Black phosphorus nanoparticles as a novel fluorescent sensing platform for nucleic acid detection
Ying Teng
Yew
a,
Zdeněk
Sofer
 b,
Carmen C.
Mayorga-Martinez
a and
Martin
Pumera
b,
Carmen C.
Mayorga-Martinez
a and
Martin
Pumera
 *a
*a
aDivision of Chemistry and Biological Chemistry, School of Physical and Mathematical Sciences, Nanyang Technological University, 21 Nanyang Link, Singapore 637371, Singapore. E-mail: pumera.research@gmail.com
bDepartment of Inorganic Chemistry, University of Chemistry and Technology Prague, Technická 5, 166 28 Prague 6, Czech Republic
First published on 13th January 2017
Abstract
Two-dimensional black phosphorus is an emerging material. For the past two years, black phosphorus has attracted the attention of the scientific community due to its promising applications in electronic devices and biosensors. In this work, black phosphorus nanoparticles (BPNPs) derived from a facile, one-step synthesis method were successfully employed as nanofluorophores in the development of a fluorescent sensing platform for DNA detection, for the first time. This detection strategy exploits the differences in affinities of labelled single-stranded and double-stranded DNA oligonucleotides towards spontaneous self-assembly with black phosphorus nanoparticles. A wide linear detection range with good linearity (r = 0.91) spanning from 4 pM to 4000 pM was achieved with the constructed nanosensor, including the ability to distinguish triple nucleotide polymorphism. Moreover, high sensitivity was also demonstrated with a low limit of detection (LOD = 5.9 pM) and limit of quantification (LOQ = 19.7 pM). This paper demonstrates a proof of concept that black phosphorus possesses promising potential for biosensing applications, in particular as DNA nanobiosensors, and the reported findings would be significant in encouraging further research work into the development of biomedical devices and technologies based on black phosphorus.
Burgeoning demand for early medical diagnosis1–4 has prompted significant research into the development of rapid, cost-efficient, highly sensitive and target recognition specific biosensors in both research and commercial healthcare sectors. Notably, fluorescent deoxyribonucleic acid (DNA) sensing platforms have gathered much research interest due to the intrinsic chemical and physical stability of genosensors5 as well as the target specificity,6 sensitivity7,8 and ease of operation6,7 associated with fluorometric detection systems. Over the past decade, nanostructures have played an increasingly imperative role in the development of genosensors owing to their unique and extraordinary properties. Examples of such nanomaterials include semiconductor quantum dots,9 carbon10 and graphene quantum dots,11,12 gold nanoparticles,13 cobalt phosphide nanowires,14 metal–organic frameworks15 and polymers.16,17
Out of the large variety of available nanostructures, black phosphorus has been especially prominent following the recent isolation of phosphorene in 2014.18 In addition to being biocompatible19 and having low cytotoxicity,20 black phosphorus displays fascinating electrical, optical and electronic properties, such as anisotropic electrical conductivity and optical response,21,22 high carrier mobility20 and a wide tunable band gap,23,24 making it a promising novel nanomaterial for the development of optoelectronic devices,25,26 nanoelectronic devices18,27 and gas sensors.28–30 Despite these promising properties, exploration of the potential biomedical applications of black phosphorus remains limited at this stage.31,32 This lack of interest predominantly stems from the susceptibility of black phosphorus towards photobleaching when exposed to air or water,23 limiting its use in biomedical applications. However, recent literature has demonstrated the successful synthesis of air- and water-stable black phosphorus nanostructures,33–36 which can overcome this major limitation allowing for potential biomedical applications.
In this study, we present a proof of concept demonstrating the application of pristine black phosphorus nanoparticles (BPNPs) as fluorophores in the development of a novel fluorescence-based nucleic acid detection platform. BPNPs were synthesized from the red allotrope using a high pressure phase transformation, which is followed by share force milling in N,N-dimethylformamide (DMF). The results obtained in this study thus aim to act as a framework for the future exploration of potential biomedical applications that black phosphorus has to offer.
