Effects of weight loss using supplementation with Lactobacillus strains on body fat and medium-chain acylcarnitines in overweight individuals
Minkyung
Kim
a,
Minjoo
Kim
a,
Miso
Kang
bc,
Hye Jin
Yoo
bc,
Min Sun
Kim
bc,
Young-Tae
Ahn
d,
Jae-Hun
Sim
d,
Sun Ha
Jee
e and
Jong Ho
Lee
 *abc
*abc
aResearch Center for Silver Science, Institute of Symbiotic Life-TECH, Yonsei University, Seoul, Korea
bNational Leading Research Laboratory of Clinical Nutrigenetics/Nutrigenomics, Department of Food and Nutrition, College of Human Ecology, Yonsei University, Seoul, Korea. E-mail: jhleeb@yonsei.ac.kr; Fax: +82-2-364-9605; Tel: +82-2-2123-3122
cDepartment of Food and Nutrition, Brain Korea 21 PLUS Project, College of Human Ecology, Yonsei University, Seoul, Korea
dKorea Yakult Co., Ltd, Yongin, Gyeonggi, Korea
eInstitute for Health Promotion, Graduate School of Public Health, Yonsei University, Seoul, Korea
First published on 12th December 2016
Abstract
Our previous study showed that supplementation with a combination of Lactobacillus curvatus (L. curvatus) HY7601 and Lactobacillus plantarum (L. plantarum) KY1032 reduced the body weight, body fat percentage, body fat mass and L1 subcutaneous fat area in overweight subjects. We aimed to evaluate whether the changes in adiposity after supplementation with Lactobacillus strains were associated with metabolic intermediates. A randomized, double-blind, placebo-controlled study was conducted on 66 non-diabetic and overweight individuals. Over a 12-week period, the probiotic group consumed 2 g of probiotic powder, whereas the placebo group consumed the same product without the probiotics. To investigate metabolic alterations, we performed plasma metabolomics using ultra-performance liquid chromatography and mass spectrometry (UPLC-LTQ/Orbitrap MS). Probiotic supplementation significantly increased the levels of octenoylcarnitine (C8:1), tetradecenoylcarnitine (C14:1), decanoylcarnitine (C10) and dodecenoylcarnitine (C12:1) compared with the levels from placebo supplementation. In the probiotic group, the changes in the body weight, body fat percentage, body fat mass and L1 subcutaneous fat area were negatively associated with changes in the levels of C8:1, C14:1, C10 and C12:1 acylcarnitines. In overweight individuals, probiotic-induced weight loss and adiposity reduction from the probiotic supplementation were associated with an increase in medium-chain acylcarnitines.
1. Introduction
Studies evaluating the influence of probiotics, prebiotics and synbiotics on body weight have been inconclusive because of the scarcity and low quality of the available studies.1–3 Recently, Madjd et al.4 found that consuming a probiotic yogurt during a 12-week hypoenergetic program resulted in no changes in body weight compared with a low-fat yogurt. However, a meta-analysis by Dror et al.5 suggested a role for probiotics in promoting weight loss in adults. In our previous study, we observed reductions in body weight, body fat percentage, and body fat mass after a 12-week treatment with two probiotic strains, Lactobacillus curvatus (L. curvatus) HY7601 and Lactobacillus plantarum (L. plantarum) KY1032, suggesting that the probiotic-induced weight loss was associated with decreased oxidative and inflammatory stress in overweight subjects.6 The mechanism behind the weight loss observed in adults likely relates to a reduction in inflammation and a strengthened intestinal epithelial barrier that has been observed to accompany probiotic administration.7–10 Because all these studies on adults involved the probiotic Lactobacillus, the observed weight loss from probiotics may be a result of anti-inflammatory effects, improved intestinal barrier integrity and increased metabolic activity associated with changes in intestinal flora. However, additional studies are required to determine the relationship between the weight loss from probiotic supplementation and metabolic changes.Metabolites as intermediates and end products of biological processes are most predictable for the phenotype of an organism.11 Therefore, metabolites may serve as biomarkers for changes in the physiological status, particularly for alterations in metabolic pathways in humans.11 However, studies on endogenous metabolic changes caused by probiotic-induced weight loss are limited. To the best of our knowledge, this is the first human intervention study to investigate the effects of probiotic supplementation on both probiotic-induced weight loss and metabolic changes. Therefore, in this study, we aimed to determine whether supplementation with a combination of L. curvatus HY7601 and L. plantarum KY1032 or placebo resulted in changes from the baseline in fasting metabolic intermediates. Furthermore, we aimed to evaluate whether changes in adiposity after probiotic or placebo supplementation were associated with changes in fasting metabolic intermediates. The plasma metabolic profiles of the probiotic group and the control group were analyzed using ultra-performance liquid chromatography and mass spectrometry (UPLC-LTQ/Orbitrap MS).
2. Materials and methods
2.1 Study subjects, study design and intervention
The details of the study subjects, study design, and interventions were published previously.6 Briefly, at the beginning of the study, 120 non-diabetic and overweight individuals were enrolled in a 12-week, double-blind, placebo-controlled, randomized study. The randomization was conducted via computer-generated block randomization (60 subjects in the control and probiotic groups). Among them, 95 individuals were included in the final analysis: a probiotic group (n = 49) that consumed 2 g of probiotic powder twice a day (after breakfast and dinner) containing L. curvatus HY7601 (2.5 × 109 colony-forming units (cfu)) and L. plantarum KY1032 (2.5 × 109 cfu) and a placebo group (n = 46) that consumed the same amount of powder daily without the probiotics (NCT02492698; http://www.clinicaltrials.gov). Only the product (powder) provider (Korea Yakult Co., Ltd, Yongin, Gyeonggi, Korea) retained the control and probiotic group list, which was disclosed after the research and all laboratory analyses were completed. Both the placebo and probiotic products had the same product weight, color, and flavor and were put into an opaque package; thus, the investigators were unaware of the individuals’ treatment allocation. Among the 95 subjects, 66 subjects (34 in the placebo group and 32 in the probiotic group) agreed to the analysis of their plasma metabolomic profiles. Written informed consent was obtained, and the study was performed according to the declaration of Helsinki. The procedures were approved by the institutional review boards of Yonsei University and Yonsei University Severance Hospital.2.2 Measurements of daily energy intake and physical activity
The details regarding daily energy intake and physical activity were published previously.6 Briefly, the subjects were instructed to maintain their existing eating habits and physical activity levels during the study period (pre-ingestion: 2 weeks; ingestion period: 12 weeks) to ensure that the changes in body weight did not result from the changes in diet or physical activity. Each participant completed a standardized 3-day diet and physical activity record at weeks 0, 6 and 12.2.3 Anthropometric parameters and blood pressure
Detailed information is provided in our previous article.6 Briefly, body weight, height and waist circumference were measured at screening, baseline and the 12-week follow-up. The body mass index (BMI) was calculated in units of kilograms per square meter (kg m−2). During each testing session, systolic and diastolic blood pressure was assessed in a supine position after a resting period.2.4 Abdominal fat area and body composition measurements
The details of the abdominal fat area and body composition measurements were published previously.6 Briefly, body fat distribution and muscle areas were measured using computed tomography (CT) at the level of the 1st lumbar (L1) and the 4th lumbar (L4) vertebrae. The body composition of the study participants was measured using dual-energy X-ray absorptiometry (DEXA) to determine the body fat percentage, body fat mass and lean body mass.2.5 Blood collection and biochemical analyses
The details of the blood collection and biochemical analyses were published previously.6 Briefly, blood samples were collected following an overnight fast of at least 12 h, and the levels of fasting triglycerides, total HDL, and LDL cholesterol, glucose, insulin and high-sensitivity C-reactive protein (hs-CRP) were measured. Insulin resistance (IR) was calculated using the homeostasis model assessment (HOMA) by the following equation: HOMA-IR = [fasting insulin (μIU mL−1) × fasting glucose (mg dL−1)]/405.2.6 Nontargeted metabolic profiling of plasma
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. A total of 820 μL of the supernatant was dried with nitrogen gas. The remaining pellet was dissolved in 10% methanol and centrifuged at 10
000 rpm for 5 min. A total of 820 μL of the supernatant was dried with nitrogen gas. The remaining pellet was dissolved in 10% methanol and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. Finally, 85 μL of the supernatant was transferred to a fresh vial.
