Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The study of complexation between dicationic surfactants and the DNA duplex using structural and spectroscopic methods†
W. Andrzejewskaa,
M. Wilkowskaa,
M. Chrabąszczewskaa and
M. Kozak *ab
*ab
aDepartment of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University, Umultowska 85, 61-614 Poznań, Poland. E-mail: mkozak@amu.edu.pl
bJoint Laboratory for SAXS Studies, Faculty of Physics, Adam Mickiewicz University, Umultowska 85, 61-614 Poznań, Poland
First published on 19th May 2017
Abstract
Dicationic (also known as gemini or dimeric) bis-alkylimidazolium surfactants belong to a group of non-viral transfection systems proposed for the successful introduction of different types of nucleic acids (i.e., siRNA, DNA oligomers, and plasmid DNA) into living cells. Our studies reveal the formation of complexes composed of dicationic (gemini) surfactants, 3,3′-[α,ω-(dioxaalkane)]bis(1-dodecylimidazolium)chlorides, and 21 base pair deoxyribonucleic acid duplexes (dsDNA). The studied dsDNA and its complex formation process was analysed by small-angle X-ray scattering (SAXS), molecular modelling (dsDNA), agarose gel electrophoresis (AGE) and circular dichroism spectroscopy (CD). We observed the formation of stable complexes for charge ratio values of p/n > 2. Moreover, we noted conformational changes similar to those observed during the transition of B-DNA to C-DNA, X-DNA, and Z-DNA in several spatial structures (i.e., micellar, hexagonal and cubic) formed in mixtures. The surfactants used in this study were investigated for the influence of dioxaalkane spacer length and the presence of an imidazolium moiety on the complexation process. The complexes formed were stable, and the complexation process was reproducible and efficient. Toxicity tests done on HeLa cells allowed for the determination of non-toxic concentrations of studied surfactants. Transfection tests have confirmed that the studied surfactant systems are effective DNA carriers.
Introduction
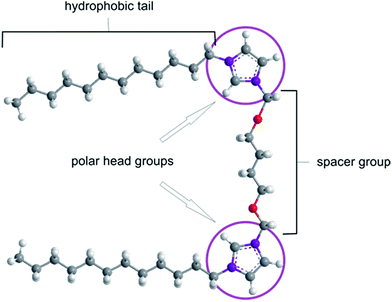
The efficient treatment of a broad spectrum of genetic diseases, cancer, and neurodegenerative diseases presents a great challenge for modern medicine. The diversity of the development and symptoms of various genetic diseases poses considerable problems for identifying satisfactory therapeutic methods.1–3 The recent development of gene therapy holds great promise for efficient treatment of these diseases. This therapeutic method relies on the delivery of a biologically active nucleotide sequence (i.e., a transgene) to target cells in order to block or eliminate pathological processes on the molecular level (for a review, see ref. 1–11). Gene therapy remains a major challenge due to the natural barriers possessed by living organisms.4 The main difficulties in implementing gene therapy are the successful introduction of therapeutic material into the cell, unloading the transgene from the carrier, carrier degradation, and specific transgene placement at the target site (e.g., tissue or cell).4,5 The natural ability of viruses to penetrate the cell membrane has led to the development of a broad range of viral vectors, but these vectors are limited with respect to the size of the plasmid that can be encapsulated.7,12 Non-viral gene delivery systems (i.e., gene carriers) based on synthetic gemini surfactants are non-toxic, non-immunogenic and show a high transfection efficiency comparable to that of viral carriers.4 The structure of these amphiphilic molecules is presented in Fig. 1. The spacer group within a gemini surfactant molecule exhibits variable chemical and geometric properties (i.e., rigid vs. flexible and polar vs. nonpolar).9,10Due to their specific construction, gemini surfactants exhibit unique properties. The high solubility, surface activity and unique physical properties of dicationic surfactants have led to their extensive study, including their application in gene therapy as a synthetic gene delivery vehicle.9–15 In therapeutic applications, the working concentration of a gemini surfactant in a gene delivery system is critical; it must be low enough so as not to induce cytotoxic effects but sufficient to create a lasting and stable carrier and to guarantee the release of the therapeutic material inside the cell.16–18 Ideally, the resultant surface charge of the complex is essentially neutral. This condition occurs when the maximum number of surfactants is complexed with a DNA molecule, resulting in a reduction of the negative charge of DNA via its electrostatic interaction with positively charged groups of the surfactant molecules. For gene therapy, the effective transfection systems typically are characterised by a charge ratio, p/n, of 1 to 10.19 Such structures exhibit the best transfection properties due to a lower tendency towards excessive aggregation than positive “transhipped” complexes.20 In addition, the micellization concentration (CMC) of a gemini surfactant is a critical parameter for determining the lowest concentration at which the formation of micelles is observed.9 Generally, the CMC for gemini surfactants is much lower (typically by more than 10-fold) than that of a monomeric surfactant due to the short-range interactions between free surfactant molecules, which increase self-aggregation.20 The process of DNA compaction by gemini surfactants has been examined in numerous studies. In one study, Wettig et al. assessed in vitro transfection using a series of m-s-m- and 12-s-12-type surfactants with the helper lipids DOPE and DMPC and calf thymus DNA.12 Transfection efficiency was found to depend on spacer group length.9,18,20 Furthermore, studies of the circular dichroism (CD) of plasmid–gemini surfactant systems demonstrated the induction of ψ-phase DNA, which is a tightly packed, condensed form of DNA.19,21 In 2013, Zhou et al. reported the in vitro transfection of human embryonic kidney 293 (HEK293) and HeLa cells using homogenous DNA/[C12-4-C12im]Br2 nanoparticles, showing that these complexes are efficient gene vectors.17
The goal of the present study was to fully characterise the interactions between short dsDNA (21 bp) and the gemini surfactants 3,3′-[α,ω-(dioxaalkane)]bis(1-dodecyllimidazolium)chlorides using small-angle X-ray scattering, molecular modelling (dsDNA), circular dichroism and electrophoresis. Additionally, the qualitative (morphological observation) and quantitative (the MTT assay) tests of the toxicity of the surfactants were performed on the model HeLa cell line.
Experimental
Materials and methods
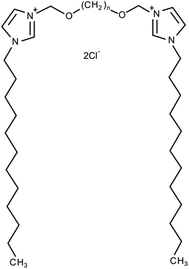 | ||
| Fig. 2 Chemical structure of 3,3′-[α,ω-(dioxaalkane)]bis(1-dodecyllimidazolium) chlorides (C12JCn, n = 2, 3, 4, 5, 6, 7, 8, 9, 10 and 12). | ||
The deoxyribonucleic acid duplex was 21 base pairs in length and was composed of the following sequence:
5′ GTTACGACAATCCTGTTTCTT 3′
3′ TTCAATGCTGTTAGGACAAAG 5′
These DNA oligomers were synthesised by Future Synthesis (Poznan, Poland) and were dissolved in 10 mM phosphate buffer, pH 7.0, for all experiments.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) of stock dsDNA solution with diluted surfactant solutions (concentrations: 10 mM, 8.4 mM, 6.3 mM, 4.2 mM, 3.15 mM, 2.1 mM, 1.58 mM, 1.05 mM, 0.525 mM and 0.273 mM). These surfactant concentrations were chosen to maintain the appropriate p/n (the gemini surfactant to DNA charge ratio). Final lipoplexes were characterised by the following p/n values: 5, 4, 3, 2, 1.5, 1, 0.75, 0.5, 0.25 and 0.125.
