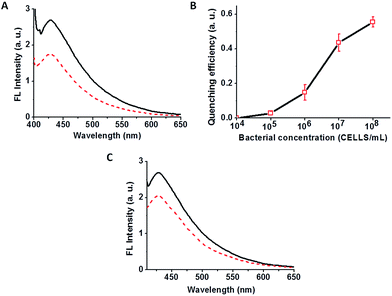
We examined the effect of bacterial suspensions upon the fluorescence properties of the C-dot–hydrogel. Fig. 3 shows the fluorescence modulation (λex = 375 nm) induced by addition of Bacillus cereus (cell concentration 108 cells per mL) to the C-dot–hydrogel. Fig. 3A reveals significant reduction of the fluorescence emission recorded 15 minutes after incubating the composite gel with the bacteria (concentration of 108 cells per mL) at 37 °C. The direct relationship between the degree of fluorescence quenching and bacterial concentration is depicted in Fig. 3B. Indeed, the dose–response curve in Fig. 3B points to a detection threshold of around 105 cells per mL reflecting excellent sensitivity of the C-dot–hydrogel platform.
While the data presented in Fig. 3A and B were recorded upon incubation of bacterial cell suspensions with the C-dot–hydrogels, we examined whether the fluorescence quenching effect was related to molecules secreted by the bacteria. Indeed, Fig. 3C reveals that the fluorescence quenching effect could be effectively induced by the supernatant (i.e. after discarding the cell precipitate), indicating that molecules secreted by the bacterial cells were responsible for fluorescence modulation. No quenching was recorded upon mixing of bacteria and soluble C-dots (data not shown), confirming that the quenching phenomenon was due to hydrogel disruption rather than direct interactions between the C-dots and bacterial cells.
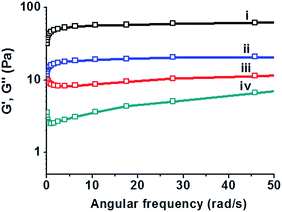
To account for the bacterially-induced modulation of hydrogel-embedded C-dots' fluorescence we carried out experiments aimed at characterization of the mechanical gel properties before and after addition of the B. cereus suspension (Fig. 4). Specifically, Fig. 4 depicts rheology analysis of the hydrogel before and after addition of B. cereus (108 cells per mL) at 37 °C. The data in Fig. 4 present frequency sweep experiments in which the storage modulus (G′) and loss modulus (G′′) were measured, reflecting the gel-phase of the material in which G′ is greater than G′′.39 The curves in Fig. 4ii and iv demonstrate that both G′ and G′′ significantly decreased after addition of the bacterial suspensions, reflecting reduction of the hydrogel's visco-elasticity (e.g. greater fluidity). The difference between G′ and G′′ was retained, however, pointing to retention of the intrinsic gel organization of the C-dot–hydrogel in the frequency range measured.
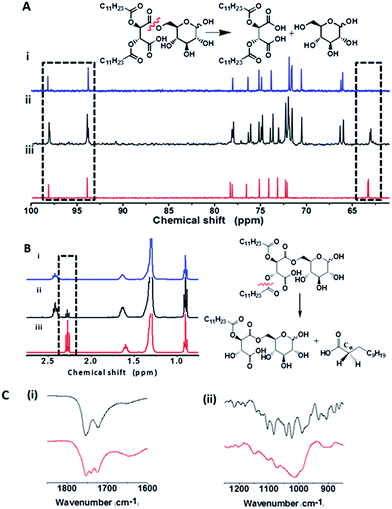
Nuclear magnetic resonance (NMR) and Fourier transform infrared (FTIR) spectroscopy experiments in Fig. 5 were designed to elucidate the molecular factors responsible for the bacterially-induced fluidization of the C-dot–hydrogel. The 13C and 1H NMR data in Fig. 5A and B reveal that the molecules secreted by B. cereus induced distinct chemical transformations within the hydrogel scaffold. Specifically, the 13C NMR spectra in Fig. 5A demonstrate the occurrence of ester bond cleavage following addition of the B. cereus suspensions, resulting on release of glucose molecules. This is reflected in the appearance of a new downfield 13C peak at 63 ppm in the B. cereus-treated sample (Fig. 6Aii) corresponding to the C6 atom of free glucose. Similarly, 13C signals corresponding to C1α and C1β carbons of free glucose appear at 94 and 98 ppm, respectively, following incubation of the hydrogel precursor with the B. cereus suspension. While the 13C spectral analysis indicates cleavage of the ester bonds linking the glucose moieties to the precursor hydrophobic backbone (Fig. 1), the 1H NMR experiments presented in Fig. 5B provide evidence for bacterially-induced hydrolysis of the ester bonds linking the lauryl side chains (Fig. 1B). Indeed, inspection of Fig. 5B reveals generation of free lauric acid, reflected in the appearance of the Cα-attached hydrogen at 2.25 ppm following addition of the bacteria.