000 rpm for 5 min. Finally, 85 μL of the supernatant was transferred to a fresh vial.
2.7 Statistical analysis
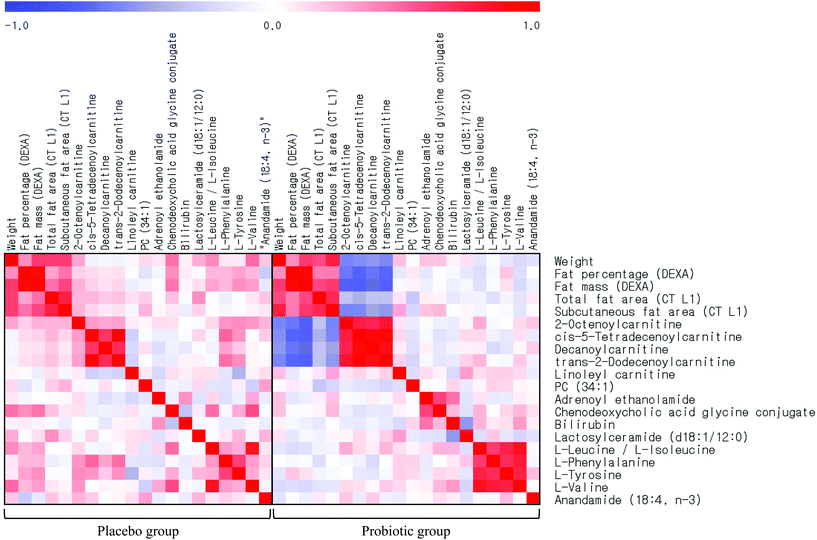
The statistical analyses were performed using the SPSS version 21.0 software (IBM/SPSS, Chicago, IL, USA). The results are expressed as the mean ± standard error (SE). Logarithmic transformation was performed on skewed variables. For descriptive purposes, the mean values are presented as median values ± standard deviation (SD) for skewed variables. A two-tailed P < 0.05 was considered statistically significant. Independent t-tests were used to compare the parameters between the two groups. Paired t-tests were performed to determine the differences between the baseline and 12-week follow-up values within each group. Pearson's correlation coefficient was used to examine the relationship between variables. False discovery rate-corrected q-values were computed using the R package ‘fdrtool’, and q-values less than 0.05 were considered to indicate significance. Heat maps were created to visualize and evaluate the relationship among metabolites and biochemical measurements in the study population.The metabolomics data were exported to SIMCA-P+ 14.0 (Umetrics, Inc., Umeå, Sweden) for multivariate analysis. The data were Pareto scaled, and an orthogonal partial least squares-discriminant analysis (OPLS-DA) was used to predict our models. The validity of the models was measured using a default cross-validation procedure of 7-fold cross-validation and R2Y and Q2Y parameters.
3. Results
3.1 The effects of 12 weeks of probiotic consumption on anthropometric parameters, clinical characteristics, body composition and abdominal fat areas
The effects of 12 weeks of probiotic consumption on anthropometric parameters, clinical characteristics, body composition, and abdominal fat areas were in agreement with our previous findings.6Table 1 shows the body weight, BMI, body fat percentage and fat mass, and the abdominal fat areas at the baseline and at 12 weeks in the placebo and probiotic groups. At the baseline, no significant differences between the two groups were observed for age, gender distribution, smoking and drinking rates (data not shown), body weight, BMI, body fat percentage, body fat mass, and the total, visceral, and subcutaneous fat areas at the L1 and L4 levels. After 12 weeks of treatment, the individuals in the probiotic group showed a significant reduction in the body weight, BMI, total fat mass (717 g), fat percentage (0.67%) and a trend toward a decrease in the L4 subcutaneous fat area (P = 0.080) from the baseline, while a significant increase in visceral fat, subcutaneous fat, and the total fat areas at the L1 level was observed in the placebo group. When we compared the changes from the baseline between the control and probiotic groups, significant differences were observed due to changes in the body weight, BMI, fat percentage, fat mass, L1 total fat area, and L1 subcutaneous fat area, with a trend toward a difference in the L4 total fat area (P = 0.063). The probiotic group showed a reduction in these parameters, whereas the control group showed an increase after the 12-week treatment.| Total (n = 66) | P a | P b | P c | ||||
|---|---|---|---|---|---|---|---|
| Placebo group (n = 34) | Probiotic group (n = 32) | ||||||
| Baseline | Follow-up | Baseline | Follow-up | ||||
| Mean ± SE. Pa-Values derived from independent t-test for baseline values. Pb-Values derived from independent t-test for follow-up values. Pc-Values derived from independent t-test for changed values. Pd-Values adjusting for baseline. †P < 0.10, *P < 0.05, **P < 0.01, ***P < 0.001 derived from paired t-test. | |||||||
| Weight (kg) | 73.2 ± 1.72 | 73.6 ± 1.82 | 71.9 ± 1.43 | 71.3 ± 1.50* | 0.581 | 0.334 | |
| Change | 0.46 ± 0.24 | −0.60 ± 0.30 | 0.008 | ||||
| Body mass index (kg m−2) | 27.1 ± 0.27 | 27.2 ± 0.28 | 26.6 ± 0.23 | 26.4 ± 0.25* | 0.173 | 0.024 | |
| Change | 0.15 ± 0.09 | −0.23 ± 0.11 | 0.008 | ||||
| DEXA evaluation | |||||||
| Fat percentage (%) | 31.2 ± 1.00 | 31.3 ± 0.96 | 30.6 ± 0.98 | 30.0 ± 1.03* | 0.712 | 0.357 | |
| Change | 0.12 ± 0.20 | −0.67 ± 0.27 | 0.020 | ||||
| Fat mass (g) | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 993.5 ± 762.7 993.5 ± 762.7 |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 236.3 ± 782.1 236.3 ± 782.1 |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 157.9 ± 585.1 157.9 ± 585.1 |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 440.6 ± 622.9** 440.6 ± 622.9** |
0.392 | 0.079 | |
| Change | 242.9 ± 200.1 | −717.3 ± 225.1 | 0.002 | ||||
| CT evaluation (L1) | |||||||
| Total fat area (cm2) | 243.8 ± 11.1 | 256.0 ± 10.0** | 247.3 ± 7.6 | 246.9 ± 8.14 | 0.797 | 0.486 | |
| Change | 12.2 ± 3.70 | −0.43 ± 4.04 | 0.024 | ||||
| Visceral fat area (cm2) | 101.8 ± 7.87 | 110.1 ± 7.38** | 115.2 ± 6.53 | 116.8 ± 7.09 | 0.199 | 0.519 | |
| Change | 8.33 ± 2.61 | 1.64 ± 3.19 | 0.108 | ||||
| Subcutaneous fat area (cm2) | 142.0 ± 6.87 | 145.84 ± 6.66* | 132.2 ± 5.65 | 130.1 ± 5.77† | 0.275 | 0.080 | |
| Change | 3.84 ± 1.88 | −2.07 ± 1.42 | 0.016 | ||||
| Visceral/subcutaneous fat ratio (%) | 0.78 ± 0.08 | 0.83 ± 0.09* | 0.94 ± 0.08 | 0.97 ± 0.08 | 0.163 | 0.251 | |
| Change | 0.05 ± 0.02 | 0.03 ± 0.03 | 0.603 | ||||
| CT evaluation (L4) | |||||||
| Total fat area (cm2) | 308.0 ± 8.60 | 314.5 ± 8.10 | 301.6 ± 7.10 | 296.6 ± 7.40 | 0.569 | 0.109 | |
| Change | 6.46 ± 4.33 | −4.99 ± 4.22 | 0.063 | ||||
| Visceral fat area (cm2) | 88.8 ± 5.51 | 90.2 ± 5.44 | 86.5 ± 5.22 | 83.1 ± 4.29 | 0.764 | 0.313 | |
| Change | 1.32 ± 2.18 | −3.43 ± 3.49 | 0.246 | ||||
| Subcutaneous fat area (cm2) | 219.2 ± 8.08 | 224.3 ± 8.12 | 215.0 ± 7.77 | 213.5 ± 7.58 | 0.714 | 0.335 | |
| Change | 5.14 ± 3.97 | −1.55 ± 2.68 | 0.172 | ||||
| Visceral/subcutaneous fat ratio (%) | 0.44 ± 0.04 | 0.43 ± 0.04 | 0.43 ± 0.04 | 0.41 ± 0.03 | 0.935 | 0.657 | |
| Change | 0.00 ± 0.02 | −0.02 ± 0.02 | 0.506 | ||||
Furthermore, no significant differences existed at the baseline, and no significant mean changes were observed after the 12-week treatment in systolic and diastolic blood pressure, levels of triglycerides, total HDL and LDL cholesterol, glucose, insulin, hs-CRP and the HOMA-IR index (data not shown) in the placebo and probiotic groups. Additionally, the estimated total calorie intake, the amount of physical activity, the percentage of protein intake, the percentage of fat intake and the percentage of carbohydrate intake did not differ significantly between the two groups at the baseline, week 6 and week 12 (data not shown).