1) of stock dsDNA solution with diluted surfactant solutions (concentrations: 10 mM, 8.4 mM, 6.3 mM, 4.2 mM, 3.15 mM, 2.1 mM, 1.58 mM, 1.05 mM, 0.525 mM and 0.273 mM). These surfactant concentrations were chosen to maintain the appropriate p/n (the gemini surfactant to DNA charge ratio). Final lipoplexes were characterised by the following p/n values: 5, 4, 3, 2, 1.5, 1, 0.75, 0.5, 0.25 and 0.125.To specify the charge ratio between gemini surfactants and DNA, we used p/n parameter (1), defined as the value of positively charged surfactant head groups (p) and the negative charges accumulated on the phosphate residues of DNA chains (n):
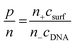 | (1) |
All steps were carried out at room temperature, and the complexation time was approximately 25 min. Before the preparation of lipoplexes, all solutions of dicationic surfactants were sonicated at 45 °C for 15 min to homogenise the solutions.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin
sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2θ/λ and wavelength λ = 0.154 nm). SAXS data were collected in 20 successive 100 ms frames. All measurements were taken in a capillary flow cell equipped with autosampling (sample volume 10 μL) at 15 °C. All recorded frames were compared for possible radiation damage, averaged, and then corrected for detector response and normalised to the incident beam intensity. The scattering pattern of the buffer was subtracted using the PRIMUS program from the ATSAS package.26
2θ/λ and wavelength λ = 0.154 nm). SAXS data were collected in 20 successive 100 ms frames. All measurements were taken in a capillary flow cell equipped with autosampling (sample volume 10 μL) at 15 °C. All recorded frames were compared for possible radiation damage, averaged, and then corrected for detector response and normalised to the incident beam intensity. The scattering pattern of the buffer was subtracted using the PRIMUS program from the ATSAS package.26Qualitative assessment. HeLa cells were cultured for experiments using a standard procedure in DMEM medium (Lonza Walkers Ville Inc., USA) supplemented with 10% FBS and appropriate antibiotic/antimycotic agents. To the cultured HeLa cells (confluence approx. 80%) 1 μL of the surfactant of the appropriate concentration was added to 99 μL DMEM medium (without FBS and antibiotics). Then, the cultures were incubated for one hour in an atmosphere of 5% CO2 at 37 °C. Morphological changes in the HeLa cells after one hour incubation were evaluated under a Zeiss Axiovert microscope (Carl Zeiss, Germany).
Quantitative assessment. Quantitative tests on previously prepared HeLa cells were performed using the MTT Cell Proliferation Assay Kit (Cayman Chemicals, USA).27 Absorption at a wavelength of 565 nm (MTT assay) was measured using a Tecan Infinite M200pro microplate reader (Männedorf, Switzerland).
Results and discussion
Complexation between C12JCn and dsDNA
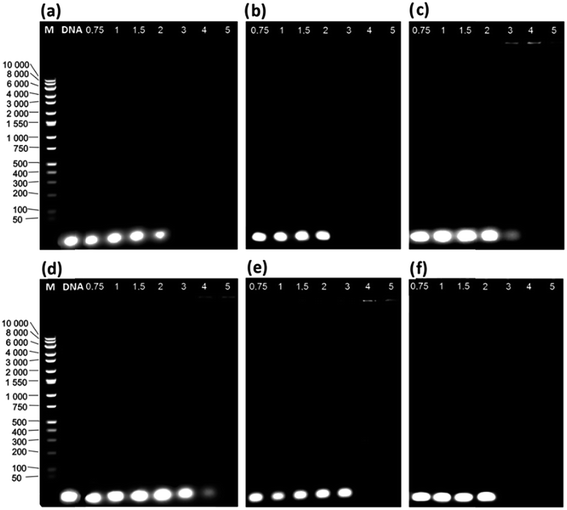
In the first stage of our study, we analysed the electrophoretic mobility of lipoplexes selected for our study of ten gemini surfactants. The agarose gel electrophoresis of the selected DNA/gemini surfactant lipoplexes is presented in Fig. 3. The creation of lipoplexes with the studied gemini surfactants is driven by the interaction of positively charged surfactant head groups, located on an imidazolium ring, and the negative charges accumulated on the phosphate residues of dsDNA chains. In the case of optimal complexation, the total charge of polyanions should be compensated for at p/n > 3, where we also observe the partial precipitation of solutions. These structures have a neutral character that is manifested by the total inhibition of the electrophoretic mobility of DNA in an electric field.11,17 As shown in Fig. 3 and S1 (ESI†), the mobility of dsDNA was reduced with increasing p/n charge ratios, indicating that C12JCn surfactants are able to effectively neutralise the dsDNA charge and participate in the complexation of nucleic acids. For gene therapy, such complexes are the most essential because high concentrations of surfactants may be toxic to cells. In some fully complexed systems, we did not observe any precipitation; moreover, at high surfactant concentrations, the total charge of the complexes was predominantly positive, as indicated by migration to the opposite side of the gel. The electrophoretic tests provide only a qualitative analysis of the complexation process and do not reveal any information regarding the structure of the analysed lipoplexes. Therefore, to better characterise the studied systems, we implemented SAXS studies and circular dichroism spectroscopy. We expected that the obtained DNA/C12JCn lipoplexes would create liposomal, lamellar or sponge-phase structures, as suggested by previous SAXS studies of similar systems.28–33Structural characterisation of dsDNA and C12JCn/dsDNA lipoplexes
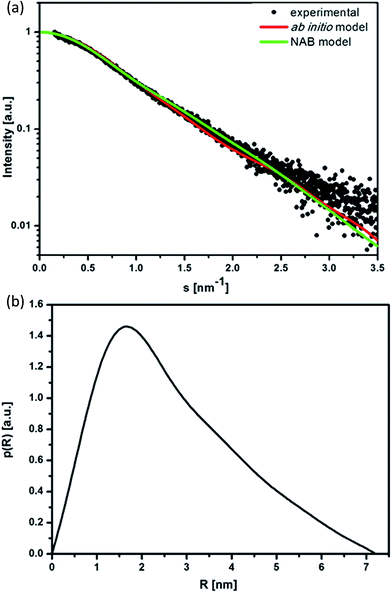
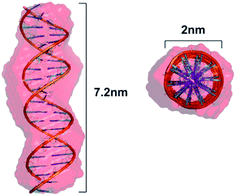
To obtain information on the structure of the studied initial dsDNA oligomer in solution and the microstructures of dsDNA complexes with gemini surfactants, we utilised the SAXS technique. We first performed shape determination studies for the dsDNA molecule in solution using the DAMMIN34 program from the ATSAS software package. Fig. 4a and b present the experimental scattering curve and the pair-distance distribution function p(R) calculated for the studied dsDNA oligomer. The maximum intramolecular distance (Dmax) calculated using the p(R) function was 7.2 nm, and the radius of gyration calculated by fitting to the Guinier equation was 2.19 nm. These structural data were used as initial parameters for shape determination studies, which were supplemented by molecular modelling of the studied DNA oligomer.Based on the nucleotide sequence of the studied dsDNA oligomer, we generated its structure in the NAB (Nucleic Acid Builder) function of the AMBER program.35 The model obtained is shown in Fig. 5. This model fits very well with the low-resolution model proposed by DAMMIN and is characterised by similar structural parameters, 7.2 nm in length and 2.2 nm in diameter, which correspond to the values obtained using the p(R) function for dsDNA of 21 base pairs in length. A direct comparison of the ab initio model with experimental data was performed using the program CRYSOL36 and exhibited a good fit represented by χ = 1.9. The fit of the theoretical scattering curve generated for a low-resolution model to experimental SAXS data is presented in Fig. 4a.