The fluorescence data, mechanical, and spectroscopic analyses in Fig. 3–5 suggest that ester-hydrolyzing enzymes, specifically esterases and lipases secreted by B. cereus are responsible for the structural and luminescent transformations observed in the C-dot–hydrogel system.40,41 Esterases constitute a diverse enzyme family secreted by both mammalian and bacterial cells, which function through hydrolysis of ester bonds in target substrates.42 Notably, the hydrogel scaffold contains a multitude of ester bonds, both positioned in polar environments (e.g. ester bonds linking the glucose residues to the modified tartaric acid-based anhydride moieties, Fig. 1A) which serve as cleavage sites for bacterially-secreted esterases, as well as ester bonds in non-polar regions of the hydrogel (e.g. between the acyl chains and the central anhydride, Fig. 1A), constituting substrates for lipases, considered as esterase subfamily which functions in non-polar lipidic environments.42
Fig. 6 presents experiments designed to provide evidence for the assignment of bacterially-secreted esterases and lipases as inducers for hydrogel reorganization and fluorescence quenching of the embedded C-dots. Fig. 6A highlights the effects of heat and ethylenediaminetetraacetic-acid (EDTA) upon fluorescence quenching. Specifically, the fluorescence emission data in Fig. 6A demonstrate that heating the B. cereus supernatant to 100 °C essentially cancelled the fluorescence quenching effect (long-dash spectrum in Fig. 6A). This result is ascribed to heat-induced denaturation of bacterially-secreted proteins, thereby eliminating enzymatic digestion of the hydrogel scaffold.
Fig. 6A also shows that addition of EDTA to the bacterial suspension reduced fluorescence quenching (short-dash spectrum in Fig. 6A); EDTA binds divalent ions such as Ca2+, which are cofactors that are necessary to lipase functioning,43,44 thus inhibiting enzymatic cleavage of the hydrogel ester bonds. It should be noted that esterases generally do not require metal cofactors for activity thus the residual quenching still recorded after EDTA addition (Fig. 6A) might provide an indication as to the relative contribution of esterases vs. lipases to the observed C-dot–hydrogel fluorescence quenching.45,46
Fig. 6B depicts quenching of the C-dot–hydrogel fluorescence following addition of commercially-obtained esterase extracted from B. subtilis (B. subtilis belongs to the same subgroup of Bacillus species as B. cereus).47 The fluorescence quenching apparent in Fig. 6B both confirms the role of esterases in hydrogel disruption, as well as the fact that the bacterially-induced quenching phenomena reported here involve contributions from both esterases and lipases.
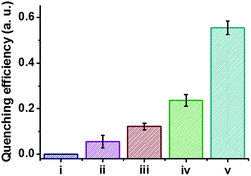
The C-dot–hydrogel fluorescence quenching assay was applied for analysis of several bacterial species (Fig. 7). Specifically, the graph depicted in Fig. 7 illustrates the degree of fluorescence quenching induced by selected bacteria (108 cells per mL) added to the C-dot–hydrogel. The significant differences in quenching magnitudes highlighted in Fig. 7 likely reflect the variability of esterase/lipase secretions by the bacteria tested.48,49 Importantly, while some esterase and lipase enzymes secreted by bacteria have been identified, the complexity and diversity of these enzyme families have been recognized and manifested in many studies;40,49 in this context, the C-dot–hydrogel fluorescence assay provides a simple platform for evaluating the overall secretion of these enzymes by bacteria. Moreover, the distinct quenching degrees could be employed for distinguishing and identification of different bacteria.
Conclusions
We have developed a new hybrid carbon-dot (C-dot)–hydrogel system capable of detecting bacteria through quenching of the C-dots fluorescence. Bacterial sensing was demonstrated in case of Bacillus and Staphylococcus strains, although the detection concept is generic and might be implemented for other bacterial species. The sensing mechanism is based upon cleavage of ester bonds within the hydrogel scaffold, induced by esterases and lipases secreted by the bacteria. Cleavage of the ester bonds disrupted the hydrogel framework, resulting in fluidization and aggregation and quenched fluorescence of the embedded C-dots. A series of physical and chemical experiments provide evidence for the gel transformation and reaction mechanism. The C-dot–hydrogel matrix enabled both high-sensitivity detection of bacteria (B. cereus) as well as distinguishing among different bacteria through the extents of fluorescence quenching.References
- N. M. Sangeetha and U. Maitra, Chem. Soc. Rev., 2005, 34, 821–836 RSC.