3.2 Plasma metabolic profiling using UPLC-LTQ Orbitrap mass spectrometry
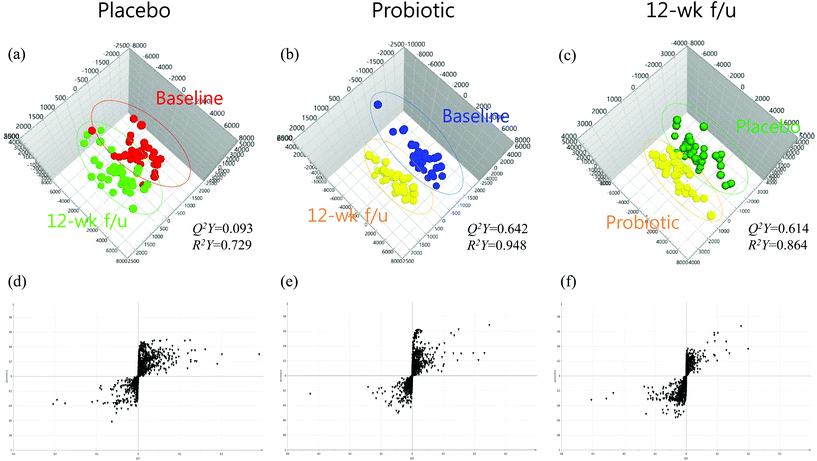
The quality of each OPLS-DA model was examined using the R2 and Q2 values to confirm that the models were not over-fitted and evaluate the predictive ability of each model. R2 represents the goodness-of-fit parameter and is defined as the proportion of variance in the data described by the model. Q2 represents the predictive ability parameter and is defined as the proportion of variance in the data predicted by the model. R2 and Q2 values greater than 0.5 indicate a high-quality OPLS-DA model.14 The two-component OPLS-DA scatter plots of the plasma metabolites for the first combination of groups did not show distinct clustering or a clear separation of participants in the placebo group at the baseline or at the 12-week follow-up assessment (R2Y = 0.729, Q2Y = 0.093). These analyses, however, revealed that the second and third models (Fig. 1) displayed greater than 86% goodness-of-fit (R2Y = 0.948 and R2Y = 0.864 for the second and third combinations of groups, respectively) and a predictive ability greater than 61% (Q2Y = 0.642 and Q2Y = 0.614 for the second and third combinations of groups, respectively), which demonstrated that the second and third OPLS-DA models in the present study were well-fitted and displayed an acceptable predictive ability. The present results clearly showed that plasma metabolomic profiles could distinguish groups based on probiotics, changes in probiotic-induced biochemical characteristics, or both. To extract potential variables contributing to the detected differences, S-plots from the OPLS-DA models were constructed. S-Plots of p(1) and p(corr)(1) were generated using centroid scaling (Fig. 1d–f). The S-plots revealed that metabolites with higher or lower p(corr) values were more relevant for discriminating between the two groups.
| Identity | Molecular formula | Mono-isotopic mass | Peak intensities | VIP | q a | q b | q c | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Placebo group (n = 34) | Probiotic group (n = 32) | Baseline vs. follow-up in test group | Placebo follow-up vs. test follow-up | ||||||||
| Baseline | Follow-up | Baseline | Follow-up | ||||||||
| Median ± SD. VIP: variable importance in projection. All q-values were tested following a logarithmic transformation; qa-value is the adjusted P-value, which is derived from an independent t-test for baseline values and controls the false discovery rate. qb-Value is the adjusted P-value, which is derived from an independent t-test for follow-up values and controls the false discovery rate. qc-Value is the adjusted P-value, which is derived from an independent t-test for changed value differences and controls the false discovery rate. | |||||||||||
| 2-Octenoylcarnitine | C15H27NO4 | 286.200 | 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 895 ± 16 895 ± 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 022 022 |
42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 ± 16 300 ± 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 991 991 |
35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 439 ± 16 439 ± 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 109 109 |
67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 961 ± 27 961 ± 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 567*** 567*** |
4.081 | 2.527 | 0.632 | 0.001 | |
| Change | 7669 ± 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 372 372 |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 443 ± 27 443 ± 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 416 416 |
0.003 | ||||||||
| Adrenoyl ethanolamide | C24H41NO2 | 376.317 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 664 ± 15 664 ± 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 997 997 |
34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 458 ± 33 458 ± 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 611** 611** |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 285 ± 8048 285 ± 8048 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 357 ± 35 357 ± 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 093** 093** |
2.891 | 0.581 | 0.376 | 0.755 | |
| Change | 8849 ± 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 046 046 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 804 ± 36 804 ± 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 139 139 |
0.975 | ||||||||
| Anandamide (18:4, n-3) | C20H33NO2 | 320.255 | 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 113 ± 115 113 ± 115![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 684 684 |
37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 985 ± 60 985 ± 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 917* 917* |
69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 127 ± 101 127 ± 101![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 656 656 |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 682 ± 140 682 ± 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 925 925 |
3.282 | 1.605 | 0.436 | 0.708 | |
| Change | −57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 216 ± 143 216 ± 143![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 517 517 |
−40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 050 ± 174 050 ± 174![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 760 760 |
0.977 | ||||||||
| Bilirubin | C33H36N4O6 | 585.268 | 8186 ± 5437 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 071 ± 9414** 071 ± 9414** |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 353 ± 5718 353 ± 5718 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 743 ± 15 743 ± 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 375 375 |
1.068 | 0.421 | 0.253 | 0.497 | |
| Change | 4819 ± 9914 | 519 ± 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 216 216 |
0.984 | ||||||||
| Chenodeoxycholic acid glycine conjugate | C26H43NO5 | 450.319 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 861 ± 29 861 ± 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 076 076 |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 169 ± 31 169 ± 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 643 643 |
8581 ± 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 756 756 |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 227 ± 28 227 ± 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 845** 845** |
2.206 | 0.543 | 0.220 | 0.697 | |
| Change | 2572 ± 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 008 008 |
6540 ± 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 414 414 |
0.947 | ||||||||
| cis-5-Tetradecenoylcarnitine | C21H39NO4 | 370.294 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536 ± 13 536 ± 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 064 064 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 224 ± 16 224 ± 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 025 025 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 161 ± 11 161 ± 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 738 738 |
42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 025 ± 24 025 ± 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 076*** 076*** |
3.426 | 2.518 | 0.466 | <0.001 | |
| Change | 1821 ± 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 554 554 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 803 ± 23 803 ± 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 697 697 |
0.001 | ||||||||
| Decanoylcarnitine | C17H33NO4 | 316.247 | 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 353 ± 23 353 ± 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 692 692 |
51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 906 ± 45 906 ± 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 196 196 |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 266 ± 31 266 ± 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 771 771 |
127![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 509 ± 58 509 ± 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 183*** 183*** |
6.787 | 4.897 | 0.373 | <0.001 | |
| Change | 7920 ± 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 471 471 |
72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 628 ± 54 628 ± 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 934 934 |
<0.001 | ||||||||
| Lactosylceramide (d18:1/12:0) | C42H79NO13 | 806.563 | 577![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 449 ± 182 449 ± 182![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 928 928 |
635![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 365 ± 218 365 ± 218![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 635* 635* |
601![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 958 ± 186 958 ± 186![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 962 962 |
600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 138 ± 250 138 ± 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 879 879 |
4.148 | 3.561 | 0.497 | 0.636 | |
| Change | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 481 ± 164 481 ± 164![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 836 836 |
48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 511 ± 237 511 ± 237![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 180 180 |
0.983 | ||||||||
| Linoleyl carnitine | C25H45NO4 | 424.340 | 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 128 ± 17 128 ± 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 619 619 |
53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 972 ± 17 972 ± 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 097 097 |
49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 633 ± 16 633 ± 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 365 365 |
59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 268 ± 18 268 ± 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 501* 501* |
1.424 | 1.174 | 0.574 | 0.386 | |
| Change | 5896 ± 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 938 938 |
5012 ± 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 913 913 |
0.964 | ||||||||
| L-Leucine/L-isoleucine | C6H13NO2 | 132.101 | 259![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 346 ± 47 346 ± 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 212 212 |
277![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 810 ± 61 810 ± 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 050*** 050*** |
257![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 218 ± 43 218 ± 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 954 954 |
254![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 271 ± 44 271 ± 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 994 994 |
1.070 | 2.095 | 0.604 | 0.200 | |
| Change | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 598 ± 39 598 ± 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 509 509 |
425 ± 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 335 335 |
0.453 | ||||||||
| L-Phenylalanine | C9H11NO2 | 166.085 | 298![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 196 ± 38 196 ± 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 248 248 |
308![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 185 ± 42 185 ± 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 274* 274* |
286![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 880 ± 35 880 ± 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 234 234 |
298![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 838 ± 37 838 ± 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 365 365 |
1.098 | 1.350 | 0.418 | 0.317 | |
| Change | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 836 ± 36 836 ± 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 555 555 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 846 ± 47 846 ± 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 793 793 |
0.978 | ||||||||
| L-Tryptophan | C11H12N2O2 | 205.096 | 420![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 022 ± 61 022 ± 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 488 488 |
405![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 622 ± 66 622 ± 66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 081 081 |
387![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 127 ± 68 127 ± 68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 946 946 |
380![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 937 ± 70 937 ± 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 598 598 |
1.453 | 2.009 | 0.444 | 0.285 | |
| Change | 6171 ± 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 997 997 |
−7153 ± 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 062 062 |
0.964 | ||||||||
| L-Tyrosine | C9H11NO3 | 182.080 | 128![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 753 ± 22 753 ± 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 084 084 |
142![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 806 ± 22 806 ± 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 605** 605** |
128![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 158 ± 24 158 ± 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 439 439 |