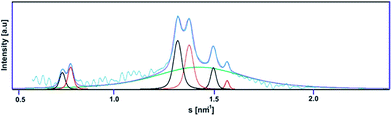
The next stage of structural analysis involved SAXS studies of lipoplexes prepared with gemini surfactants. The experimental SAXS data are summarised in Fig. 6 and S2 (ESI†), and it is evident from this comparison that complexes of C12JCn/dsDNA form a variety of spatial structures. The scattering data were deconvoluted using the PEAKFIT37 program, and the fit to the experimental data is presented in Fig. 7. The analysis of the diffraction peak positions and the probable structural phases is summarised in Table 1.
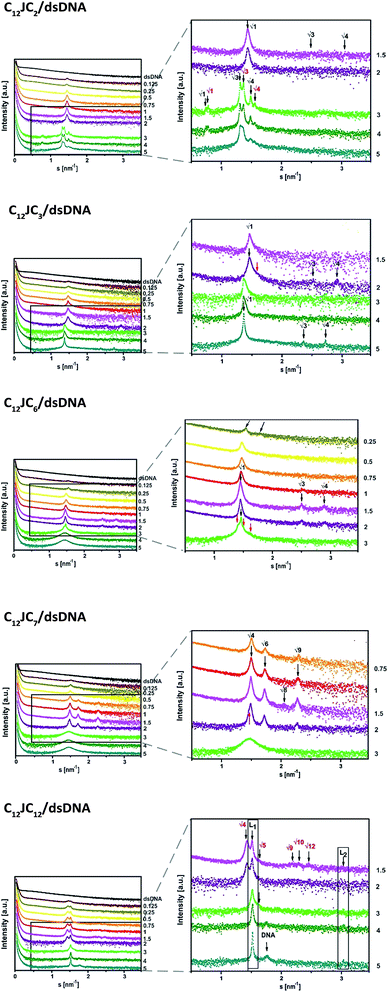 | ||
| Fig. 6 The SAXS patterns recorded for various lipoplexes. SAXS data were shifted for clarity. The p/n values are marked on the right side. The reciprocal spacings ratios are presented for each phase.38 | ||
| Surfactant | p/n | Observed phase | d-Spacing of the observed phase [nm] |
|---|---|---|---|
| C12JC2 | 0.1–1 | Micellar | 5.00 |
| 1.5–2 | Inverted hexagonal | 4.33 | |
| 3–5 | Double inverted hexagonal | 9.18 | |
| 9.67 | |||
| C12JCn, n = 3–4 | 0.1–1 | Micellar | 5.00 |
| 1.5–2 | Inverted hexagonal | 4.95 | |
| 3–5 | Inverted hexagonal | 5.26 | |
| C12JCn, n = 5–6 | 0.1–0.5 | Micellar | 5.00 |
| 0.75–3 | Inverted hexagonal | 4.96 | |
| 4–5 | Liposomes/sponge | 4.36 | |
| C12JCn, n = 7–10 | 0.5–0.1 | Micellar | 5.00 |
| 0.75–2 | Cubic | 8.38 | |
| 3–5 | Liposomes/sponge | 4.25 | |
| C12JC12 | 0.1–2 | Cubic | 8.80 |
| 3–5 | Lamellar | 4.08 |
The dsDNA oligomers can be surrounded by bilayers of gemini surfactant molecules or can be concentrically arranged inside the lipoplex (the hydrophobic tails are turned inward, whereas the hydrophilic heads are exposed to the solution). DNA lipoplexes can also form structures characterised by a higher ordering degree, such as a lamellar (LCI)-complex with DNA rods sandwiched between the gemini surfactant bilayers. In the hexagonal or inverted hexagonal (HCI/HCII) structures, DNA rods are coated with surfactant tubular micelles arranged on a hexagonal network12–14 (Fig. 8). Taking into account these properties, we observed several structural phases in all examined lipoplexes that often coexisted; we will divide them into five groups for discussion. For lipoplexes prepared using C12JC2 and increasing concentrations of amphiphilic compound, we observed the micellar phase (d001 = 5 nm), hexagonal phase (d10 = 4.33 nm) and what appeared to be two separate double-layered hexagonal (d10 = 9.18 nm and d10 = 9.67 nm) phases. The phenomenon of micro-separation of two very similar hexagonal phases occurring at the a certain p/n charge ratio can be explained as the result of different packing densities of the surfactant molecules in the presence of dsDNA.
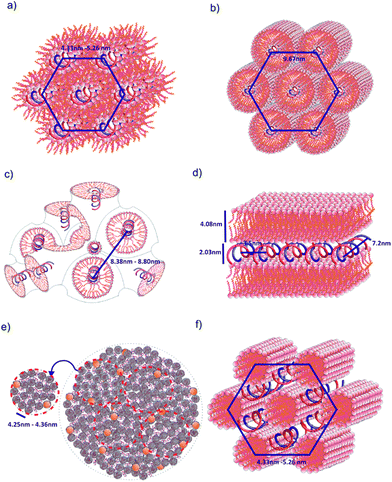 | ||
| Fig. 8 Models of spatial structures formed by C12JCn/dsDNA lipoplexes. (a) Hexagonal HCII phase, (b) double inverted hexagonal phase, (c) cubic Pn3m phase, (d) lamellar phase LCI, (e) liposomes/sponge phase, (f) hexagonal HCI phase. This representation is based on previously proposed models.13,14,28,29,40 | ||
The second group of studied lipoplexes contains systems prepared using C12JC3 and C12JC4 surfactants. With increasing concentrations of amphiphilic compound, we observed the formation of a micellar phase and then hexagonal phases: at p/n 1.5–2, characterised by basal spacing d10 = 4.95 nm (a0 = 5.73 nm), and at 3–5, characterised by d10 = 5.26 nm (a0 = 6.07 nm). For C12JC4 lipoplexes, we also observed an additional peak, which can be ascribed to stacked DNA (sDNA = 1.59 nm−1). For the next group of lipoplexes, prepared using C12JC5 and C12JC6 surfactants, the highest concentrations of surfactant (p/n 3–5) produced a liposomal-like or even sponge phase28–33 (Fig. 8e). This phase is characterised by d001 = 4.36 nm (Fig. 6c p/n = 3). With decreasing surfactant concentration, the inverted hexagonal phase (p/n = 0.75–3, d10 = 4.96 nm) and the micellar phase occurred (p/n = 0.1–0.5, d001 = 5 nm).