- R. V. Ulijn, N. Bibi, V. Jayawarna, P. D. Thornton, S. J. Todd, R. J. Mart, A. M. Smith and J. E. Gough, Mater. Today, 2007, 10, 40–48 CrossRef CAS.
- T. R. Hoare and D. S. Kohane, Polymer, 2008, 49, 1993–2007 CrossRef CAS.
- Z. Yang, G. Liang and B. Xu, Acc. Chem. Res., 2008, 41, 315–326 CrossRef CAS PubMed.
- L. Yu and J. Ding, Chem. Soc. Rev., 2008, 37, 1473–1481 RSC.
- A. Guiseppi-Elie, Biomaterials, 2010, 31, 2701–2716 CrossRef CAS PubMed.
- C. Aimé and T. Coradin, J. Polym. Sci., Part B: Polym. Phys., 2012, 50, 669–680 CrossRef.
- P. P. Joshi, S. A. Merchant, Y. Wang and D. W. Schmidtke, Anal. Chem., 2005, 77, 3183–3188 CrossRef CAS PubMed.
- J. H. Kim, S. Y. Lim, D. H. Nam, J. Ryu, S. H. Ku and C. B. Park, Biosens. Bioelectron., 2011, 26, 1860–1865 CrossRef CAS PubMed.
- Z. Zhu, C. Wu, H. Liu, Y. Zou, X. Zhang, H. Kang, C. J. Yang and W. Tan, Angew. Chem., 2010, 122, 1070–1074 CrossRef.
- N. Massad-Ivanir, G. Shtenberg, T. Zeidman and E. Segal, Adv. Funct. Mater., 2010, 20, 2269–2277 CrossRef CAS.
- I. Katakis and A. Heller, Anal. Chem., 1992, 64, 1008–1013 CrossRef CAS PubMed.
- A. F. Revzin, K. Sirkar, A. Simonian and M. V. Pishko, Sens. Actuators, B, 2002, 81, 359–368 CrossRef CAS.
- R. M. Unruh, J. R. Roberts, S. P. Nichols, S. Gamsey, N. A. Wisniewski and M. J. McShane, J. Diabetes Sci. Technol., 2015, 9, 985–992 CrossRef PubMed.
- K. Gawel, D. Barriet, M. Sletmoen and B. T. Stokke, Sensors, 2010, 10, 4381 CrossRef CAS PubMed.
- R. Wang and Y. Li, Biosens. Bioelectron., 2013, 42, 148–155 CrossRef CAS PubMed.
- X. Xu, R. Ray, Y. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS PubMed.
- Y.-P. Sun, B. Zhou, Y. Lin, W. Wang, K. A. S. Fernando, P. Pathak, M. J. Meziani, B. A. Harruff, X. Wang, H. Wang, P. G. Luo, H. Yang, M. E. Kose, B. Chen, L. M. Veca and S.-Y. Xie, J. Am. Chem. Soc., 2006, 128, 7756–7757 CrossRef CAS PubMed.
- S.-T. Yang, L. Cao, P. G. Luo, F. Lu, X. Wang, H. Wang, M. J. Meziani, Y. Liu, G. Qi and Y.-P. Sun, J. Am. Chem. Soc., 2009, 131, 11308–11309 CrossRef CAS PubMed.
- A. B. Bourlinos, R. Zbořil, J. Petr, A. Bakandritsos, M. Krysmann and E. P. Giannelis, Chem. Mater., 2012, 24, 6–8 CrossRef CAS.
- L. Cao, M. J. Meziani, S. Sahu and Y.-P. Sun, Acc. Chem. Res., 2013, 46, 171–180 CrossRef CAS PubMed.
- H. Nie, M. Li, Q. Li, S. Liang, Y. Tan, L. Sheng, W. Shi and S. X.-A. Zhang, Chem. Mater., 2014, 26, 3104–3112 CrossRef CAS.
- S. Y. Lim, W. Shen and Z. Gao, Chem. Soc. Rev., 2015, 44, 362–381 RSC.
- L. Pan, S. Sun, A. Zhang, K. Jiang, L. Zhang, C. Dong, Q. Huang, A. Wu and H. Lin, Adv. Mater., 2015, 27, 7782–7787 CrossRef CAS PubMed.
- L. Cao, X. Wang, M. J. Meziani, F. Lu, H. Wang, P. G. Luo, Y. Lin, B. A. Harruff, L. M. Veca, D. Murray, S.-Y. Xie and Y.-P. Sun, J. Am. Chem. Soc., 2007, 129, 11318–11319 CrossRef CAS PubMed.