142![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 174 ± 24 174 ± 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 097 097 |
1.062 | 0.823 | 0.550 | 0.602 | |
| Change | 8322 ± 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 579 579 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 705 ± 26 705 ± 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 690 690 |
0.980 | ||||||||
| L-Valine | C5H11NO2 | 118.086 | 106![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 915 ± 13 915 ± 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 888 888 |
113![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 757 ± 17 757 ± 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 977*** 977*** |
107![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 229 ± 14 229 ± 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 443 443 |
109![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 364 ± 13 364 ± 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 897 897 |
0.453 | 1.167 | 0.546 | 0.200 | |
| Change | 9254 ± 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 011 011 |
4818 ± 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 304 304 |
0.441 | ||||||||
| LysoPC (14:0) | C22H46NO7P | 468.306 | 191![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 259 ± 62 259 ± 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 602 602 |
197![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 769 ± 69 769 ± 69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 984 984 |
160![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 679 ± 59 679 ± 59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 919 919 |
138![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 275 ± 41 275 ± 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 111 111 |
1.371 | 3.692 | 0.220 | 0.010 | |
| Change | −2249 ± 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 314 314 |
−4864 ± 71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 319 319 |
0.961 | ||||||||
| LysoPC (15:0) | C23H48NO7P | 482.322 | 136![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 071 ± 34 071 ± 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 911 911 |
135![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 438 ± 42 438 ± 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 891 891 |
125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 379 ± 49 379 ± 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 937 937 |
123![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 569 ± 35 569 ± 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 075 075 |
1.299 | 1.720 | 0.426 | 0.288 | |
| Change | 1279 ± 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 434 434 |
−700 ± 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 029 029 |
0.980 | ||||||||
| LysoPC (16:0) | C24H50NO7P | 496.337 | 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 911 911![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 302 ± 981 302 ± 981![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 548 548 |
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 353 ± 1 353 ± 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 088 088 |
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 186 186![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 667 ± 1 667 ± 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 651 651![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 882 882 |
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 401 401![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 677 ± 987 677 ± 987![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 676 676 |
8.505 | 9.633 | 0.320 | 0.282 | |
| Change | −309![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 358 ± 1 358 ± 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 383 383![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 345 345 |
340![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 441 ± 1 441 ± 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 836 836![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 755 755 |
0.981 | ||||||||
| LysoPC (16:1) | C24H48NO7P | 494.321 | 358![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 836 ± 92 836 ± 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 212 212 |
352![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 522 ± 115 522 ± 115![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 315 315 |
285![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 779 ± 119 779 ± 119![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 295 295 |
320![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 807 ± 68 807 ± 68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 597 597 |
1.449 | 2.485 | 0.284 | 0.255 | |
| Change | −25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 519 ± 128 519 ± 128![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 269 269 |
35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 786 ± 123 786 ± 123![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 142 142 |
0.983 | ||||||||
| LysoPC (17:0) | C25H52NO7P | 510.353 | 169![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 493 ± 39 493 ± 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 705 705 |
169![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 256 ± 55 256 ± 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 360 360 |
163![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 555 ± 70 555 ± 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 853 853 |
168![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 666 ± 54 666 ± 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 476 476 |
1.732 | 1.369 | 0.516 | 0.746 | |
| Change | 8430 ± 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 157 157 |
2026 ± 84![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 403 403 |
0.981 | ||||||||
| LysoPC (18:0) | C26H54NO7P | 524.368 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 524 524![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 370 ± 444 370 ± 444![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 664 664 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 523 523![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 021 ± 569 021 ± 569![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 624 624 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 308 308![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 683 ± 788 683 ± 788![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 463 463 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 394 394![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 730 ± 596 730 ± 596![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 285 285 |
5.657 | 6.066 | 0.516 | 0.496 | |
| Change | −72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 701 ± 623 701 ± 623![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 538 538 |
96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 537 ± 893 537 ± 893![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 514 514 |
0.979 | ||||||||
| LysoPC (18:1) | C26H52NO7P | 522.353 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 255 255![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 002 ± 430 002 ± 430![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 202 202 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 106 106![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 593 ± 523 593 ± 523![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 438 438 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 008 008![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 890 ± 566 890 ± 566![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 552 552 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 959 959![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 515 ± 461 515 ± 461![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 353 353 |
4.929 | 5.711 | 0.439 | 0.392 | |
| Change | −13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 978 ± 519 978 ± 519![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 148 148 |
−81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 397 ± 690 397 ± 690![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 692 692 |
0.982 | ||||||||
| LysoPC (18:2) | C26H50NO7P | 520.337 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 858 858![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 939 ± 744 939 ± 744![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 384 384 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 764 764![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 738 ± 933 738 ± 933![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 905 905 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 568 568![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 005 ± 589 005 ± 589![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 188 188 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 473 473![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 871 ± 762 871 ± 762![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 047 047 |
5.104 | 6.841 | 0.278 | 0.304 | |
| Change | −27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 714 ± 724 714 ± 724![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 738 738 |
−11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 376 ± 766 376 ± 766![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 262 262 |
0.982 | ||||||||
| LysoPC (18:3) | C26H48NO7P | 518.319 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 366 366![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 847 ± 247 847 ± 247![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 361 361 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 321 321![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 522 ± 278 522 ± 278![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 890 890 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 153 153![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 260 ± 334 260 ± 334![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 090 090 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 223 223![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 654 ± 226 654 ± 226![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 409 409 |
4.552 | 4.522 | 0.254 | 0.307 | |
| Change | −27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 577 ± 348 577 ± 348![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 379 379 |
77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 923 ± 397 923 ± 397![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 424 424 |
0.975 | ||||||||
| LysoPC (20:2) | C28H54NO7P | 548.368 | 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 650 ± 14 650 ± 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 453 453 |
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 271 ± 18 271 ± 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 219 219 |
43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780 ± 14 780 ± 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320 320 |
43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 290 ± 12 290 ± 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 385 385 |
0.738 | 1.047 | 0.280 | 0.333 | |
| Change | −5237 ± 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 679 679 |
−1080 ± 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 676 676 |
0.983 | ||||||||
| LysoPC (20:3) | C28H52NO7P | 546.353 | 261![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 010 ± 91 010 ± 91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 967 967 |
215![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 766 ± 109 766 ± 109![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 297 297 |
226![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 020 ± 79 020 ± 79![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 881 881 |
216![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 478 ± 49 478 ± 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 161 161 |
1.995 | 3.072 | 0.265 | 0.198 | |
| Change | −8869 ± 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 560 560 |
−18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 003 ± 87 003 ± 87![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 498 498 |
0.974 | ||||||||
| LysoPC (20:4) | C28H50NO7P | 544.337 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 309 309![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 520 ± 226 520 ± 226![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 881 881 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 252 252![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 848 ± 318 848 ± 318![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 502 502 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 188 188![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 819 ± 287 819 ± 287![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 103 103 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 273 273![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 091 ± 209 091 ± 209![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 477 477 |