Experimental section
Materials
Red phosphorus (99.999%) was obtained from Sigma-Aldrich (Prague, Czech Republic). N,N-Dimethylformamide (DMF) was purchased from PENTA (Prague, Czech Republic). Tris(hydroxymethyl)aminomethane, sodium chloride and hydrochloric acid (37%, w/w) were purchased from Sigma-Aldrich (Singapore). Magnesium chloride hexahydrate was purchased from Quality Reagent Chemical (Auckland, New Zealand). Tris-HCl buffer was prepared by dissolving NaCl (150 mM) and MgCl2 (5 mM) in milli-Q water (18.2 MΩ cm), and was used for all fluorescence spectroscopy experiments. DNA sequences were procured from Sigma-Aldrich (Singapore). The oligonucleotide sequences are listed as follows:Dabcyl-Lprobe: 5′ [dabcyl] ACC AGG CGG CCG CAC ACG TCC TCC AT 3′;
Complementary DNA, cDNA: 5′ ATG GAG GAC GTG TGC GGC CGC CTG GT 3′;
Three-base mismatched DNA, mDNA-3: 5′ ATG GAG GAC GTG ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GGC CGC CTG GT 3′;
GGC CGC CTG GT 3′;
Non-complementary DNA, ncDNA: 5′ AAA AAA AAA AAA AAA AAA AAA AAA AA 3′.
Equipment
High-resolution transmission electron microscopy (HR-TEM) was performed with an EFTEM Jeol 2200 FS microscope (Jeol, Japan). An acceleration voltage of 200 keV was used for measurement. The sample was prepared by drop casting the suspension (1 mg mL−1 in water) on a TEM grid (Cu; 200 mesh; Formvar/carbon) and drying at 60 °C for 12 h.X-ray powder diffraction (XRD) data were collected at room temperature on a Bruker D8 Discoverer powder diffractometer with parafocusing Bragg–Brentano geometry using CuKα radiation (λ = 0.15418 nm, U = 40 kV, I = 40 mA). The data was scanned over the angular range of 10–80° (2θ) with a step size of 0.02° (2θ). Data evaluation was performed in the software package EVA.
X-ray photoelectron spectroscopy (XPS) was performed with an ESCAProbeP spectrometer (Omicron Nanotechnology Ltd, Taunusstein, Germany) using a monochromatic aluminium X-ray radiation source (1486.7 eV). The samples were prepared for XPS measurements by drop-casting the BPNPs on gold coated silicon wafers and dried in a vacuum oven (50 °C).
Raman spectroscopy spectrum was recorded using a CCD detector using an inVia Raman microscope (Renishaw, England), under backscattering geometry mode using DPSS laser (532 nm, 50 mW), through a 50× objective and using 2.5 mW laser power.
Photoluminescence spectroscopy scans were recorded at room temperature using a Varian Cary Eclipse fluorescence spectrophotometer. The λex value of the nucleic acid detection experiments involving BPNPs was fixed at 200 nm, and the fluorescence emission spectra were measured across the range of 200–900 nm. All measurements were conducted using a quartz glass cuvette.
Synthesis of BPNPs
Red phosphorus (10 g) was wrapped in graphite foil and placed in a high pressure/high temperature uniaxial pressing apparatus of 1′′ size. The sample was compressed at 6 GPa and heated at 600 °C for 30 min, at a rate of 100 °C min−1, then allowed to cool to room temperature at a rate of 100 °C min−1. The graphite foil was removed by polishing. Pulverization of the obtained black phosphorus in an agate mortar followed by sieving was carried out to separate particles with sizes below 0.5 mm. Black phosphorus powder was then dispersed in DMF by ultrasonication (9 mg mL−1; 400 W; 15 min). Milling using share force milling apparatus at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm was then performed in a glass jacketed vessel at 15 °C for 1 h, under an argon atmosphere.
000 rpm was then performed in a glass jacketed vessel at 15 °C for 1 h, under an argon atmosphere.
Preparation of a DNA biosensing system based on BPNPs as fluorophores
For assay development, cDNA prepared at various concentrations was added to 160 nM of dabcyl-Lprobe in 1 mL of Tris-HCl buffer. The mixture was heated at 50 °C for 30 min with continuous agitation at 400 rpm, then cooled to room temperature over 30 min. The solution mixture was then incubated with 75 μL of BPNPs for 20 min at room temperature. Spectroscopic analysis was performed using 1 mL of the prepared sample.For selectivity evaluation, 75 μL of BPNPs was incubated with 160 nM of dabcyl-Lprobe in 1 mL of Tris-HCl buffer for 20 min. 400 pM of the DNA of interest (cDNA, mDNA-3 or ncDNA) was added into the mixture, which was then heated at 50 °C for 30 min with continuous agitation at 250 rpm. Spectroscopic analysis was performed using 1 mL of the prepared sample upon cooling the sample mixture to room temperature over 30 min.