- S. K. Bhunia, A. R. Maity, S. Nandi, D. Stepensky and R. Jelinek, ChemBioChem, 2016, 17, 614–619 CrossRef CAS PubMed.
- W. Wei, C. Xu, J. Ren, B. Xu and X. Qu, Chem. Commun., 2012, 48, 1284–1286 RSC.
- A. Zhu, Q. Qu, X. Shao, B. Kong and Y. Tian, Angew. Chem., Int. Ed., 2012, 51, 7185–7189 CrossRef CAS PubMed.
- A. Barati, M. Shamsipur and H. Abdollahi, Biosens. Bioelectron., 2015, 71, 470–475 CrossRef CAS PubMed.
- W. Shi, Q. Wang, Y. Long, Z. Cheng, S. Chen, H. Zheng and Y. Huang, Chem. Commun., 2011, 47, 6695–6697 RSC.
- E. Ju, Z. Liu, Y. Du, Y. Tao, J. Ren and X. Qu, ACS Nano, 2014, 8, 6014–6023 CrossRef CAS PubMed.
- Y. Song, S. Zhu, S. Xiang, X. Zhao, J. Zhang, H. Zhang, Y. Fu and B. Yang, Nanoscale, 2014, 6, 4676–4682 RSC.
- S. Nandi, M. Ritenberg and R. Jelinek, Analyst, 2015, 140, 4232–4237 RSC.
- A. Cayuela, S. R. Kennedy, M. L. Soriano, C. D. Jones, M. Valcarcel and J. W. Steed, Chem. Sci., 2015, 6, 6139–6146 RSC.
- N. Gogoi, M. Barooah, G. Majumdar and D. Chowdhury, ACS Appl. Mater. Interfaces, 2015, 7, 3058–3067 CAS.
- S. Nandi, H.-J. Altenbach, B. Jakob, K. Lange, R. Ihizane, M. P. Schneider, Ü. Gün and A. Mayer, Org. Lett., 2012, 14, 3826–3829 CrossRef CAS PubMed.
- S. Nandi, R. Malishev, K. Parambath Kootery, Y. Mirsky, S. Kolusheva and R. Jelinek, Chem. Commun., 2014, 50, 10299–10302 RSC.
- S. Melamed Yerushalmi, M. E. Buck, D. M. Lynn, N. G. Lemcoff and M. M. Meijler, Chem. Commun., 2013, 49, 5177–5179 RSC.
- J. Nanda, A. Biswas and A. Banerjee, Soft Matter, 2013, 9, 4198–4208 RSC.
- F. Hasan, A. A. Shah and A. Hameed, Biotechnol. Adv., 2009, 27, 782–798 CrossRef CAS PubMed.
- I. R. Montella, R. Schama and D. Valle, Mem. Inst. Oswaldo Cruz, 2012, 107, 437–449 CrossRef CAS PubMed.
- J. Skjold-Jørgensen, J. Vind, A. Svendsen and M. J. Bjerrum, Biochemistry, 2016, 55, 146–156 CrossRef PubMed.
- I. Posner and A. Morales, J. Biol. Chem., 1972, 247, 2255–2265 CAS.
- M. M. Momsen, M. Dahim and H. L. Brockman, Biochemistry, 1997, 36, 10073–10081 CrossRef CAS PubMed.
- U. T. Bornscheuer, FEMS Microbiol. Rev., 2002, 26, 73–81 CrossRef CAS PubMed.
- L. F. Godinho, C. R. Reis, P. G. Tepper, G. J. Poelarends and W. J. Quax, Appl. Environ. Microbiol., 2011, 77, 6094–6099 CrossRef CAS PubMed.
- O. A. Økstad, I. Hegna, T. Lindbäck, A.-L. Rishovd and A.-B. Kolstø, Microbiology, 1999, 145, 621–631 CrossRef PubMed.
- K.-E. Jaeger and T. Eggert, Curr. Opin. Biotechnol., 2002, 13, 390–397 CrossRef CAS PubMed.
- R. Gupta, N. Gupta and P. Rathi, Appl. Microbiol. Biotechnol., 2004, 64, 763–781 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Photoluminescence spectra of C-dots in gel and in solution (Fig. S1). See DOI: 10.1039/c6ra25148j |
| ‡ Present address: Ruhr Universitat Bochum, Universitaetsstrasse 150, D-44780, Bochum, Germany. |
| This journal is © The Royal Society of Chemistry 2017 |

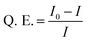
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) spectral region; (ii) ester and carboxylic acid (–C–O) region.
O) spectral region; (ii) ester and carboxylic acid (–C–O) region.