Another group of lipoplexes includes the systems prepared from four surfactants (C12JC7–C12JC10). In this group, we observed at the highest surfactant concentrations (p/n 3–5) the liposomal-like or sponge phase (d001 = 4.25 nm). At surfactant concentrations with a charge ratio p/n ranging from 0.75 to 2, a cubic structure11,33,39 with lattice parameter a0 = 8.38 nm is likely formed. At the highest concentrations of C12JC12 surfactant (p/n = 3–5), the lamellar phase is clearly visible, manifested by strong peaks at d001 = 4.08 nm and d002 = 2.03 nm. The cubic phase was also observed for C12JC12 lipoplexes as a secondary phase with a0 = 8.80 nm (p/n = 0.1–2).
When analysing the influence of individual surfactant geometry on the ability to form certain structural phases, it is important to recognize that the long and flexible spacer groups in the surfactant molecules can undergo compaction (self-coiling). Furthermore, there is no rule for binding surfactants to dsDNA to create specific spatial forms. For example, the hexagonal phase occurred both at medium and high charge ratio values (p/n), whereas the electrophoretic mobility is completely blocked only for the latter. In addition, the DNA oligomer is not necessarily bound inside the tubular micelles in the hexagonal phase but may also be present between cylinders (Fig. 8). The d-spacing parameter would then reach practically the same value. Such a situation has been predicted for monomeric cationic surfactants40 as well as cationic lipids.13,14
Conformational changes of dsDNA complexed with C12JCn surfactants
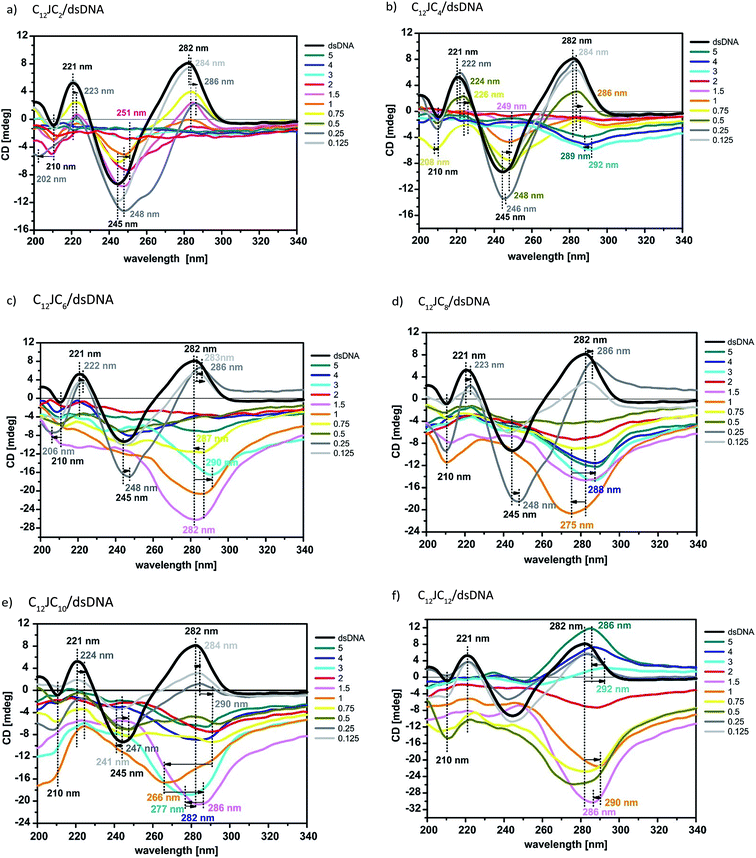
The next step in our analysis was to perform circular dichroism studies of a reference dsDNA oligomer and its lipoplexes with the studied dicationic surfactants. Due to the high turbidity of the studied solutions, circular dichroism measurements for DNA lipoplexes at high p/n values did not yield any CD signal. We therefore applied centrifugation prior to CD measurements. Additional electrophoresis testing of these samples showed identical electrophoretic mobility of samples without centrifugation, indicating that the centrifugation process does not impact the stability of the complexes. The obtained CD data are presented in Fig. 9 and S3 (ESI†). The circular dichroism data obtained for the reference solution dsDNA oligomer exhibit a characteristic positive peak at approximately 282 nm and a minimum at approximately 245 nm. The similar intensity of both bands and the opposite sign and symmetry of the CD curve with respect to the intersecting point (261 nm) attest to its semi-conserved character. These features indicate that the duplex in solution assumes the form of B-DNA, which is the most typical form of the double-stranded native, fully hydrated DNA.41 Moreover, the solutions of surfactants alone does not absorb differently left and right circularly polarized light (no circular dichroism signal).CD spectra were measured for solutions of the DNA/C12JCn lipoplexes freshly prepared from the ten tested surfactants and dsDNA, indicating significant changes in the conformation of the DNA oligomer depending on the length of the spacer and the concentration of the amphiphilic molecule studied. With the extension of the linker group, we observed a progressive deformation of the CD spectra of the DNA duplex. This process can be attributed to both changes throughout the helix and local interference in the geometry of nucleobases.42–45 According to previously reported CD analyses of DNA under different conditions,13,21,46–61 the studied C12JCn/DNA lipoplexes may induce conformational changes typical of the B-DNA conformation to the geometry observed for the C-DNA, X-DNA and Z-DNA forms. These changes are discussed in detail later. It should also be emphasised that the contribution to the recorded CD signal was only from chromophores derived from nucleic acid because surfactant molecules themselves do not participate in CD signal.42,43
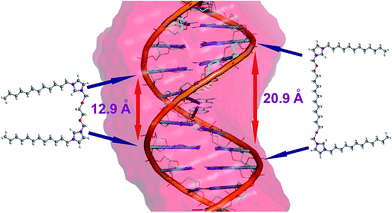
At low concentrations, all tested surfactants (charge ratios p/n = 0.125 and 0.25) caused fluctuations in the intensity of the whole CD spectrum as well as a slight (1–2 nm) shift of the positive band towards longer wavelengths. The reason for these changes is the influence of electrostatic interactions between the positively charged imidazolium groups of the surfactant and negatively charged phosphate groups of dsDNA. Consequently, the oligomers are surrounded by the surfactant molecules, with the partial exposure of surfactant hydrophobic chains towards the solvent. Such a binding pattern results in changes in the solvation shell of the nucleic acid molecule and changes the polarity of the local environment of nucleobases on a somewhat smaller scale than previously noted.43 The hydrophobic chains of surfactants are combined, in the later stage of the self-assembly process, with further surfactant molecules via hydrophobic interactions until the thermodynamic stabilisation of the whole system is reached. According to SAXS studies at these concentrations, the surfactants tend to form micelles (in fact, all concentrations of surfactants used in this study were greater than the CMC value). Electrophoretic tests indicate that in these systems, however, dsDNA is not permanently complexed because it retains some electrophoretic mobility. For systems with p/n = 0.125 and 0.25, we also observed an increase in the intensity and a broadening of the negative CD band at 245 nm for gemini surfactants with a shorter spacer group (i.e., surfactants from C12JC2 to C12JC9). These characteristics may arise because molecules with shorter spacer groups more easily and effectively interact with dsDNA. These surfactants may even intercalate between the small and the large groove of the DNA oligomer (Fig. 10) such that conformational changes of the duplex are induced and are apparent in the CD spectrum.
As noted above, dehydration decreases the number of base pairs on a single turn of the DNA double helix.62 On the basis of the geometry of such a system, a single molecule of dsDNA may be covered by more molecules of surfactant with a short spacer group (C12JC2) than with the long spacer (C12JC12). Therefore, the water molecules will have also a more difficult access to the DNA molecule. The local disorder of the hydration shell of dsDNA at these charge ratios (p/n) will thus diminish with an increasing length of the linker group of the surfactant, which is evident in the CD spectra.