Results and discussion
Morphological and chemical characterization of BPNPs
In order to prepare BPNPs, black phosphorus crystals were first synthesized by the high pressure phase transformation of the red phosphorus allotrope. The obtained crystals were pulverized using an agate mortar, sieved to retain only the microparticles, and then milled by share force milling in DMF solvent to afford nanoparticles of approximately 100 nm in size. The synthesis protocol was adapted from a synthesis method that was previously reported.37Comprehensive characterization was then performed to analyze the structural and chemical properties of BPNPs. BPNP characterization was performed using transmission electron microscopy (TEM), X-ray powder diffraction (XRD), X-ray photoelectron spectroscopy (XPS), and Raman spectroscopy.
TEM was performed to study the structural morphology38 and crystallinity39 of the black phosphorus material. Few layered nanoparticles with lateral dimensions of approximately 100 nm are observed (Fig. 1A). The orthorhombic crystalline nature of the as-synthesized black phosphorus can be observed using high-resolution TEM (HR-TEM), as shown in Fig. 1B and is further confirmed by the selected area electron diffraction (SAED) pattern shown in the inset of Fig. 1A.
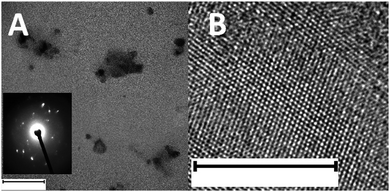 | ||
| Fig. 1 (A) TEM and (B) HR-TEM images of BPNPs. Inset: SAED pattern of BPNPs. Scale bar: (A) 100 nm and (B) 10 nm. | ||
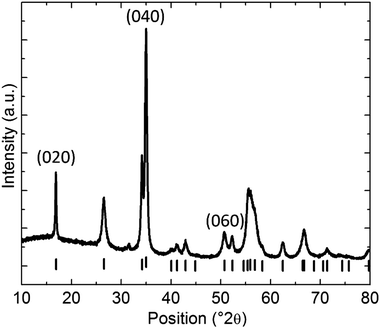
XRD was performed to study the crystal structure of the BPNPs.40 The obtained diffractogram further affirms the orthorhombic crystal structure of the BPNPs. The weak reflections present in the diffractogram are attributed to the (0k0) reflections (Fig. 2).28
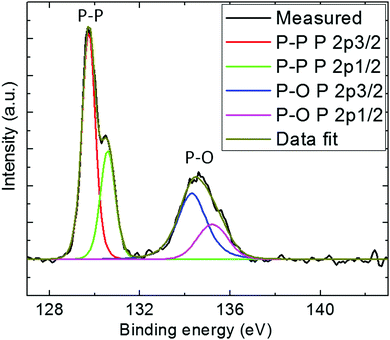
High-resolution XPS of P 2p was conducted to determine the types of chemical bonds present in the BPNPs (Fig. 3).41 Two peaks at 129.7 eV and 134.6 eV were observed, and are assigned to elemental P0 and oxidized P, respectively. The peak at 134.6 eV may be attributed to the partial surface oxidation of the BPNPs.37 Moreover, a predominant peak at 129.7 eV corresponding to 2p energy binding was also observed. The obtained high-resolution XPS spectrum is in good agreement with the literature.42,43
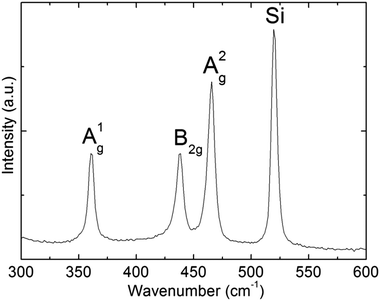
Further structural characterization of BPNPs was conducted using Raman spectroscopy (Fig. 4). The A1g, B2g and A2g optical photons characteristic of black phosphorus materials were observed at 360.7 cm−1, 438.6 cm−1 and 465.7 cm−1. The locations of the three Raman active modes correspond well with the literature values.44
Application of black phosphorus nanoparticles in fluorometric DNA analysis
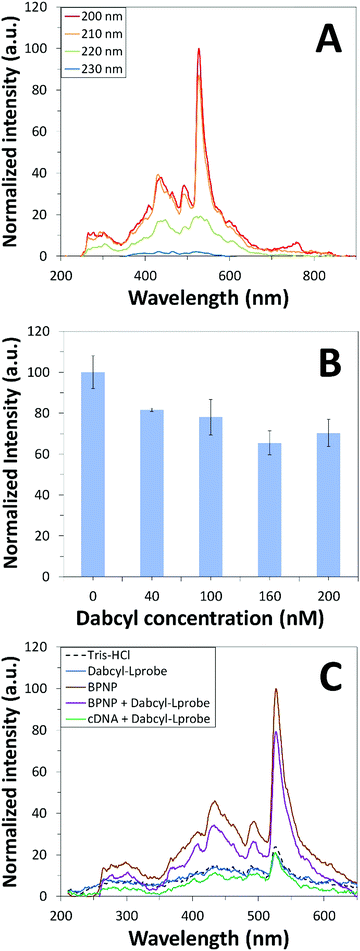