3.374 | 3.635 | 0.413 | 0.610 | |
| Change | −19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 723 ± 285 723 ± 285![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 236 236 |
93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 833 ± 329 833 ± 329![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 350 350 |
0.979 | ||||||||
| LysoPC (20:5) | C28H48NO7P | 542.322 | 900![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 448 ± 149 448 ± 149![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 034 034 |
885![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 285 ± 195 285 ± 195![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 560 560 |
830![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 623 ± 150 623 ± 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 060 060 |
807![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 360 ± 162 360 ± 162![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 594 594 |
2.503 | 4.029 | 0.425 | 0.202 | |
| Change | 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 099 ± 172 099 ± 172![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 802 802 |
−1727 ± 172![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 024 024 |
0.937 | ||||||||
| LysoPC (22:5) | C30H52NO7P | 570.353 | 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 338 ± 23 338 ± 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 881 881 |
78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258 ± 32 258 ± 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 512 512 |
71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 864 ± 27 864 ± 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 196 196 |
72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 223 ± 19 223 ± 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 298 298 |
0.951 | 1.454 | 0.472 | 0.237 | |
| Change | 2394 ± 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 480 480 |
−3078 ± 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 556 556 |
0.956 | ||||||||
| LysoPC (22:6) | C30H50NO7P | 568.337 | 347![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 641 ± 104 641 ± 104![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 527 527 |
364![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 518 ± 158 518 ± 158![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 401 401 |
361![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 487 ± 119 487 ± 119![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 798 798 |
328![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 312 ± 83 312 ± 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 996 996 |
2.024 | 2.858 | 0.631 | 0.286 | |
| Change | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 429 ± 150 429 ± 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 946 946 |
−7672 ± 117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 998 998 |
0.935 | ||||||||
| LysoPC (P-16:0) | C24H50NO6P | 480.343 | 112![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 472 ± 26 472 ± 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 724 724 |
106![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 976 ± 32 976 ± 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 924 924 |
103![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 047 ± 37 047 ± 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 472 472 |
109![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 072 ± 34 072 ± 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 122 122 |
1.505 | 1.141 | 0.614 | 0.687 | |
| Change | −3702 ± 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 989 989 |
8568 ± 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 280 |
0.978 | ||||||||
| LysoPE (16:0) | C21H44NO7P | 454.291 | 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 587 ± 18 587 ± 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 587 587 |
59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 544 ± 22 544 ± 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 020 020 |
49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 518 ± 29 518 ± 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 359 359 |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 262 ± 15 262 ± 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 759 759 |
1.034 | 1.372 | 0.380 | 0.221 | |
| Change | −4162 ± 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 985 985 |
5346 ± 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 519 519 |
0.981 | ||||||||
| LysoPE (18:1) | C23H46NO7P | 480.306 | 59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151 ± 22 151 ± 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 856 856 |
53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 238 ± 28 238 ± 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 464 464 |
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 245 ± 17 245 ± 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 341 341 |
37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 270 ± 13 270 ± 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 029 029 |
0.941 | 1.930 | 0.219 | 0.023 | |
| Change | 689 ± 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 555 555 |
−5829 ± 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 298 298 |
0.957 | ||||||||
| LysoPE (18:2) | C23H44NO7P | 478.291 | 149![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 773 ± 80 773 ± 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 948 948 |
132![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 497 ± 85 497 ± 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 664 664 |
96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 380 ± 40 380 ± 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 395 395 |
97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 739 ± 49 739 ± 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 668 668 |
1.325 | 2.816 | 0.220 | 0.135 | |
| Change | −389 ± 68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 207 207 |
−12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 733 ± 57 733 ± 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 233 233 |
0.974 | ||||||||
| LysoPE (20:3) | C25H46NO7P | 504.304 | 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 831 ± 11 831 ± 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 582 582 |
39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 069 ± 13 069 ± 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 861 861 |
34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 437 ± 12 437 ± 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 637 637 |
31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 771 ± 8892 771 ± 8892 |
0.626 | 1.076 | 0.375 | 0.218 | |
| Change | −570 ± 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 767 767 |
−783 ± 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 822 822 |
0.974 | ||||||||
| LysoPE (20:4) | C25H44NO7P | 502.290 | 101![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 015 ± 32 015 ± 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 492 492 |
99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 479 ± 42 479 ± 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 240 240 |
86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 886 ± 27 886 ± 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 057 057 |
85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 829 ± 26 829 ± 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 674 674 |
0.914 | 1.546 | 0.223 | 0.229 | |
| Change | −9526 ± 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 282 282 |
−1367 ± 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 385 385 |
0.985 | ||||||||
| LysoPE (20:5) | C25H42NO7P | 500.275 | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 974 ± 18 974 ± 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 239 239 |
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 421 ± 17 421 ± 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 863 863 |
32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 976 ± 11 976 ± 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 085 085 |
32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 072 ± 11 072 ± 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 452 452 |
0.740 | 1.774 | 0.223 | 0.010 | |
| Change | 1247 ± 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 455 455 |
−1354 ± 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 689 689 |
0.937 | ||||||||
| LysoPE (22:6) | C27H44NO7P | 526.291 | 117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 205 ± 46 205 ± 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 081 081 |
141![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 015 ± 53 015 ± 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 875 875 |
122![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 738 ± 43 738 ± 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 119 119 |
119![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 554 ± 36 554 ± 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 333 333 |
1.049 | 1.862 | 0.482 | 0.213 | |
| Change | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 636 ± 52 636 ± 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 949 949 |
−1734 ± 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 114 114 |
0.939 | ||||||||
| Oleamide | C18H35NO | 282.278 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258 258![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 806 ± 677 806 ± 677![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 934 934 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 196 196![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 507 ± 875 507 ± 875![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 468 468 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 108 108![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 012 ± 452 012 ± 452![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 940 940 |
949![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 756 ± 437 756 ± 437![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 199 199 |
5.943 | 9.144 | 0.264 | 0.222 | |
| Change | −73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 276 ± 892 276 ± 892![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 773 773 |
430 ± 543![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 359 359 |
0.976 | ||||||||
| Palmitamide | C16H33NO | 256.262 | 283![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 497 ± 105 497 ± 105![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 409 409 |
255![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 684 ± 179 684 ± 179![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 302 302 |
226![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 556 ± 77 556 ± 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 557 557 |
195![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 703 ± 119 703 ± 119![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 138 138 |
2.309 | 4.296 | 0.234 | 0.226 | |
| Change | 8205 ± 169![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 982 982 |
−27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 776 ± 119 776 ± 119![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 511 511 |
0.972 | ||||||||
| PC (34:1) | C42H82NO8P | 760.581 | 209![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 456 ± 134 456 ± 134![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 065 065 |
145![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 127 ± 65 127 ± 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 678* 678* |
167![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 808 ± 65 808 ± 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 642 642 |
197![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 479 ± 74 479 ± 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 849* 849* |
2.689 | 3.688 | 0.459 | 0.019 | |
| Change | −23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 560 ± 118 560 ± 118![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 192 192 |
33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 155 ± 77 155 ± 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 617 617 |
0.158 | ||||||||
| PC (34:3) | C42H78NO8P | 756.550 | 101![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 569 ± 47 569 ± 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 108 108 |
104![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 238 ± 52 238 ± 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 437 437 |
114![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 304 ± 39 304 ± 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 540 540 |
129![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 799 ± 58 799 ± 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 007 007 |
1.320 | 2.079 | 0.392 | 0.224 | |
| Change | 1167 ± 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 344 344 |
9921 ± 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 901 901 |
0.952 | ||||||||
| PC (36:2) | C44H84NO8P | 786.597 | 349![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 783 ± 152 783 ± 152![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 011 011 |