All these regularities illustrate the structural changes of the dsDNA oligomer to the geometry similar to the C-DNA form.63–65 However, the obtained CD spectrum suggests the existence in solution of only certain forms of the intermediates similar to C-DNA, but we did not observe a typical curve of the C-DNA form. According to structural studies, mainly involving the X-ray diffraction of oligonucleotide crystals and solution and solid-state NMR,50,51,62,63,66 this form is characterised by the reduced helix radius in relation to the classic B-DNA helix. This form was observed under relatively low humidity and has 8.8–9.7 base pairs per turn.62 The deviation of the base pairs' plane from the axis of the C-DNA helix is approximately −8°,52 and the DNA molecule is elongated. However, in C-DNA form, the fundamental geometric parameters of the helix (the conformation of the deoxyribose position C2′-endo conformation and the anti-torsion angle χ of the glycoside bond) are not changed in relation to the B-DNA. In addition, the decrease in the intensity of the positive band on the CD spectrum in the region of 280 nm is generally proportional to the reduction in the number of base pairs per turn of the helix.51 This condition leads to a decrease in the interaction of the chromophore with its closest neighbours and an increased interaction with 2nd and 3rd closest neighbours, which is manifested mainly in the form of the abolition of positive bands in the region of 280 nm, and an increase in the intensity of the negative band at approximately 245 nm. As a result, the contribution to the resultant dipole moment produces exciton-type transitions π → π* between a nucleobase and pentose in the chain of dsDNA.42,43 The interaction of polar groups of surfactants with DNA induces conformational effects similar to the counter-ion influence (e.g., Mg2+, Li+) and results in the transition of nucleotides to the BI and BII conformations.53,58,62,66 It is assumed, however, that this structure is still a conformational variant of B-DNA because the majority of nucleotides are in the BI conformation.58
Similar changes in the CD spectrum were also observed in the case of the interaction of the plasmid DNA with cationic lipids (with a charge ratio p/n = 2), which are interpreted as the formation of the ψ-DNA form.54 Additionally, other studies 21,67 have shown a putative connection between ψ-DNA and C-DNA forms. For DNA from calf thymus (3000–4000 bp) as well as from E. coli, the C-DNA form in the solution containing Li+ cations was observed, which is likely a transitional form in the transformation to the ψ-DNA form. Similar results were obtained for calf thymus DNA complexed with polyhistidine.47 In the case of the studied dsDNA oligomer, this molecule is too short (only 21 bp) to adopt a conformation similar to the ψ-DNA conformation.47–49,67 With increasing concentrations of surfactants in the investigated systems (up to a value of p/n = 5), a significant deformation of the CD spectra is observed. Based on the results of SAXS and gel electrophoresis, it appears advisable to analyse them separately, i.e., first, the lipoplexes with p/n from 0.5 to 2 and then the systems characterised by p/n from 3 to 5. It is clear that the concentration of gemini surfactants at approximately p/n = 2 is a type of critical value for phase transitions in these solutions. The CD spectra reveal a flattening of the curve (even signal decay) for short linker groups (C12JC2–C12JC6) or changes in the shape of the curve showing the behaviour of dsDNA conformational changes that have taken place at lower concentrations of surfactant (C12JC7–C12JC12).
At values of p/n = 0.5–2 of surfactants from C12JC5 to C12JC12, a reversed CD spectrum above 260 nm is observed. The rest of the spectrum exhibits a disturbance in the form of a flattened or reduced intensity. This cause of this behaviour is unclear. In addition to the inversion of the CD spectrum in most cases, the resulting signal decreases to a minimum as a function of wavelength. The reason for this effect is likely a disorder of the geometry of the dsDNA molecule, which results in a change in the geometry of the helix. However, it is not known how these changes affect the interaction with the nearest neighbours. For the C12JC2, C12JC3 and C12JC4 surfactants, a reduction in the intensity of positive and negative bands was observed in the circular dichroism spectrum. Typically, the inversion of CD spectra shows an inversion of helicity,42,43 whereas at a partial signal inversion, this effect cannot be explicitly confirmed. Similar CD studies were performed for various DNA oligomers (TA)n in the presence of monovalent cations (CsCl and CsF).55–57 The nucleic acid takes the form of X-DNA, which is characterised by an increased degree of condensation compared to B-DNA, because of the extreme propeller-twist and the bend of nucleotides (adenine and thymine), whereas the conformation of the deoxyribose (C2-endo) and the “anti” conformation of torsion angle χ of the glycosidic bond are preserved. This condition is the reason for the formation of the negative band near 280 nm and the absence of significant changes in the CD spectrum below 260 nm. The X-DNA form is unstable in the presence of cytosine and guanine in the polynucleotide chain and in situations when the chain is extended over eight TA repeats.55 It is possible that the DNA duplex examined in this study, which contains 31.8% CG pairs, exhibits only an incomplete transition to the X-DNA form or to the other related structural forms because of the described instability. The studied systems included no caesium salts; however, certain similarities of the CD spectra were obtained in these studies. Additionally, the SAXS results show that lipoplexes of these surfactants with the dsDNA form hexagonal phases (C12JC2–C12JC6) and phases of cubic symmetry (C12JC7–C12JC12), and the packing state in these phases can directly affect the oligomer conformation.
In contrast to the low concentrations of surfactants (e.g., p/n = 0.125, 0.25) in the discussed lipoplexes characterised by p/n ratios = 0.5–2, the surfactants with a longer spacer group caused greater changes in the conformation of dsDNA. Surfactants C12JC5–C12JC12 more significantly deformed the CD spectrum even at p/n = 0.5. The reason for this behaviour may be the self-organisation of amphiphilic molecules in highly symmetric structures (hexagonal or cubic) or the self-coiling process of the surfactant spacer group, leading to the formation of additional steric hindrance during the association of surfactant molecules with DNA. In these concentrations of gemini surfactants for these lipoplexes, we also did not observe full DNA complexation, as the electrophoretic mobility of DNA did not cease. Only in the case of the C12JC2 surfactant did we observe a significant decrease in the intensity of the electrophoretic band at p/n = 2. Gemini surfactants in aqueous solutions are able to form spatial structures in solution (e.g., micelles) without the presence of DNA.10 These structures likely bind acid only partially, which is sufficient for the production of diffraction peaks.