Fluorescence emission spectroscopy was performed to evaluate the optical properties of the BPNPs (Fig. 5A). A maximum PL emission was observed at 527 nm, corresponding to an excitation wavelength (λex) of 200 nm. This verifies the fluorescent nature of the BPNPs and supports their potential application as nanofluorophores in fluorescence-based biosensing platforms. BPNPs were excited at 200 nm for subsequent experiments in this study.Scheme 1 illustrates the schematic representation (not to scale) of a DNA biosensor system based on BPNPs as nanofluorophores: (A) dabcyl-Lprobe adsorbs onto the BPNP surface, likely via dispersive forces and hydrophobic interactions.45 The favoured adsorption process of single-stranded DNA onto the BPNPs results in photoluminescence (PL) quenching of the BPNPs by the dark quencher label, dabcyl. In (B), the initial addition of the cDNA target to dabcyl-Lprobe promotes the hybridization of both single-stranded oligonucleotide sequences, resulting in the formation of a double-stranded DNA complex (ds-cDNA-dabcyl-Lprobe) which is then incubated with the BPNP fluorophore. The ds-cDNA-dabcyl-Lprobe assembly has reduced affinity for BPNPs and does not adsorb onto the fluorophore surface, consequently retaining the PL signal of the BPNPs as opposed to (A).
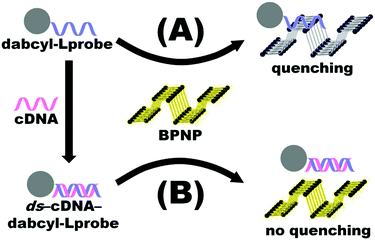 | ||
| Scheme 1 Schematic representation (not to scale) of the proposed DNA detection strategy based on BPNPs as fluorophores. | ||
BPNPs were then incubated with dabcyl-Lprobe to confirm the successful adsorption of the quencher molecules onto the fluorescent nanoparticle surface. The quencher concentration was optimized for a utilized volume of 75 μL BPNPs by recording changes in the fluorescence signal of the nanofluorophore upon exposure to increasing concentrations of dabcyl-Lprobe for 20 min (Fig. 5B). A gradual increase in the quenching efficiency of dabcyl on the BPNPs was observed with the incubation of higher concentrations of dabcyl-Lprobe, indicating successful fluorescence quenching of the BPNPs, which stabilizes at around 160 nM. Consequently, an optimum concentration of 160 nM was applied for subsequent fluorometric assays.
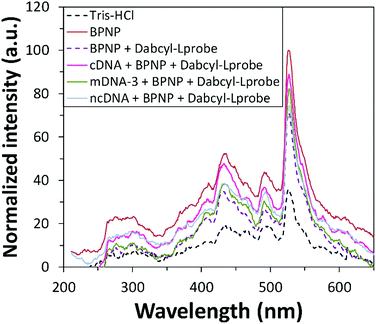
Prior to investigating the sensing performance of BPNPs for cDNA target detection, a control study was first conducted in order to investigate the effects of interference by the cDNA target on the detected fluorescence signal (Fig. 5C). No apparent fluorescence emission was observed by cDNA following hybrid formation with dabcyl-Lprobe (compare the blue line with the green line). Conversely, fluorescence quenching was evident following the adsorption of dabcyl-Lprobe onto BPNPs (compare the brown line with the purple line). This therefore confirms that the PL emission intensity is solely influenced by the BPNPs, and that cDNA has no direct effect on the detected PL intensity.
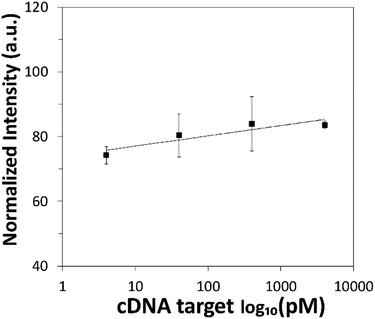
The sensing performance of the proposed DNA detection strategy was then evaluated over a range of cDNA target concentrations spanning across 4 pM to 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pM. The corresponding calibration graph based on the fluorescence intensity of BPNPs was plotted as a logarithmic function of cDNA concentration (Fig. 6). Higher fluorescence intensity of BPNPs was achieved with higher concentrations of cDNA, tapering off at 4000 pM. This corresponds to the formation of larger amounts of the ds-cDNA-dabcyl-Lprobe hybrid complex which exhibits poor physical adsorption onto BPNPs, thereby resulting in a lower extent of fluorescence quenching and giving rise to a greater extent of fluorescence intensity retention. The linear dynamic range of detection (r = 0.91) extends from less than 4 pM to 4000 pM. Low limits of detection (LOD) and quantification (LOQ) of 5.9 pM and 19.7 pM are attained, respectively.
000 pM. The corresponding calibration graph based on the fluorescence intensity of BPNPs was plotted as a logarithmic function of cDNA concentration (Fig. 6). Higher fluorescence intensity of BPNPs was achieved with higher concentrations of cDNA, tapering off at 4000 pM. This corresponds to the formation of larger amounts of the ds-cDNA-dabcyl-Lprobe hybrid complex which exhibits poor physical adsorption onto BPNPs, thereby resulting in a lower extent of fluorescence quenching and giving rise to a greater extent of fluorescence intensity retention. The linear dynamic range of detection (r = 0.91) extends from less than 4 pM to 4000 pM. Low limits of detection (LOD) and quantification (LOQ) of 5.9 pM and 19.7 pM are attained, respectively.
In order to evaluate the selectivity of the proposed DNA sensing platform, the fluorescence responses of BPNPs towards complementary (cDNA), three-base mismatched (mDNA-3) and non-complementary (ncDNA) nucleic acid sequences were examined and compared, following incubation with dabcyl-Lprobe for 20 min. Fluorescence recovery of up to 45% was achieved with the introduction of cDNA, as compared to only 37% and 27% in the presence of mDNA-3 and ncDNA, respectively (Fig. 7). The results obtained illustrate the ability of the detection assay to distinguish complementary DNA from a mismatched or non-complementary sequence, as well as the ability to discriminate multiple nucleotide length polymorphisms from the cDNA of interest.
Conclusions
Herein, for the first time, we report a proof of concept detailing the application of BPNPs as a novel nanomaterial in fluorescence-based nucleic acid detection. Employing BPNPs as fluorophores, the proposed detection assay was able to detect DNA concentrations with LOD and LOQ values as low as 5.9 pM and 19.7 pM, respectively, and demonstrated a large linear dynamic detection range from less than 4 pM to 4000 pM with good linearity (r = 0.91). The DNA probe was also capable of realizing triple nucleotide polymorphism determination; nevertheless further experimentation using different quencher systems may be beneficial towards improving the target specificity performance of the detection assay. The results obtained from this study extend the application of BPNPs to nucleic acid detection, which may further promote the investigation of BPNPs as a fluorescence-based detection platform for other targets including proteins, enzymes and inorganic ions.Author contributions
The manuscript was written with contributions from all authors. All of the authors have approved the final version of the manuscript.Competing financial interests
The authors declare no competing financial interests.Acknowledgements
M. P. acknowledges a Tier 2 grant (MOE2013-T2-1-056; ARC 35/13) from the Ministry of Education, Singapore. Z. S. was supported by Czech Science Foundation (GACR No. 15-09001S).References
- T. G. Drummond, M. G. Hill and J. K. Barton, Electrochemical DNA sensors, Nat. Biotechnol., 2003, 21(10), 1192–1199 CrossRef CAS PubMed.