306![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 883 ± 137 883 ± 137![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 471 471 |
302![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 397 ± 136 397 ± 136![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 950 950 |
301![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 049 ± 153 049 ± 153![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 631 631 |
2.534 | 1.839 | 0.274 | 0.659 | |
| Change | −25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 685 ± 130 685 ± 130![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 773 773 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 086 ± 164 086 ± 164![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 738 738 |
0.817 | ||||||||
| PC (36:3) | C44H82NO8P | 784.581 | 244![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 534 ± 89 534 ± 89![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 903 903 |
268![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 692 ± 79 692 ± 79![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 760 760 |
256![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 467 ± 103 467 ± 103![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 724 724 |
254![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 206 ± 144 206 ± 144![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 209 209 |
2.360 | 2.070 | 0.592 | 0.677 | |
| Change | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 843 ± 91 843 ± 91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 335 335 |
5295 ± 107![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 697 697 |
0.969 | ||||||||
| PC (36:5) | C44H78NO8P | 780.549 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 528 528![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 074 ± 546 074 ± 546![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 480 480 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 705 705![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 305 ± 511 305 ± 511![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 360 360 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 603 603![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 113 ± 619 113 ± 619![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 869 869 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 524 524![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 850 ± 702 850 ± 702![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 578 578 |
6.076 | 5.518 | 0.596 | 0.703 | |
| Change | 117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 492 ± 507 492 ± 507![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258 258 |
83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 735 ± 594 735 ± 594![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 039 039 |
0.974 | ||||||||
| PC (38:5) | C46H82NO8P | 808.581 | 161![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 745 ± 47 745 ± 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 093 093 |
165![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 948 ± 53 948 ± 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 864 864 |
152![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 075 ± 37 075 ± 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 523 523 |
163![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 267 ± 53 267 ± 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 008* 008* |
2.067 | 1.111 | 0.372 | 0.654 | |
| Change | −7615 ± 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 883 883 |
9464 ± 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 071 071 |
0.886 | ||||||||
| PC (40:6) | C48H84NO8P | 834.597 | 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 173 ± 23 173 ± 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 647 647 |
81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 893 ± 23 893 ± 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 530 530 |
73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 703 ± 22 703 ± 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 060 060 |
68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 659 ± 37 659 ± 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 805 805 |
1.007 | 0.826 | 0.492 | 0.727 | |
| Change | 5324 ± 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 740 740 |
7111 ± 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 919 919 |
0.982 | ||||||||
| PC (P-38:6) | C46H80NO7P | 790.570 | 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 623 ± 23 623 ± 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 721 721 |
57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 387 ± 28 387 ± 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 949* 949* |
47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 040 ± 25 040 ± 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 569 569 |
54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 003 ± 32 003 ± 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 395* 395* |
1.530 | 1.143 | 0.486 | 0.587 | |
| Change | 5753 ± 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 034 034 |
8692 ± 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 566 566 |
0.981 | ||||||||
| SM (d18:0/16:1) | C39H79N2O6P | 703.572 | 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 898 ± 26 898 ± 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 461 461 |
59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 176 ± 29 176 ± 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 207 207 |
57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 906 ± 26 906 ± 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 876 876 |
66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 527 ± 49 527 ± 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 231 231 |
1.822 | 1.194 | 0.561 | 0.591 | |
| Change | 44 ± 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 137 137 |
2789 ± 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 890 890 |
0.960 | ||||||||
| Taurocholic acid | C26H45NO7S | 516.303 | 109![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 376 ± 31 376 ± 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 186 186 |
106![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027 ± 39 027 ± 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 733 733 |
88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 557 ± 38 557 ± 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 499 499 |
99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 801 ± 23 801 ± 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 475 475 |
0.796 | 1.381 | 0.318 | 0.276 | |
| Change | −7591 ± 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 832 832 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 509 ± 40 509 ± 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 732 732 |
0.985 | ||||||||
| trans-2-Dodecenoylcarnitine | C19H35NO4 | 342.262 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 290 ± 11 290 ± 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 877 877 |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 674 ± 13 674 ± 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 824 824 |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 695 ± 10 695 ± 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 693 693 |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 513 ± 34 513 ± 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 370*** 370*** |
3.716 | 2.852 | 0.427 | <0.001 | |
| Change | 2764 ± 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 198 198 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 585 ± 34 585 ± 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 310 310 |
0.002 | ||||||||
| Uric acid | C5H4N4O3 | 169.035 | 146![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 921 ± 29 921 ± 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 685 685 |
152![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 417 ± 33 417 ± 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 376 376 |
127![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 808 ± 38 808 ± 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 889 889 |
138![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640 ± 33 640 ± 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 804 804 |
0.809 | 1.181 | 0.437 | 0.220 | |
| Change | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 476 ± 25 476 ± 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 637 637 |
−989 ± 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 350 350 |
0.944 | ||||||||
3.3 The relationship between the changes in the body weight, body fat percentage, body fat mass and the total L1 subcutaneous fat areas and the major metabolites
Fig. 2 shows the correlations among the changes from the baseline in body weight, total fat percentage and total body fat mass, the total L1 subcutaneous fat areas and the major metabolites that showed significant differences between the baseline and the 12-week follow-up in the placebo or probiotic groups. The changes in the body weight, total body fat percentage, total body fat mass and L1 subcutaneous fat area were strongly and negatively correlated with the changes in octenoylcarnitine, tetradecenoylcarnitine, decanoylcarnitine and dodecenoylcarnitine in the probiotic group. No significant associations between the changes in body weight and body adiposity and the changes in the levels of plasma medium-chain acylcarnitines (ACs) were observed in the placebo group. However, changes in body weight were positively correlated with changes in leucine/isoleucine and valine levels in the placebo group but not in the probiotic group. Positive interrelations were observed among the changes in the medium-chain ACs with positive interrelations among the changes in the levels of leucine/isoleucine, valine, phenylalanine and tyrosine in both the placebo and probiotic groups (Fig. 2).4. Discussion
We showed that supplementation with the two probiotic strains resulted in significant differences in octenoylcarnitine (C8:1), tetradecenoylcarnitine (C14:1), decanoylcarnitine (C10) and dodecenoylcarnitine (C12:1) compared with the placebo group (n = 34). Additionally, we found that in the 32 overweight men and women in the probiotic group, the changes in body weight, whole body adiposity and abdominal fat were strongly and negatively associated with the changes in C8:1, C14:1, C10 and C12:1 ACs; however, no significant association between these measures was observed in the placebo group. Therefore, the probiotic-induced weight loss and adiposity reduction in healthy overweight subjects consuming a weight maintenance diet is associated with an increase in medium-chain ACs.ACs are derived from mitochondrial and peroxisomal acyl-CoA metabolites by the substitution of the carnitine moiety for CoA. They are the product of incomplete fatty acid oxidation in the mitochondria and are elevated in obesity and type 2 diabetes; they are also positively associated with insulin resistance15–18 even after adjusting the BMI.19 However, in this study, no significant differences were observed from the baseline in fasting glucose, insulin, C-peptide levels or the HOMA-IR index in the probiotic group at the 12-week follow-up. We observed that the probiotic-induced weight and adiposity loss were coupled with an increase in lipid-derived medium-chain ACs. Therefore, the current study provides new findings on the effects of probiotic-induced weight loss on metabolic intermediates and particularly on increases in plasma medium-chain ACs. Brady et al.20 previously found an increase in ACs after starvation in both lean and obese Zucker rats. Additionally, Redman et al.21 observed that the weight loss with calorie restriction in overweight subjects resulted in an increase in serum ACs. Huffman et al.22 also reported that fasting ACs were increased after 3 months of calorie restriction. In a recent study, the assessment of plasma acylcarnitines before and after the weight loss in obese subjects showed increased levels of acylcarnitines and a strong correlation between non-esterified fatty acids (NEFA) and several acylcarnitines during the weight loss. Since plasma NEFA are indicative of lipolysis,23 increased lipolysis might drive the generation of acylcarnitines.24 Similarly, in the present study, increased ACs in the probiotic group may have resulted from increased lipolysis during body fat reduction. Additionally, tetradecenoylcarnitine (C14:1) increases in altered fatty acid oxidation states, such as very long chain acyl-CoA dehydrogenase deficiency.25 Indeed, Yoo et al.26 found that the combination of the probiotics L. curvatus and L. plantarum was effective in inhibiting the gene expression of various fatty acid synthesis enzymes in the liver in parallel with the decrease in fatty acid oxidation-related enzymatic activity and gene expression in mice. Therefore, increased lipolysis together with decreased fatty acid oxidation may result in elevations in medium-chain acylcarnitines during the weight loss after probiotic consumption.
