Synergistic antiviral effect of curcumin functionalized graphene oxide against respiratory syncytial virus infection†
Abstract
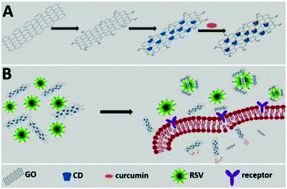
The diseases attributable to viruses remain a global burden. The respiratory syncytial virus (RSV), which is considered as the major viral pathogen of the lower respiratory tract of infants, has been implicated in severe lung disease. In this contribution, we developed a β-cyclodextrin (CD) functionalized graphene oxide (GO) composite, which displayed excellent antiviral activity and could load curcumin efficiently. RSV, a negative-sense single-stranded enveloped RNA virus, was employed as a model virus to investigate the antiviral activity of multifunctional GO. Proved by the tissue culture infectious dose assay and immunofluorescence assay, the curcumin loaded functional GO was confirmed with highly efficient inhibition for RSV infection and great biocompatibility to the host cells. The results showed that the composite could prevent RSV from infecting the host cells by directly inactivating the virus and inhibiting the viral attachment, and possessed prophylactic and therapeutic effects towards the virus. Our data indicate that the composite may provide new insights into antiviral therapy for RSV infection.
- This article is part of the themed collection: Coronavirus articles - free to access collection


 Please wait while we load your content...
Please wait while we load your content...