The highest concentration of surfactants used resulted in the most desirable effect on the formation of a stable complex with the oligomer dsDNA. SAXS studies indicated the presence of a hexagonal structure in solution (C12JC2–C12JC4) as well as liposomal or sponge phases (C12JC5–C12JC10) and the lamellar structure (C12JC12). The CD signal for these mixtures can be divided into 3 groups. Surfactants with a short spacer group (C12JC2–C12JC4) are likely flattened; the results of electrophoresis indicate complete binding of the oligomer surfactants. The CD signal can also be disrupted by the micro-separation of newly formed phases.59
At the medium spacer group length (surfactants C12JC5–C12JC10), we observed a significant conformational deformation of circular dichroism curves, similar to that observed for lipoplexes at p/n = 0.5–2. In this case, however, with increasing length of the spacer group, the CD signal shows a strong minimum and moves significantly towards the longer wavelengths of up to 293 nm. However, these changes are similar to the effects associated with the transition of B-DNA to Z-DNA rather than X-DNA.47–49,67 These two DNA forms, however, are closely related, resulting in similarly shaped CD spectra. Crystallographic studies have shown that both forms contain well-characterised Hoogsteen's pairs.56,60,61,68–70 Z-DNA is a left-handed form of DNA, which may explain the inversion of the spectrum, and is characterised by a CD spectrum minimum at approximately 295 nm, likely due to perpendicular low-energy n → π* transitions between successive conjugated oxygen atoms of the carbonyl group in protonated cytosine. Moreover, it is thought that Hoogsteen DNA may be a transitional phase between B-DNA and Z-DNA, where nucleotides (A) are reversed so that the carbonyl group assumes a conformation antitypical of Z-DNA.60 Part of the CD signal below 260 nm no longer has characteristics similar to the spectrum of Z-DNA. This behaviour may be explained by a decrease in stacking interactions caused by the additional protonation of cytosine N3 and by stretching of the double helix.44,70 This type of protonation destabilises Z-DNA.60 For C12JC12 lipoplexes at a charge ratio of p/n = 3–5, we noted a very interesting effect, unique to this type of amphiphilic molecule. The positive band in the CD spectrum at approximately 282 nm was shifted towards the longer wavelengths and slowly disappeared; then, with increasing surfactant concentration, it grew in intensity and shifted towards shorter wavelengths. These changes indicate a probable inversion of helicity back to a right-handed form and disorder of interaction within the closest few neighbours.
The results of circular dichroism do not allow us to clearly state that the tested system has undergone a complete conversion in the conformation of native B-DNA to C-DNA, X-DNA or Z-DNA. However, we believe that the observed deformation of CD spectra is the result of changes induced by gemini surfactants in the studied dsDNA oligomer to the conformations observed for these forms of DNA. Gemini surfactants, as well as other factors, may cause transitions between different DNA conformational forms by altering properties such as monovalent cations, ionic strength, pH or humidity. In general, structural differences are induced in DNA due mainly to the adjustment of the spatial structure of native DNA to a conformation required for its biological function.56,60,61,70
Toxicity of the surfactants to HeLa cells
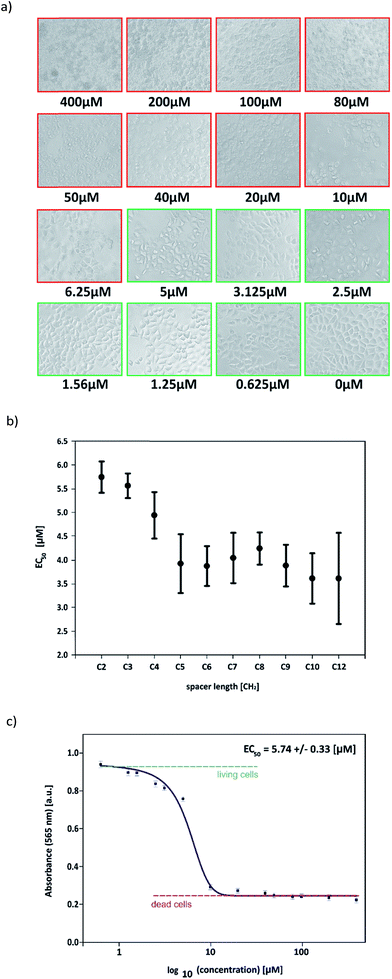
Evaluation of the impact of the cytotoxic effect of a series of dicationic surfactants, differing in the length of the linker, on the culture HeLa tumor cells included the qualitative and quantitative tests. Differences in the C12JCn surfactant toxicity were observed associated with increasing length of the alkyl chain in the linker. Microscopic observations carried out on HeLa cells, after incubation under defined conditions, showed significant morphological changes in the cell number in cultures to which a surfactant had been in different concentrations (Fig. 11a). Microscopic observations did not show any significant morphological changes in the cell number for all tested surfactants in concentration from 0.625 μM to 2.5 μM. At higher concentrations (6.25–3.125 μM), a tendency to slower the growth rate of HeLa cells and a small morphological changes of the cells were observed, although the magnitude of the changes was different for different surfactants. The cells tolerance towards C12JCn surfactants with shorter spacer groups was greater. The concentrations of surfactant in the range from 6.25 μM to 80 μM resulted in significant morphological changes in the cells, such as appearing granularity, and the low proliferative potential. For very high concentrations of surfactants, in the range of 80 μM to 400 μM complete destruction of cell membranes was noted, which means an extremely high mortality rate in the HeLa cultures tested.During the next stages of the experiment, the quantitative assessment of cytotoxicity of the studied gemini surfactants was performed. A colorimetric MTT assay was chosen. This assay based on capturing the MTT molecule by the cells studied which is then reduced to the formazan molecule by intracellular oxidoreductases NAD(P)H. This assay allows the measurement of the activity of mitochondrial energy conversion processes, and thus the number of living cells (metabolically active) in a tested population. In this assay the effective concentration (EC50), corresponding to the concentration of the tested compound in which proliferation of cells cultured in vitro is inhibited by 50% relative to control culture is determined.
MTT tests showed that the presence of C12JCn surfactants in high concentrations in the culture medium has a great influence on the number of living cells. As a consequence, different values of the EC50 parameter for the surfactants tested were noted (Table 2). The increasing length of alkyl chain in the spacer group resulted in decreasing of the effective concentration of surfactant affecting the biological activity of cells (see Fig. 11b and c).
| EC50 [μM] | ΔEC50 [μM] | |
|---|---|---|
| C12JC2 | 5.74 | 0.33 |
| C12JC3 | 5.56 | 0.26 |
| C12JC4 | 4.94 | 0.49 |
| C12JC5 | 3.92 | 0.62 |
| C12JC6 | 3.87 | 0.42 |
| C12JC7 | 4.04 | 0.53 |
| C12JC8 | 4.24 | 0.34 |
| C12JC9 | 3.88 | 0.44 |
| C12JC10 | 3.61 | 0.53 |
| C12JC12 | 3.61 | 0.96 |
Transfection tests
Considering the results of electrophoretic tests, for the transfection test were selected, as the most optimal, the complexes characterized by p/n = 2. The results obtained show the various efficacy of penetration of the test DNA into fibroblast cells. Two of the complexes tested, based on C12JC10 and C12JC12 surfactants, resulted in complete degradation of the cells at selected concentrations. These results agree with previous surfactant toxicity tests on HeLa cells and average cell survival in the surfactant environment (exemplary fluorescent image is presented in Fig. 12), other tests are presented in and Fig. S4 and S5 (ESI†). For the other surfactants tested, we observed effective transfection into fibroblast cells. In conclusion, most of the tested DNA complexes with selected surfactants exhibit good transfection properties at p/n = 2. It is also worth noting, that we have previously characterized the interaction of selected C12JCn surfactants71,72 with bovine serum albumin (BSA), which is a component of FBS used in transfection tests.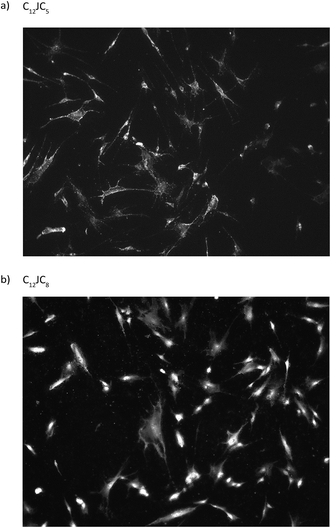 | ||
| Fig. 12 Exemplary photos from fluorescent microscope of fibroblast cultures transfected by the use of fluorescently labelled ssDNA oligomer and dicationic surfactants C12JC5 (a) and C12JC8 (b). | ||
Conclusions
Based on the results of gel electrophoresis, SAXS and CD studies, we determined that stable complexes of dsDNA oligomers and gemini surfactants of the C12JCn type are formed at charge ratio (p/n) values greater than 2. We found that the length of the spacer group influences this process; the best complexing surfactants possessed a short spacer group (C12JC4) or a very flexible one (C12JC12). However, this relationship was not constant and was dependent on the geometry of the surfactant molecule. The length of the spacer group is critical for the sustainable oligomer binding by the studied dicationic surfactants because this geometry corresponded most closely to the structural parameters set by the large and small grooves of dsDNA. Considering the probability of creating specific spatial structures in the studied mixtures of surfactants and dsDNA, we must also take into account the possibility of self-organisation of both the components themselves (i.e., the micellization of surfactants) and the studied lipoplexes. As a result, lipoplexes that are hexagonal (both single and double layered), lamellar and liposomal/sponge type should be tested. Furthermore, the analysis of circular dichroism spectra supported the conclusion that conformational changes of oligomer dsDNA in surfactant solutions resulted in the formation of DNA forms resembling the geometry of C-DNA, X-DNA and Z-DNA. Because of their geometry, the dsDNA molecules are unlikely to resemble the structure of ψ-DNA. Toxicity tests have shown that the concentration of surfactants used are safe for the HeLa cell after one hour incubation. Finally, the transfection tests on the fibroblasts cultures have also confirmed, that the studied surfactant systems can be used as effective DNA carriers.Acknowledgements
This research project has been financed by the funds from the National Science Centre (Poland) granted on the basis of decision no. DEC-2011/01/B/ST5/00846. We thank the P12 beamline25 of the EMBL Hamburg Outstation at DESY (Hamburg, Germany) for beamtime allocation and particularly the beamline staff for technical support. SAXS measurements, performed on P12 Beamline of EMBL Hamburg Outstation on PETRA III storage ring at DESY have received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under BioStruct-X (grant agreement no. 283570).References
- E. C. Lattime and S. L. Gerson, Gene Therapy of Cancer, Elsevier, 3rd edn, 2013 Search PubMed.
- M. Giacca, Gene Therapy, Springer, 2010 Search PubMed.
- D. V. Schaffer and W. Zhou, Gene Therapy and Gene Delivery Systems, Springer, 2010 Search PubMed.
- S. Szala, Terapia genowa, PWN, 2003 Search PubMed.
- A. Józkowicz and J. Dulak, BioTechnologia, 2007, 3, 7 Search PubMed.
- C. Amoruso, T. Lagache and D. Holcman, SIAM J. Appl. Math., 2011, 71, 2334 CrossRef CAS.
- S. D. Patil, D. G. Rhodes and D. J. Burgess, AAPS J., 2005, 7, E61 CrossRef CAS PubMed.
- P. C. Bell, M. Bergsma, I. P. Dolbnya, W. Bras, M. C. A. Stuart, A. E. Rowan, M. C. Feiters and J. B. F. N. Engberts, J. Am. Chem. Soc., 2003, 125, 1551 CrossRef CAS PubMed.
- S. M. C. Silva, J. J. S. Sousa, E. F. Marques, A. A. C. C. Pais and B. B. Michniak-Kohn, AAPS J., 2013, 15, 1119 CrossRef CAS PubMed.
- R. Zieliński, Surfaktanty: budowa, właściwości, zastosowania, Poznan University of Economics Press, 2009 Search PubMed.
- V. D. Sharma, J. Lees, N. E. Hoffman, E. Brailoiu, M. Madesh, S. L. Wunder and M. A. Ilies, Mol. Pharm., 2014, 11, 545 CrossRef CAS PubMed.
- S. D. Wettig, R. E. Verrall and M. Foldvari, Curr. Gene Ther., 2008, 8, 9 CrossRef CAS PubMed.
- B. Ma, S. Zhang, H. Jiang, B. Zhao and H. Lv, J. Controlled Release, 2007, 123, 184 CrossRef CAS PubMed.
- K. K. Ewert, H. M. Evans, A. Zidovska, N. F. Bouxsein, A. Ahmad and C. R. Safinya, J. Am. Chem. Soc., 2006, 128, 3998 CrossRef CAS PubMed.
- N. Dan, Biophys. J., 1997, 73, 1842 CrossRef CAS PubMed.
- T. Piskorska and E. Obłak, Postepy. Hig. Med. Dosw., 2010, 64, 161 Search PubMed.
- T. Zhou, A. Llizo, P. Li, C. Wang, Y. Guo, M. Ao, L. Bai, C. Wang, Y. Yang and G. Xu, J. Phys. Chem. C, 2013, 117, 26573 CAS.
- S. D. Wettig, I. Badea, M. Donkuru, R. E. Verrall and M. J. Foldvari, Genet. Med., 2007, 9, 649 CAS.
- I. Badea, Gemini cationic surfactant-based delivery systems for non-invasive cutaneous gene therapy, Ph. D. thesis, Univ. of Sask., Saskatoon, 2006.
- L. Karlsson, M. C. P. van Eijk and O. J. Söderman, J. Colloid Interface Sci., 2002, 252, 290 CrossRef CAS PubMed.
- N. J. Zuidam, Y. Barenholz and A. Minsky, FEBS Lett., 1999, 457, 419 CrossRef CAS PubMed.
- W. Andrzejewska, Z. Pietralik, M. Taube, A. Skrzypczak and M. Kozak, Polimery, 2014, 59, 569 CrossRef CAS.
- V. N. Emel'yanenko, S. V. Portnova, S. P. Verevkin, A. Skrzypczak and T. J. Schubert, Chem. Thermodyn., 2011, 43, 1500 CrossRef.
- Z. Pietralik, M. Taube, A. Skrzypczak and M. Kozak, Acta Phys. Pol., A, 2010, 117, 311 CrossRef CAS.
- C. E. Blanchet, A. Spilotros, F. Schwemmer, M. A. Graewert, A. G. Kikhney, C. M. Jeffries, D. Franke, D. Mark, R. Zengerle, F. Cipriani, S. Fiedler, M. Roessle and D. I. Svergun, J. Appl. Crystallogr., 2015, 48, 431 CrossRef CAS PubMed.
- P. V. Konarev, V. V. Volkov, A. V. Sokolova, M. H. J. Koch and D. I. Svergun, J. Appl. Crystallogr., 2003, 36, 1277 CrossRef CAS.
- T. J. Mosmann, Immunol. Methods, 1983, 65, 55 CrossRef CAS PubMed.
- S. Ristori, L. Ciani, G. Candiani, C. Battistini, A. Frati, I. Grillo and M. In, Soft Matter, 2011, 8, 749 RSC.
- S. Falsini, S. Ristori, L. Ciani, E. D. Cola, C. T. Supuran, A. Arcangeli and M. In, Soft Matter, 2014, 10, 2226 RSC.
- M. Kleman, Pramana, 1999, 53, 107 CrossRef CAS.
- S. H. Bhansali, A. S. Malik, J. M. Jarvis, I. Akartuna, D. M. Dabbs, J. D. Carbeck and I. A. Aksay, Langmuir, 2006, 22, 4060 CrossRef CAS PubMed.
- A. Angelova, Prog. Colloid Polym. Sci., 2011, 138, 1 CAS.
- B. Angelov, A. Angelova, R. Mutafchieva, S. Lesieur, U. Vainio, V. M. Garamus, G. V. Jensen and J. S. Pedersen, Phys. Chem. Chem. Phys., 2011, 13, 3073 RSC.
- M. V. Petoukhov, D. Franke, A. V. Shkumatov, G. Tria, A. G. Kikhney, M. Gajda, C. Gorba, H. D. T. Mertens, P. V. Konarev and D. I. Svergun, J. Appl. Crystallogr., 2012, 45, 342 CAS.
- D. A. Case, V. Babin, J. T. Berryman, R. M. Betz, Q. Cai, D. S. Cerutti, T. E. Cheatham, III, T. A. Darden, R. E. Duke, H. Gohlke, A. W. Goetz, S. Gusarov, N. Homeyer, P. Janowski, J. Kaus, I. Kolossváry, A. Kovalenko, T. S. Lee, S. LeGrand, T. Luchko, R. Luo, B. Madej, K. M. Merz, F. Paesani, D. R. Roe, A. Roitberg, C. Sagui, R. Salomon-Ferrer, G. Seabra, C. L. Simmerling, W. Smith, J. Swails, R. C. Walker, J. Wang, R. M. Wolf, X. Wu and P. A. Kollman, AMBER 14, University of California, San Francisco, 2014 Search PubMed.
- D. Svergun, C. Barberato and M. H. J. Koch, J. Appl. Crystallogr., 1995, 28, 768 CrossRef CAS.
- http://www.systatsoftware.com.
- D. Marsh, Handbook of Lipid Bilayers, CRC Press, Taylor & Francis Group, 2nd edn, 2013 Search PubMed.
- D. McLoughlin, M. Impéror-Clerc and D. Langevin, ChemPhysChem, 2004, 5, 1619 CrossRef CAS PubMed.
- A. Mezei, R. Pons and M. C. Morán, Colloids Surf., B, 2013, 111, 663 CrossRef CAS.
- Z. Pietralik, R. Krzysztoń, W. Kida, W. Andrzejewska and M. Kozak, Int. J. Mol. Sci., 2013, 14, 7642 CrossRef PubMed.
- M. Boczkowska, Wiad. Chem., 2002, 56, 39 CAS.
- M. Boczkowska, Wiad. Chem., 2002, 56, 203 CAS.
- B. Wanik and M. Lidwin, Zagadnienia Biofizyki Współczesnej, 1976, 1, 39 Search PubMed.
- P. Crabbé, An Introduction to the Chiroptical Methods in Chemistry, Impr. Offsali-G, 1971 Search PubMed.
- T. Zhou, G. Xu, M. Ao, Y. Yang and C. Wang, Colloids Surf., B, 2012, 414, 33 CrossRef CAS.
- G. Burchkardt, G. Zimmer and G. Luck, Nucleic Acids Res., 1976, 3, 561 CrossRef CAS PubMed.
- C. Bombelli, F. Faggioli, P. Luciani, G. Mancini and M. G. Sacco, J. Med. Chem., 2005, 48, 5378 CrossRef CAS PubMed.
- S. M. Cheng and S. C. Mohr, FEBS Lett., 1974, 49, 37 CrossRef CAS PubMed.
- N. J. Rhodes, A. Mahendrasingam, W. J. Pigram, W. Fuller, J. Brahms, J. Vergne and R. A. J. Warren, Nature, 1982, 296, 267 CrossRef CAS PubMed.
- W. A. Baase and W. C. Johnson, Nucleic Acids Res., 1979, 6, 797 CrossRef CAS PubMed.
- W. Sanger, Principles of Nucleic Acids Structure, Springer, 1984 Search PubMed.
- A. V. Fratini, M. L. Kopka, H. R. Drew and R. E. J. Dickerson, J. Biol. Chem., 1982, 257, 14686 CAS.
- D. Simberg, D. Danino, Y. Talmon, A. Minsky, M. E. Ferrari, C. J. Wheeler and Y. J. Barenholz, J. Biol. Chem., 2001, 276, 47453 CrossRef CAS PubMed.
- J. Kypr, J. Chladkova, L. Arnold, J. Sagi, A. Szemzo and M. J. Vorlickova, J. Biomol. Struct. Dyn., 1996, 13, 999 CAS.
- J. Kypr, I. Kejnovská, D. Renčiuk and M. Vorlíčková, Nucleic Acids Res., 2009, 37, 1713 CrossRef CAS PubMed.
- M. Vorlíčková, I. Kejnovská, K. Bednářová, D. Renčiuk and J. Kypr, Chirality, 2012, 24, 691 CrossRef PubMed.
- L. van Dam and M. H. Levitt, J. Mol. Biol., 2000, 304, 541 CrossRef CAS PubMed.
- L. Xu, L. Feng, R. Dong, J. Hao and S. Dong, Biomacromolecules, 2013, 14, 2781 CrossRef CAS PubMed.
- N. G. A. Abrescia, A. Thompson, T. Huynh-Dinh and J. A. Subirana, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 2806 CrossRef CAS PubMed.
- E. Cubero, F. J. Luque and M. Orozco, Biophys. J., 2006, 90, 1000 CrossRef CAS PubMed.
- J. Portugal and J. A. Subirana, EMBO J., 1985, 4, 2403 CAS.
- D. A. Marvin, M. Spencer, M. H. F. Wilkins and L. D. Hamilton, J. Mol. Biol., 1961, 3, 547 CrossRef CAS PubMed.
- M. J. B. Tunis-Schneider and M. F. Maestre, J. Mol. Biol., 1970, 52, 521 CrossRef CAS PubMed.
- D. A. Marvin, M. Spencer, M. H. F. Wilkins and L. D. Hamilton, Nature, 1958, 182, 387 CrossRef CAS PubMed.
- I. I. Skuratovskii and V. N. Bartenev, Mol. Biol., 1978, 12, 1359–1376 CAS.
- W. C. Brunner and M. F. Maestre, Biopolymers, 1974, 13, 345 CrossRef CAS PubMed.
- A. H. Wang, R. V. Gessner, G. A. van der Marel, J. H. van Boom and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 1985, 82, 3611 CrossRef CAS.
- K. Hoogsteen, Acta Crystallogr., 1959, 12, 822 CrossRef CAS.
- J. Aishima, R. K. Gitti, J. E. Noah, H. H. Gan, T. Schlick and C. Wolberger, Nucleic Acids Res., 2002, 30, 5244 CrossRef CAS PubMed.
- W. Gospodarczyk, K. Szutkowski and M. Kozak, J. Phys. Chem. B, 2014, 118, 8652 CrossRef CAS PubMed.
- W. Gospodarczyk and M. Kozak, Colloid Polym. Sci., 2015, 293, 2855 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra24978g |
| This journal is © The Royal Society of Chemistry 2017 |