Several mechanisms regarding the effects of Lactobacillus strains on adiposity reduction have been proposed. In a study reporting the effect of L. plantarum KY1032 on fat mass reduction, L. plantarum KY1032 cell extracts were suggested as directly decreasing fat mass, by modulating the adipogenesis of 3T3-L1 cells.27 Yoo et al.26 demonstrated that two Lactobacillus strains, L. plantarum KY1032 and L. curvatus HY7601, have an adverse effect on fat accumulation in adipose tissue in diet-induced obese mice by changing the gut microbiome composition or modulating cholesterol metabolism. Additionally, another study reported on the roles of L. plantarum KY1032 and L. curvatus HY7601 in the gut; the two probiotics may reduce fat accumulation by inducing competition for nutrients within the gut or by reducing gut microbiota diversity.26 In the present study, we observed weight loss in the probiotic group while they consumed a weight maintenance diet. Therefore, we hypothesize that the administration of the Lactobacillus strains may have partly affected adipogenesis in adipose tissue or the gut microbiome in our study subjects, resulting in decreased fat accumulation.
Previous studies have reported elevated C3 and C5 ACs, which are the by-products of leucine, isoleucine and valine (branched chain amino acid, BCAA) catabolism, and higher levels of BCAAs and other amino acids including phenylalanine, tryptophan and tyrosine in obese individuals than in lean individuals, which is possibly because of higher insulin resistance.17,28 Prez-Cornago et al.29 showed a significant reduction in serum isoleucine in overweight adults following a weight loss intervention. In the present study, the subjects were overweight and had slightly higher HOMA-IR indexes (mean HOMA-IR index at baseline = 2.7) compared to the reference range for Koreans.30 Since an association exists between the HOMA-IR index and BCAAs,17,28 we expected that possible associations might exist among the probiotic-induced weight loss, the HOMA-IR index, and the changes of metabolic intermediates, such as BCAAs. However, in the probiotic group of this study, changes in amino acids were not associated with changes in body weight and body adiposity. Only the placebo group showed significant elevations from the baseline in BCAAs, phenylalanine and tyrosine at the 12-week follow-up and positive correlations between the changes in BCAAs and changes in body weight. No significant change was observed in the HOMA-IR index or the fasting glucose, insulin and C-peptide levels after the 12-week follow-up with no correlation observed between the HOMA-IR index and BCAAs in the placebo or probiotic groups. These findings appear to show a lack of association and interaction between the HOMA-IR index and BCAAs and may be inconsistent with previous studies.17,28 However, the significant increase of BCAAs observed in the placebo group at the 12-week follow-up may imply that the maintenance of an overweight state may accelerate the increase of BCAAs, whereas the consumption of the probiotics may inhibit their increase. Although we did not observe a correlation between the HOMA-IR index and BCAAs, increased BCAAs may influence insulin resistance in overweight individuals as previously described in several studies.17,28 Therefore, taken together, we propose that the intake of probiotics may help inhibit rising BCAAs in overweight individuals.
This study has some limitations. We specifically focused on overweight individuals without diabetes. Therefore, our data cannot be generalized to obese individuals or diabetic patients. Additionally, numerous metabolic intermediates were observed using UPLC-LTQ/Orbitrap MS in this study; however, most of these metabolites remain unidentified because the databases of endogenous biomolecules have not yet been constructed for this technique.31 Therefore, we identified only a limited number of metabolites among the 238 variables that contributed to the separation between the placebo and probiotic groups. Moreover, since the metabolomics analyses were conducted in some of the participants from the previous study, the statistical power to detect changes in metabolites may be reduced. Regarding gut microbiome alterations in our study subjects, we cannot characterize such changes because we did not measure the amount of the probiotics present in the feces of our study participants. Finally, we cannot determine whether the changes in the metabolic intermediates observed in the present study are derived from the direct effects of the probiotics themselves or the probiotic-induced adiposity reduction.
Despite these limitations, we found significant differences in octenoylcarnitine (C8:1), tetradecenoylcarnitine (C14:1), decanoylcarnitine (C10) and dodecenoylcarnitine (C12:1) in the group of healthy overweight individuals who consumed a weight maintenance diet supplemented with the two probiotic strains compared with the placebo-supplemented group. Furthermore, in the probiotic group, a significant decrease in body weight, whole body adiposity (percentage body fat or body fat mass) and the L1 subcutaneous fat area was strongly associated with an increase in C8:1, C14:1, C10 and C12:1 ACs. However, the precise mechanisms underlying the metabolic changes in the probiotic-induced weight loss should be examined in further studies.
Abbreviations
| AC | Acylcarnitine |
| BCAA | Branched chain amino acid |
| BMI | Body mass index |
| CT | Computed tomography |
| DEXA | Dual-energy X-ray absorptiometry |
| L1 | 1st lumbar |
| L4 | 4th lumbar |
| lysoPC | Lysophosphatidylcholine |
| lysoPE | Lysophosphatidylethanolamine |
| HOMA-IR | Homeostasis model assessment of insulin resistance |
| hs-CRP | High-sensitivity C-reactive protein |
| OPLS-DA | Orthogonal partial least squares-discriminant analysis |
| PC | Phosphatidylcholine |
| SE | Standard error |
| UPLC-LTQ/Orbitrap MS | Ultra-performance liquid chromatography-linear-trap quadrupole-Orbitrap mass spectrometry |
| VIP | Variable importance in projection |
Conflict of interest
The authors declare no conflicts of interest.Acknowledgements
This study was funded by grants from the Korean Health Technology R&D Project, the Ministry of Health & Welfare, Republic of Korea (HI14C2686010115 and HI14C2686); and the Bio-Synergy Research Project (NRF-2012M3A9C4048762) and the Mid-career Researcher Program (NRF-2016R1A2B4011662) of the Ministry of Science, ICT and the Future Planning through the National Research Foundation, Republic of Korea.References
- M. N. Mugambi, A. Musekiwa, M. Lombard, T. Young and R. Blaauw, Synbiotics, probiotics or prebiotics in infant formula for full term infants: a systematic review, Nutr. J., 2012, 11, 81 CrossRef PubMed.
- S. Park and J. H. Bae, Probiotics for weight loss: a systematic review and meta-analysis, Nutr. Res., 2015, 35, 566–575 CrossRef CAS PubMed.
- S. Rao, R. Srinivasjois and S. Patole, Prebiotic supplementation in full-term neonates: a systematic review of randomized controlled trials, Arch. Pediatr. Adolesc. Med., 2009, 163, 755–764 CrossRef PubMed.
- A. Madjd, M. A. Taylor, N. Mousavi, A. Delavari, R. Malekzadeh, I. A. Macdonald and H. R. Farshchi, Comparison of the effect of daily consumption of probiotic compared with low-fat conventional yogurt on weight loss in healthy obese women following an energy-restricted diet: a randomized controlled trial, Am. J. Clin. Nutr., 2016, 103, 323–329 CrossRef CAS PubMed.
- T. Dror, Y. Dickstein, G. Dubourg and M. Paul, Microbiota manipulation for weight change, Microb. Pathog. DOI:10.1016/j.micpath.2016.01.002 , in press.
- H. Y. Ahn, M. Kim, J. S. Chae, Y. T. Ahn, J. H. Sim, I. D. Choi, S. H. Lee and J. H. Lee, Supplementation with two probiotic strains, Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032, reduced body adiposity and Lp-PLA2 activity in overweight subjects, J. Funct. Foods, 2015, 19, 744–752 CrossRef.
- M. Le Barz, F. F. Anhê, T. V. Varin, Y. Desjardins, E. Levy, D. Roy, M. C. Urdaci and A. Marette, Probiotics as complementary treatment for metabolic disorders, J. Diabetes Metab., 2015, 39, 291–303 CrossRef PubMed.
- A. Moya-Pérez, M. Romo-Vaquero, F. Tomás-Barberán, Y. Sanz and M. T. García-Conesa, Hepatic molecular responses to Bifidobacterium pseudocatenulatum CECT 7765 in a mouse model of diet-induced obesity, Nutr., Metab. Cardiovasc. Dis., 2014, 24, 57–64 CrossRef PubMed.