- Y. Du, S. Guo, S. Dong and E. Wang, An integrated sensing system for detection of DNA using new parallel-motif DNA triplex system and graphene–mesoporous silica–gold nanoparticle hybrids, Biomaterials, 2011, 32(33), 8584–8592 CrossRef CAS PubMed.
- A. H. Loo, A. Bonanni and M. Pumera, Impedimetric thrombin aptasensor based on chemically modified graphenes, Nanoscale, 2012, 4(1), 143–147 RSC.
- S. Aiyer, R. Prasad, M. Kumar, K. Nirvikar, B. Jain and O. S. Kushwaha, Fluorescent carbon nanodots for targeted in vitro cancer cell imaging, Appl. Mater. Today, 2016, 4, 71–77 CrossRef.
- F. Teles and L. Fonseca, Trends in DNA biosensors, Talanta, 2008, 77(2), 606–623 CrossRef CAS.
- S. Shi, X. Wang, W. Sun, X. Wang, T. Yao and L. Ji, Label-free fluorescent DNA biosensors based on metallointercalators and nanomaterials, Methods, 2013, 64(3), 305–314 CrossRef CAS PubMed.
- Y. Du, B. Li and E. Wang, “Fitting” makes “sensing” simple: label-free detection strategies based on nucleic acid aptamers, Acc. Chem. Res., 2012, 46(2), 203–213 CrossRef PubMed.
- X.-B. Zhang, R.-M. Kong and Y. Lu, Metal ion sensors based on DNAzymes and related DNA molecules, Annu. Rev. Anal. Chem., 2011, 4(1), 105 CrossRef CAS PubMed.
- Z. Liu and X. Su, A novel fluorescent DNA sensor for ultrasensitive detection of Helicobacter pylori, Biosens. Bioelectron., 2017, 87, 66–72 CrossRef CAS PubMed.
- A. H. Loo, Z. Sofer, D. Bouša, P. Ulbrich, A. Bonanni and M. Pumera, Carboxylic carbon quantum dots as a fluorescent sensing platform for DNA detection, ACS Appl. Mater. Interfaces, 2016, 8(3), 1951–1957 CAS.
- Z. S. Qian, X. Y. Shan, L. J. Chai, J. J. Ma, J. R. Chen and H. Feng, DNA nanosensor based on biocompatible graphene quantum dots and carbon nanotubes, Biosens. Bioelectron., 2014, 60, 64–70 CrossRef CAS PubMed.
- (a) Z. Qian, X. Shan, L. Chai, J. Chen and H. Feng, Simultaneous detection of multiple DNA targets by integrating dual-color graphene quantum dot nanoprobes and carbon nanotubes, Chem. – Eur. J., 2014, 20(49), 16065–16069 CrossRef CAS PubMed; (b) A. Bonanni, C. K. Chua, G. Zhao, Z. Sofer and M. Pumera, Inherently Electroactive Graphene Oxide Nanoplatelets As Labels for Single Nucleotide Polymorphism Detection, ACS Nano, 2012, 6, 8546 CrossRef CAS PubMed; (c) A. H. Loo, A. Bonanni and M. Pumera, Inherently electroactive graphene oxide nanoplatelets as labels for specific protein-target recognition, Nanoscale, 2013, 5, 7844 RSC.
- (a) D. Zheng, D. S. Seferos, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Aptamer nano-flares for molecular detection in living cells, Nano Lett., 2009, 9(9), 3258–3261 CrossRef CAS PubMed; (b) M. Pumera, M. T. Castaneda, M. I. Pividori, R. Eritja, A. Merkoci and S. Alegret, Magnetically Trigged Direct Electrochemical Detection of DNA Hybridization Using Au67 Quantum Dot as Electrical Tracer, Langmuir, 2015, 21, 9625 CrossRef PubMed.
- J. Tian, N. Cheng, Q. Liu, W. Xing and X. Sun, Cobalt phosphide nanowires: efficient nanostructures for fluorescence sensing of biomolecules and photocatalytic evolution of dihydrogen from water under visible light, Angew. Chem., Int. Ed., 2015, 54(18), 5493–5497 CrossRef CAS PubMed.
- J. Tian, Q. Liu, J. Shi, J. Hu, A. M. Asiri, X. Sun and Y. He, Rapid, sensitive, and selective fluorescent DNA detection using iron-based metal–organic framework nanorods: Synergies of the metal center and organic linker, Biosens. Bioelectron., 2015, 71, 1–6 CrossRef CAS PubMed.
- L. Wang, Y. Zhang, J. Tian, H. Li and X. Sun, Conjugation polymer nanobelts: a novel fluorescent sensing platform for nucleic acid detection, Nucleic Acids Res., 2011, 39(6), e37 CrossRef CAS PubMed.
- H. Li and X. Sun, Fluorescence-enhanced nucleic acid detection: using coordination polymer colloids as a sensing platform, Chem. Commun., 2011, 47(9), 2625–2627 RSC.
- L. Li, Y. Yu, G. J. Ye, Q. Ge, X. Ou, H. Wu, D. Feng, X. H. Chen and Y. Zhang, Black phosphorus field-effect transistors, Nat. Nanotechnol., 2014, 9(5), 372–377 CrossRef CAS PubMed.
- H. U. Lee, S. Y. Park, S. C. Lee, S. Choi, S. Seo, H. Kim, J. Won, K. Choi, K. S. Kang and H. G. Park, Black phosphorus (BP) nanodots for potential biomedical applications, Small, 2016, 12(2), 214–219 CrossRef CAS PubMed.
- N. M. Latiff, W. Z. Teo, Z. Sofer, A. C. Fisher and M. Pumera, The cytotoxicity of layered black phosphorus, Chem. – Eur. J., 2015, 21(40), 13991–13995 CrossRef CAS PubMed.
- R. Fei and L. Yang, Strain-engineering the anisotropic electrical conductance of few-layer black phosphorus, Nano Lett., 2014, 14(5), 2884–2889 CrossRef CAS PubMed.
- V. Tran, R. Soklaski, Y. Liang and L. Yang, Layer-controlled band gap and anisotropic excitons in few-layer black phosphorus, Phys. Rev. B: Condens. Matter Mater. Phys., 2014, 89(23), 235319 CrossRef.
- A. Castellanos-Gomez, Black phosphorus: narrow gap, wide applications, J. Phys. Chem. Lett., 2015, 6(21), 4280–4291 CrossRef CAS PubMed.
- J. Kim, S. S. Baik, S. H. Ryu, Y. Sohn, S. Park, B.-G. Park, J. Denlinger, Y. Yi, H. J. Choi and K. S. Kim, Observation of tunable band gap and anisotropic Dirac semimetal state in black phosphorus, Science, 2015, 349(6249), 723–726 CrossRef CAS PubMed.
- (a) M. Engel, M. Steiner and P. Avouris, Black phosphorus photodetector for multispectral, high-resolution imaging, Nano Lett., 2014, 14(11), 6414–6417 CrossRef CAS PubMed; (b) D. Li, A. E. D. R. Castillo, H. Jussila, G. Ye, Z. Ren, J. Bai, X. Chen, H. Lipsanen, Z. Sun and F. Bonaccorso, Black phosphorus polycarbonate polymer composite for pulsed fibre lasers, Appl. Mater. Today, 2016, 4, 17 CrossRef.
- N. Youngblood, C. Chen, S. J. Koester and M. Li, Waveguide-integrated black phosphorus photodetector with high responsivity and low dark current, Nat. Photonics, 2015, 9, 247 CAS.
- J. Miao, S. Zhang, L. Cai, M. Scherr and C. Wang, Ultrashort channel length black phosphorus field-effect transistors, ACS Nano, 2015, 9(9), 9236–9243 CrossRef CAS PubMed.
- C. C. Mayorga-Martinez, Z. Sofer and M. Pumera, Layered black phosphorus as a selective vapor sensor, Angew. Chem., 2015, 127(48), 14525–14528 CrossRef.
- A. N. Abbas, B. Liu, L. Chen, Y. Ma, S. Cong, N. Aroonyadet, M. Köpf, T. Nilges and C. Zhou, Black phosphorus gas sensors, ACS Nano, 2015, 9(5), 5618–5624 CrossRef CAS PubMed.
- S. Y. Cho, Y. Lee, H. J. Koh, H. Jung, J. S. Kim, H. W. Yoo, J. Kim and H. T. Jung, Superior Chemical Sensing Performance of Black Phosphorus: Comparison with MoS2 and Graphene, Adv. Mater., 2016, 28, 7020–7028 CrossRef CAS PubMed.
- V. Kumar, J. R. Brent, M. Shorie, H. Kaur, G. Chadha, A. G. Thomas, E. A. Lewis, A. P. Rooney, L. Nguyen and X. L. Zhong, Nanostructured aptamer-functionalized black phosphorus sensing platform for label-free detection of myoglobin, a cardiovascular disease biomarker, ACS Appl. Mater. Interfaces, 2016, 8(35), 22860–22868 CAS.
- C. C. Mayorga-Martinez, N. Mohamad Latiff, A. Y. S. Eng, Z. k. Sofer and M. Pumera, Black Phosphorus Nanoparticle Labels for Immunoassays via Hydrogen Evolution Reaction Mediation, Anal. Chem., 2016, 88(20), 10074–10079 CrossRef CAS PubMed.
- J. D. Wood, S. A. Wells, D. Jariwala, K.-S. Chen, E. Cho, V. K. Sangwan, X. Liu, L. J. Lauhon, T. J. Marks and M. C. Hersam, Effective passivation of exfoliated black phosphorus transistors against ambient degradation, Nano Lett., 2014, 14(12), 6964–6970 CrossRef CAS PubMed.
- P. Li, D. Zhang, J. Liu, H. Chang, Y. e. Sun and N. Yin, Air-stable black phosphorus devices for ion sensing, ACS Appl. Mater. Interfaces, 2015, 7(44), 24396–24402 CAS.
- Y. Zhao, H. Wang, H. Huang, Q. Xiao, Y. Xu, Z. Guo, H. Xie, J. Shao, Z. Sun and W. Han, Surface coordination of black phosphorus for robust air and water stability, Angew. Chem., 2016, 128(16), 5087–5091 CrossRef.
- H. U. Lee, S. C. Lee, J. Won, B.-C. Son, S. Choi, Y. Kim, S. Y. Park, H.-S. Kim, Y.-C. Lee and J. Lee, Stable semiconductor black phosphorus (BP)@ titanium dioxide (TiO2) hybrid photocatalysts, Sci. Rep., 2015, 5, 8691 CrossRef CAS PubMed.
- Z. Sofer, D. Bouša, J. Luxa, V. Mazanek and M. Pumera, Few-layer black phosphorus nanoparticles, Chem. Commun., 2016, 52(8), 1563–1566 RSC.
- S. Stankovich, D. A. Dikin, G. H. Dommett, K. M. Kohlhaas, E. J. Zimney, E. A. Stach, R. D. Piner, S. T. Nguyen and R. S. Ruoff, Graphene-based composite materials, Nature, 2006, 442(7100), 282–286 CrossRef CAS PubMed.
- P. Yasaei, B. Kumar, T. Foroozan, C. Wang, M. Asadi, D. Tuschel, J. E. Indacochea, R. F. Klie and A. Salehi-Khojin, High-quality black phosphorus atomic layers by liquid-phase exfoliation, Adv. Mater., 2015, 27(11), 1887–1892 CrossRef CAS PubMed.
- H. E. Swanson, E. Tatge and R. K. Fuyat, Standard X-ray diffraction powder patterns, 1953 Search PubMed.
- N. M. Stark and L. M. Matuana, Characterization of weathered wood–plastic composite surfaces using FTIR spectroscopy, contact angle, and XPS, Polym. Degrad. Stab., 2007, 92(10), 1883–1890 CrossRef CAS.
- X. Zhang, H. Xie, Z. Liu, C. Tan, Z. Luo, H. Li, J. Lin, L. Sun, W. Chen and Z. Xu, Black phosphorus quantum dots, Angew. Chem., Int. Ed., 2015, 54(12), 3653–3657 CrossRef CAS PubMed.
- L. Wang, Z. Sofer and M. Pumera, Voltammetry of Layered Black Phosphorus: Electrochemistry of Multilayer Phosphorene, ChemElectroChem, 2015, 2(3), 324–327 CrossRef CAS.
- S. Sugai and I. Shirotani, Raman and infrared reflection spectroscopy in black phosphorus, Solid State Commun., 1985, 53(9), 753–755 CrossRef CAS.
- W. Zhang, T. Huynh, P. Xiu, B. Zhou, C. Ye, B. Luan and R. Zhou, Revealing the importance of surface morphology of nanomaterials to biological responses: Adsorption of the villin headpiece onto graphene and phosphorene, Carbon, 2015, 94, 895–902 CrossRef CAS.
| This journal is © the Partner Organisations 2017 |