- P. D. Cani, J. Amar, M. A. Iglesias, M. Poggi, C. Knauf, D. Bastelica, A. M. Neyrinck, F. Fava, K. M. Tuohy, C. Chabo, A. Waget, E. Delmée, B. Cousin, T. Sulpice, B. Chamontin, J. Ferrières, J. F. Tanti, G. R. Gibson, L. Casteilla, N. M. Delzenne, M. C. Alessi and R. Burcelin, Metabolic endotoxemia initiates obesity and insulin resistance, Diabetes, 2007, 56, 1761–1772 CrossRef CAS PubMed.
- J. Amar, C. Chabo, A. Waget, P. Klopp, C. Vachoux, L. G. Bermúdez-Humarán, N. Smirnova, M. Bergé, T. Sulpice, S. Lahtinen, A. Ouwehand, P. Langella, N. Rautonen, P. J. Sansonetti and R. Burcelin, Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment, EMBO Mol. Med., 2011, 3, 559–572 CrossRef CAS PubMed.
- H. H. Chen, Y. J. Tseng, S. Y. Wang, Y. S. Tsai, C. S. Chang, T. C. Kuo, W. J. Yao, C. C. Shieh, C. H. Wu and P. H. Kuo, The metabolome profiling and pathway analysis in metabolic healthy and abnormal obesity, Int. J. Obes., 2015, 39, 1241–1248 CrossRef CAS PubMed.
- S. Y. Lee, M. Kim, S. Jung, S. H. Lee and J. H. Lee, Altered plasma lysophosphatidylcholines and amides in non-obese and non-diabetic subjects with borderline-to-moderate hypertriglyceridemia: A case-control study, PLoS One, 2015, 10, e0123306 Search PubMed.
- S. H. Lee, S. Park, H. Kim and B. H. Jung, Metabolomic approaches to the normal aging process, Metabolomics, 2014, 10, 1268–1292 CAS.
- M. Bylesjö, D. Eriksson, A. Sjödin, S. Jansson, T. Moritz and J. Trygg, Orthogonal projections to latent structures as a strategy for microarray data normalization, BMC Bioinf., 2007, 8, 207 CrossRef PubMed.
- M. J. Patel, B. C. Batch, L. P. Svetkey, J. R. Bain, C. B. Turer, C. Haynes, M. J. Muehlbauer, R. D. Stevens, C. B. Newgard and S. H. Shah, Race and sex differences in small-molecule metabolites and metabolic hormones in overweight and obese adults, OMICS, 2013, 17, 627–635 CrossRef CAS PubMed.
- K. M. Huffman, S. H. Shah, R. D. Stevens, J. R. Bain, M. Muehlbauer, C. A. Slentz, C. J. Tanner, M. Kuchibhatla, J. A. Houmard, C. B. Newgard and W. E. Kraus, Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women, Diabetes Care, 2009, 32, 1678–1683 CrossRef CAS PubMed.
- C. B. Newgard, J. An, J. R. Bain, M. J. Muehlbauer, R. D. Stevens, L. F. Lien, A. M. Haqq, S. H. Shah, M. Arlotto, C. A. Slentz, J. Rochon, D. Gallup, O. Ilkayeva, B. R. Wenner, W. S. Yancy Jr., H. Eisenson, G. Musante, R. S. Surwit, D. S. Millington, M. D. Butler and L. P. Svetkey, A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance, Cell Metab., 2009, 9, 311–326 CrossRef CAS PubMed.
- S. H. Adams, C. L. Hoppel, K. H. Lok, L. Zhao, S. W. Wong, P. E. Minkler, D. H. Hwang, J. W. Newman and W. T. Garvey, Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women, J. Nutr., 2009, 139, 1073–1081 CrossRef CAS PubMed.
- E. S. Tai, M. L. Tan, R. D. Stevens, Y. L. Low, M. J. Muehlbauer, D. L. Goh, O. R. Ilkayeva, B. R. Wenner, J. R. Bain, J. J. Lee, S. C. Lim, C. M. Khoo, S. H. Shah and C. B. Newgard, Insulin resistance is associated with a metabolic profile of altered protein metabolism in Chinese and Asian-Indian men, Diabetologia, 2010, 53, 757–767 CrossRef CAS PubMed.
- L. J. Brady, P. S. Brady, L. Albers, A. T. Davis and C. L. Hoppel, Carnitine metabolism in lean and obese Zucker rats during starvation, J. Nutr., 1986, 116, 668–674 CAS.
- L. M. Redman, K. M. Huffman, L. R. Landerman, C. F. Pieper, J. R. Bain, M. J. Muehlbauer, R. D. Stevens, B. R. Wenner, V. B. Kraus, C. B. Newgard, W. E. Kraus and E. Ravussin, Effect of caloric restriction with and without exercise on metabolic intermediates in nonobese men and women, J. Clin. Endocrinol. Metab., 2011, 96, E312–E321 CrossRef CAS PubMed.
- K. M. Huffman, L. M. Redman, L. R. Landerman, C. F. Pieper, R. D. Stevens, M. J. Muehlbauer, B. R. Wenner, J. R. Bain, V. B. Kraus, C. B. Newgard, E. Ravussin and W. E. Kraus, Caloric restriction alters the metabolic response to a mixed-meal: results from a randomized, controlled trial, PLoS One, 2012, 7, e28190 CAS.
- S. Klein, Y. Sakurai, J. A. Romijn and R. M. Carroll, Progressive alterations in lipid and glucose metabolism during short-term fasting in young adult men, Am. J. Physiol., 1993, 265, E801–E806 CAS.
- M. G. Schooneman, A. Napolitano, S. M. Houten, G. K. Ambler, P. R. Murgatroyd, S. R. Miller, C. E. Hollak, C. Y. Tan, S. Virtue, A. Vidal-Puig, D. J. Nunez and M. R. Soeters, Assessment of plasma acylcarnitines before and after weight loss in obese subjects, Arch. Biochem. Biophys., 2016, 606, 73–80 CrossRef CAS PubMed.
- C. G. Costa, E. A. Struys, A. Bootsma, H. J. ten Brink, L. Dorland, I. Tavares de Almeida, M. Duran and C. Jakobs, Quantitative analysis of plasma acylcarnitines using gas chromatography chemical ionization mass fragmentography, J. Lipid Res., 1997, 38, 173–182 CAS.
- S. R. Yoo, Y. J. Kim, D. Y. Park, U. J. Jung, S. M. Jeon, Y. T. Ahn, C. S. Huh, R. McGregor and M. S. Choi, Probiotics L. plantarum and L. curvatus in combination alter hepatic lipid metabolism and suppress diet-induced obesity, Obesity, 2013, 21, 2571–2578 CrossRef CAS PubMed.
- D. Y. Park, Y. T. Ahn, C. S. Huh, S. M. Jeon and M. S. Choi, The inhibitory effect of Lactobacillus plantarum KY1032 cell extract on the adipogenesis of 3T3-L1 Cells, J. Med. Food, 2011, 14, 670–675 CrossRef CAS PubMed.
- J. Y. Kim, J. Y. Park, O. Y. Kim, B. M. Ham, H. J. Kim, D. Y. Kwon, Y. Jang and J. H. Lee, Metabolic profiling of plasma in overweight/obese and lean men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC-Q-TOF MS), J. Proteome Res., 2010, 9, 4368–4375 CrossRef CAS PubMed.
- A. Perez-Cornago, L. Brennan, I. Ibero-Baraibar, H. H. Hermsdorff, A. O'Gorman, M. A. Zulet and J. A. Martínez, Metabolomics identifies changes in fatty acid and amino acid profiles in serum of overweight older adults following a weight loss intervention, J. Physiol. Biochem., 2014, 70, 593–602 CrossRef CAS PubMed.
- K. J. Yun, K. Han, M. K. Kim, Y. M. Park, K. H. Baek, K. H. Song and H. S. Kwon, Insulin Resistance Distribution and Cut-Off Value in Koreans from the 2008-2010 Korean National Health and Nutrition Examination Survey, PLoS One, 2016, 11, e0154593 Search PubMed.
- C. Ezzili, K. Otrubova and D. L. Boger, Fatty acid amide signaling molecules, Bioorg. Med. Chem. Lett., 2010, 20, 5959–5968 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |