Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Crystal structure of a DNA duplex containing four Ag(I) ions in consecutive dinuclear Ag(I)-mediated base pairs: 4-thiothymine–2Ag(I)–4-thiothymine†
Jiro
Kondo
 *a,
Toru
Sugawara
*a,
Toru
Sugawara
 b,
Hisao
Saneyoshi
b,
Hisao
Saneyoshi
 b and
Akira
Ono
b and
Akira
Ono
 *b
*b
aDepartment of Materials and Life Sciences, Faculty of Science and Technology, Sophia University, 7-1 Kioi-cho, Chiyoda-ku, Tokyo 102-8554, Japan. E-mail: j.kondo@sophia.ac.jp
bDepartment of Materials & Life Chemistry, Faculty of Engineering, Kanagawa University, 3-27-1 Rokkakubashi, Kanagawa-ku, Yokohama, 221-8686 Kanagawa, Japan. E-mail: akiraono@kanagawa-u.ac.jp
First published on 9th October 2017
Abstract
Herein, we determined a high-resolution crystal structure of a B-form DNA duplex containing consecutive dinuclear metal ion-mediated base pairs, namely, 4-thiothymine–2Ag(I)–4-thiothymine (S–2Ag(I)–S), and four Ag(I) ions form a rectangular network and the distances between the Ag(I) ions are 2.8–3.2 Å, which may indicate the existence of metallophilic attractions.
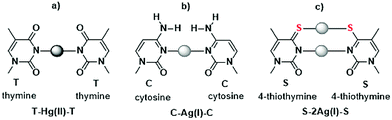
Strategies to use DNA as templates to generate functional molecules, precisely designed molecular architecture, and nanoscale devices have been widely investigated since nucleic acids can be assembled in a sequence-directed manner to form multidimensional structures.1 Recently, duplexes containing metal-mediated base pairs (metallo-base pairs), in which Watson–Crick base pairs are replaced by metal–base coordination complexes, have been identified. In addition, extensive studies on metal-mediated pairs of natural and artificial bases have expanded the design possibilities of functional DNA molecules.2 Metallo-base pairs have been applied to DNA molecular devices, such as ion-sensors,3 electric transport nano-wires,4 and DNA magnets.5 Many metallo-base pairs consist of two bases (heterocycles) and a metal ion placed between the bases to connect them.2 For example, in the presence of Hg(II) ions, a thymine–thymine (T–T) pair in a duplex forms a metallo-base pair, T–Hg(II)–T (Fig. 1a),6 and in the presence of Ag(I) ions, a cytosine–cytosine (C–C) pair forms C–Ag(I)–C (Fig. 1b).7 Recently, metallo-base pairs containing two metal ions between bases have been identified. A 4-thiothymine–4-thiothymine (S–S) in a duplex captures two Ag(I) ions to form a dinuclear Ag(I)-mediated base pair, S–2Ag(I)–S; thus, the duplex was thermally stabilized in the presence of Ag(I) ions.8 The predicted structure of S–2Ag(I)–S is shown in Fig. 1c. Metal ion binding to the pyrimidine base pairs is highly selective; thus, Ag(I) ions do not bind to T–T pairs and Hg(II) ions do not bind to C–C pairs under ordinary circumstances.9 Therefore, a minor modification of the pyrimidine base strongly affects the metal ion binding properties, such as the 4-carbonyl group being converted to a thiocarbonyl group8 and substitutions at the 5-position of uracil residues.10
To date, several dinuclear metal ion-mediated base pairs have been reported.11 Müller and colleagues used an artificial base to form metallo-base pairs containing two Ag(I) ions12 and two Hg(II) ions13 in antiparallel- and parallel-oriented duplexes. In addition, Seela and colleagues used ring-expanded pyrimidine bases to form extremely stable dinuclear Ag(I)-mediated base pairs in antiparallel- and parallel-oriented duplexes.14
Only a few studies have provided structural information on nucleic acid duplexes containing metallo-base pairs.2e Recently, we solved the solution structure of a DNA duplex containing two consecutive T–Hg(II)–T base pairs using NMR spectroscopy6f and X-ray crystallography.6g In the present study, we performed X-ray analysis of a DNA duplex containing consecutive dinuclear Ag(I)-mediated base pairs, S–2Ag(I)–S (Fig. 2). Four Ag(I) ions metallophilically interacted in a B-form duplex. Data acquired in crystalline and solution states provide a reliable structural basis for advanced design and further development of metallo-DNAs.
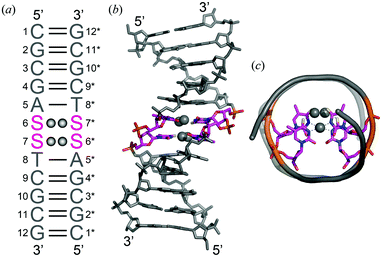
The DNA dodecamer d(CGCGASSTCGCG) (S = 4-thiothymine), hereafter referred to as 12SS, was designed to fold as a self-complementary duplex containing tandem S–S mispairs at the centre (Fig. 2a). These DNA fragments are commonly used as models in crystallographic studies of B-form DNAs containing natural bases, artificial bases, and metallo-base pairs. Recently, the crystal structure of a DNA duplex containing consecutive T–Hg(II)–T base pairs has been solved using d(CGCGATTTCGCG), 12TT.6g
The DNA dodecamer 12SS was synthesized on a DNA synthesizer using commercially available phosphoramidite units, after which the dodecamer was deprotected using protocols recommended by the provider. The deprotected dodecamer was purified using HPLC with a reverse-phase silica gel column.8 The 12SS was co-crystallized with Ag(I) ions. The crystal structures of 12SS/Ag(I) were deposited in the Protein Data Bank (PDB) under ID codes 5XUV. A detailed description of the Materials and methods is included in the ESI.†
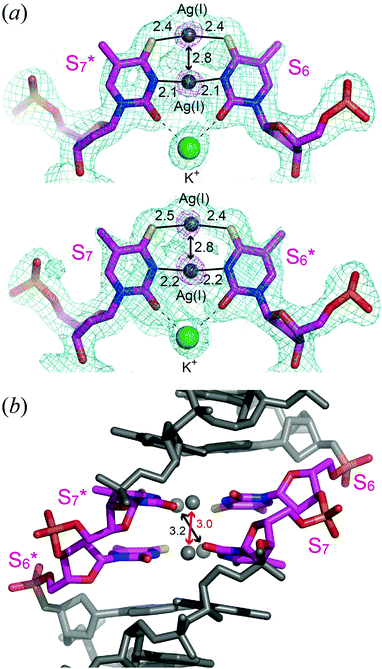
In the presence of Ag(I) ions, 12SS formed an antiparallel right-handed double helix (Fig. 2a and b). At both ends of the double helix, canonical Watson–Crick G–C and A–T pairs were formed. In the center of the double helix, two Ag(I) ions were placed in each S–S mispair and formed S–2Ag(I)–S metallo-base pairs (Fig. 1c); thus, the four Ag(I) ions were placed in proximity. Additionally, a K+ ion bridges two O2 atoms of the S–S mispair (Fig. 3a). The local helical parameters, intra-base pair parameters, and pseudorotation phase angles clearly indicate that the duplex adopts a standard B-conformation (ESI,† Tables S2 and S3). Furthermore, the overall duplex structure was similar to that of a duplex containing consecutive T–Hg(II)–T metallo-base pairs, 12TT/Hg.6g The formation of S–2Ag(I)–S base pairs did not significantly affect the overall B-form conformation.
The electron density maps and geometries of the S–2Ag(I)–S base pairs are shown in Fig. 3a. The 2|Fo| − |Fc| maps for S–2Ag(I)–S pairs are broadened at the center, where the Ag(I) ions are found. Based on the omit map calculated after removing the four Ag(I) ions, the metal ion occupies the center between the two S residues. N3⋯Ag(I) and S4⋯Ag(I) distances are around 2.1 Å and 2.4 Å, respectively; thus, N3⋯N3 and S4⋯S4 distances in each metallo-base pair are around 4.2 Å and 4.8 Å, respectively, and the numerical values are indicative of the disappearance of hydrogens (imino protons). Overall, the imino protons dissociated to generate anionic pyrimidine bases, between which Ag(I) ions are incorporated to form the metallo-base pair, S–2Ag(I)–S.
The propeller twist angles of the S–2Ag(I)–S base pairs (−19° and −21°) are larger than that of the canonical Watson–Crick base pairs in the B-form DNA (−1°) (Fig. 3b and ESI,† Table S2). This property is similar to that of T–Hg(II)–T6g and C–Ag(I)–C7b pairs, in which propeller twist occurs since there is no extra bond (excluding the N3–Hg(II)–N3 and N3–Ag(I)–N3 bonds between two pyrimidine residues) and due to the repulsion of carbonyl groups and amino groups, respectively (Fig. 1). Although 4-thiothymine residues are connected by two silver-mediated bonds, N3–Ag(I)–N3 and S4–Ag(I)–S4, and a potassium-mediated bond, O2–K+–O2, the propeller twist angles of the S–2Ag(I)–S pairs are as large as those of T–Hg(II)–T and C–Ag(I)–C pairs. The C1′⋯C1′ distances of the S–2Ag(I)–S base pairs (9.2 Å and 9.4 Å) are more than 1 Å shorter than those in the canonical Watson–Crick base pairs (∼10.7 Å), but the S–2Ag(I)–S base pairs fit into the B-form conformation (ESI,† Tables S2 and S3).
The intra-base pair Ag(I)⋯Ag(I) distance in each S–2Ag(I)–S is 2.8 Å (Fig. 3a), much less than the sum of their van der Waals radii (3.44 Å).15 The numerical value, 2.8 Å, is suggestive of a metallophilic interaction between Ag(I) ions.16 Similar values for Ag(I)⋯Ag(I) distances in metallo-base pairs were estimated based on DFT calculations.12a,c,e
In the B-form DNA, the helical axis runs through the center of the base pairs, and the Ag(I) ions in the S–2Ag(I)–S base pairs are aligned along the helical axis (Fig. 2c). The inter-base pair Ag(I)⋯Ag(I) distances are 3.0 Å and 3.2 Å (Fig. 3b). The distance between the two Ag(I) ions between the N3 positions (shown in red, in the center of the helix) is slightly shorter than that of the two Ag(I) ions between the 4-thiocarbonyl groups (shown in black, slightly strayed off the center of the axis). The numerical values, 3.0 Å and 3.2 Å, are shorter than the distances between Watson–Crick base pairs in B-form DNA (3.4 Å), which is indicative of a metallophilic attraction between Ag(I) inter-base pairs. A similar metallophilic attraction has been observed in the above-mentioned crystals of the DNA duplex containing consecutive T–Hg(II)–T base pairs.6g A Hg(II)⋯Hg(II) distance of 3.3 Å was indicative of a metallophilic attraction for Hg(II). The NMR solution structure of a B-form DNA duplex containing three consecutive imidazole–Ag(I)–imidazole base pairs17 showed an average Ag(I)⋯Ag(I) distance of 3.45 Å. Thus, it is important to compare the Hg(II)⋯Hg(II) distance6g and Ag(I)⋯Ag(I) distances in this study since the DNA sequences and whole duplex structures in the two crystals are similar. The Ag(I)⋯Ag(I) distances (3.0 Å and 3.2 Å) are smaller than the Hg(II)⋯Hg(II) distance (3.3 Å), which may indicate that S–2Ag(I)–S pairs are more tightly fixed into the duplex structure. The short distances and the relatively large propeller twist angles of the S–2Ag(I)–S base pairs might be explained by metallophilic attractions.
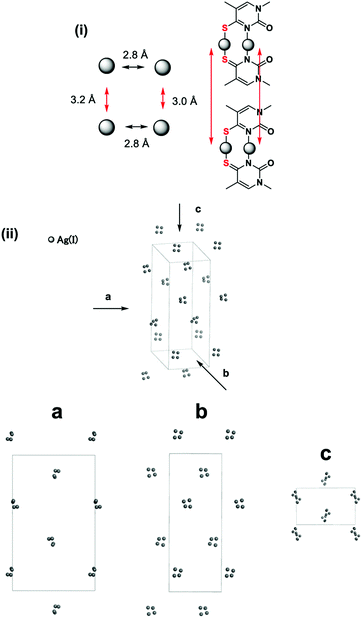
A schematic representation of the steric arrangement of four Ag(I) ions is shown in Fig. 4(i). The four Ag(I) ions were arranged in a rectangle (approximately square)-shaped network and the distances between the Ag(I) ions were 2.8–3.2 Å, which may indicate the existence of metallophilic attractions. In addition, four Ag(I) ion clusters are regularly arranged in a three-dimensional space in the crystal (Fig. 4(ii)). The distances between the four Ag(I) ion clusters are 25–30 Å.
Recently, a strategy to knit DNA strands and obtain nanoscale architecture (known as “DNA origami”) has been developed.18 Crystals consisting of functional oligonucleotides may be another strategy to construct DNA-based architecture, in which the functional groups are precisely arranged in a three-dimensional space.9b,19 For instance, crystals consisting of metal–DNA nanowires with uninterrupted one-dimensional silver arrays have been identified.9b In the crystals, silver atom arrays are precisely arranged to form the three-dimensional nanoscale architecture.
In conclusion, crystal structures of DNA duplexes revealed the following: (i) Ag(I) ions stabilize the B-form conformation by forming S–2Ag(I)–S base pairs, (ii) the dinuclear Ag(I)-mediated base pair is structurally replaceable with normal Watson–Crick base pairs based on the size and position within the B-form DNA duplex, (iii) four Ag(I) ions form a rectangular network and the relatively short Ag(I)⋯Ag(I) distances observed inside DNA are suggestive of the stabilizing metallophilic attraction between Ag(I) ions in consecutive S–2Ag(I)–S base pairs. The crystal structure of the metallo–DNA duplex containing the consecutive dinuclear Ag(I)-mediated base pairs provides the basis for the structure-based design of metal-conjugated nucleic acid nanomaterials.
This work was supported by a Grant in Aid for Scientific Research (B) (No. 17H03033), a Strategic Development of Research Infrastructure for Private Universities from the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT). We thank the Photon Factory for provision of synchrotron radiation facilities (No. 2015G533) and acknowledge the staff of the BL-1A beamline.
Conflicts of interest
There are no conflicts to declare.Notes and references
- DNA in Supramolecular Chemistry and Nanotechnology, ed. E. Stulz, GH. Clever, Wiley, Chichester, 2015 Search PubMed.
- (a) G. H. Clever, C. Kaul and T. Carell, Angew. Chem., 2007, 119, 6340–6350 ( Angew. Chem., Int. Ed. , 2007 , 46 , 6226–6236 ) CrossRef; (b) A. Ono, H. Torigoe, Y. Tanaka and I. Okamoto, Chem. Soc. Rev., 2011, 40, 5855–5866 RSC; (c) Y. Takezawa and M. Shionoya, Acc. Chem. Res., 2012, 45, 2066–2076 CrossRef CAS PubMed; (d) P. Scharf and J. Müller, ChemPlusChem, 2013, 78, 20–34 CrossRef CAS; (e) Y. Tanaka, J. Kondo, V. Sychrovský, J. Šebera, T. Dairaku, H. Saneyoshi, H. Urata, H. Torigoe and A. Ono, Chem. Commun., 2015, 51, 17343–17360 RSC; (f) Y. Takezawa, J. Müller and M. Shionoya, Chem. Lett., 2017, 46, 622–633 CrossRef CAS.
- (a) A. Ono and H. Togashi, Angew. Chem., 2004, 116, 4400–4402 ( Angew. Chem., Int. Ed. , 2004 , 43 , 4300–4302 ) CrossRef; (b) E. M. Nolan and S. J. Lippard, Chem. Rev., 2008, 108, 3443–3480 CrossRef CAS PubMed; (c) D. L. Ma, D. S. Chan, B. Y. Man and C. H. Leung, Chem. – Asian J., 2011, 6, 986–1003 CrossRef CAS PubMed; (d) Y. Song, W. Wei and X. Qu, Adv. Mater., 2011, 23, 4215–4236 CrossRef CAS PubMed.
- (a) T. Carell, C. Behrens and J. Gierlich, Org. Biomol. Chem., 2003, 1, 2221–2228 RSC; (b) T. Ito, G. Nikaido and S. Nishimoto, J. Inorg. Biochem., 2007, 101, 1090–1093 CrossRef CAS PubMed; (c) J. Joseph and G. B. Schuster, Org. Lett., 2007, 9, 1843–1846 CrossRef CAS PubMed; (d) L. Guo, N. Yin and G. Chen, J. Phys. Chem. C, 2011, 115, 4837–4842 CrossRef CAS; (e) H. Isobe, N. Yamazaki, A. Asano, T. Fujino, W. Nakanishi and S. Seki, Chem. Lett., 2011, 40, 318–319 CrossRef CAS; (f) S. Liu, G. H. Clever, Y. Takezawa, M. Kaneko, K. Tanaka, X. Guo and M. Shionoya, Angew. Chem., 2011, 123, 9048–9052 ( Angew. Chem., Int. Ed. , 2011 , 50 , 8886–8890 ) CrossRef.
- (a) K. Tanaka, A. Tengeiji, T. Kato, N. Toyama and M. Shionoya, Science, 2003, 299, 1212–1213 CrossRef CAS PubMed; (b) S. S. Mallajosyula and S. K. Pati, Angew. Chem., 2009, 121, 5077–5081 ( Angew. Chem., Int. Ed. , 2009 , 48 , 4977–4981 ) CrossRef; (c) G. H. Clever, S. J. Reitmeier, T. Carell and O. Schiemann, Angew. Chem., 2010, 122, 5047–5049 ( Angew. Chem., Int. Ed. , 2010 , 49 , 4927–4929 ) CrossRef.
- (a) S. Katz, Nature, 1962, 195, 997–998 CrossRef CAS PubMed; (b) S. Katz, Biochim. Biophys. Acta, 1963, 68, 240–253 CrossRef CAS PubMed; (c) Z. Kuklenyik and L. G. Marzilli, Inorg. Chem., 1996, 35, 5654–5662 CrossRef CAS PubMed; (d) Y. Miyake, H. Togashi, M. Tashiro, H. Yamaguchi, S. Oda, M. Kudo, Y. Tanaka, Y. Kondo, R. Sawa, T. Fujimoto, T. Machinami and A. Ono, J. Am. Chem. Soc., 2006, 128, 2172–2173 CrossRef CAS PubMed; (e) Y. Tanaka, S. Oda, H. Yamaguchi, Y. Kondo, C. Kojima and A. Ono, J. Am. Chem. Soc., 2007, 129, 244–245 CrossRef CAS PubMed; (f) H. Yamaguchi, J. Šebera, J. Kondo, S. Oda, T. Komuro, T. Kawamura, T. Dairaku, Y. Kondo, I. Okamoto, A. Ono, J. V. Burda, C. Kojima, V. Sychrovský and Y. Tanaka, Nucleic Acids Res., 2014, 42, 4094–4099 CrossRef CAS PubMed; (g) J. Kondo, T. Yamada, C. Hirose, I. Okamoto, Y. Tanaka and A. Ono, Angew. Chem., 2014, 126, 2417–2420 ( Angew. Chem., Int. Ed. , 2014 , 53 , 2385–2388 ) CrossRef.
- (a) A. Ono, S. Cao, H. Togashi, M. Tashiro, T. Fujimoto, T. Machinami, S. Oda, Y. Miyake, I. Okamoto and Y. Tanaka, Chem. Commun., 2008, 4825–4827 RSC; (b) J. Kondo, Y. Tada, T. Dairaku, H. Saneyoshi, I. Okamoto, Y. Tanaka and A. Ono, Angew. Chem., 2015, 127, 13521–23524 ( Angew. Chem., Int. Ed. , 2015 , 54 , 13323–13326 ) CrossRef; (c) T. Dairaku, K. Furuita, H. Sato, J. Šebera, K. Nakashima, J. Kondo, D. Yamanaka, Y. Kondo, I. Okamoto, A. Ono, V. Sychrovský, C. Kojima and Y. Tanaka, Chem. – Eur. J., 2016, 22, 13028–13031 CrossRef CAS PubMed; (d) H. Liu, C. Cai, P. Haruehanroengra, Q. Yao, Y. Chen, C. Yang, Q. Luo, B. Wu, J. Li, J. Ma, J. Sheng and J. Gan, Nucleic Acids Res., 2017, 45, 2910–2918 Search PubMed.
- I. Okamoto, T. Ono, R. Sameshima and A. Ono, Chem. Commun., 2012, 48, 4347–4349 RSC.
- For the first time, we have observed the formation of thymine–Ag(I)–thymine pairs in crystals consisting of all metallo-base pairs. (a) J. Kondo, Y. Tada, T. Dairaku, H. Saneyoshi, Y. Tanaka and A. Ono, Crystal Structure of Silver–DNA Hybrid Nanowire, The 43th International Symposium on Nucleic Acids Chemistry (ISNAC2016), 100th Anniversary Hall, Kumamoto University, Sept. 27–29, 2016, pp. 108–109; (b) J. Kondo, Y. Tada, T. Dairaku, Y. Hattori, H. Saneyoshi, A. Ono and Y. Tanaka, Nat. Chem., 2017 DOI:10.1038/nchem.2808 , published online.
- (a) I. Okamoto, K. Iwamoto, Y. Watanabe, Y. Miyake and A. Ono, Angew. Chem., 2009, 121, 1676–1679 ( Angew. Chem., Int. Ed. , 2009 , 48 , 1648–1651 ) CrossRef; (b) X. Guo, S. A. Ingale, H. Yang, Y. He and F. Seela, Org. Biomol. Chem., 2017, 15, 870–883 RSC.
- S. Mandal and J. J. Müller, Curr. Opin. Chem. Biol., 2017, 37, 71–79 CrossRef CAS PubMed.
- (a) D. A. Megger, C. F. Guerra, J. Hoffmann, B. Brutschy, F. Matthias Bickelhaupt and J. Müller, Chem. – Eur. J., 2011, 17, 6533–6544 CrossRef CAS PubMed; (b) S. Mandal, A. Hepp and J. Müller, Dalton Trans., 2015, 44, 3540–3543 RSC; (c) S. Litau and J. Müller, Z. Anorg. Allg. Chem., 2015, 641, 2169–2173 CrossRef CAS; (d) S. Mandal, C. Wang, R. K. Prajapati, J. Kösters, S. Verma, L. Chi and J. J. Müller, Inorg. Chem., 2016, 55, 7041–7050 CrossRef CAS PubMed; (e) S. Mandal, M. Hebenbrock and J. Müller, Chem. – Eur. J., 2017, 23, 5962–5965 CrossRef CAS PubMed.
- S. Mandal, M. Hebenbrock and J. Müller, Angew. Chem., 2016, 128, 15747–15750 ( Angew. Chem., Int. Ed. , 2016 , 55 , 15520–15523 ) CrossRef.
- (a) H. Mei, I. Röhl and F. Seela, J. Org. Chem., 2013, 78, 9457–9463 CrossRef CAS PubMed; (b) H. Mei, S. A. Ingale and F. Seela, Chem. – Eur. J., 2014, 20, 16248–16257 CrossRef CAS PubMed; (c) H. Mei, H. Yang, I. Röhl and F. Seela, ChemPlusChem, 2014, 79, 914–918 CrossRef CAS; (d) H. Yang, H. Mei and F. Seela, Chem. – Eur. J., 2015, 21, 10207–10219 CrossRef CAS PubMed; (e) S. K. Jana, X. Guo and H. Mei. F. Seela, Chem. Commun., 2015, 51, 17301–17304 RSC.
- A. Bondi, J. Phys. Chem., 1964, 68, 441–451 CrossRef CAS.
- H. Schmidbaur and A. Schier, Angew. Chem., 2015, 127, 756–797 ( Angew. Chem., Int. Ed. , 2015 , 54 , 746–784 ) CrossRef.
- (a) S. Johannsen, N. Megger, D. Böhme, R. K. O. Sigel and J. Müller, Nat. Chem., 2010, 2, 229–234 CrossRef CAS PubMed; (b) S. Kumbhar, S. Johannsen, R. K. O. Sigel, M. P. Waller and J. Müller, J. Inorg. Biochem., 2013, 127, 203–210 CrossRef CAS PubMed.
- (a) N. Seeman, Nature, 2003, 421, 427–431 CrossRef PubMed; (b) A. R. Chandrasekaran and O. Levchenko, Chem. Mater., 2016, 28, 5569–5581 CrossRef CAS.
- H. Liu, F. Shen, P. Haruehanroengra, Q. Yao, Y. Cheng, Y. Chen, C. Yang, J. Zhang, B. Wu, Q. Luo, R. Cui, J. Li, J. Ma and J. Sheng, Angew. Chem., 2017, 129, 9558–9562 ( Angew. Chem., Int. Ed. , 2017 , 56 , 9430–9434 ) CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7cc06153f |
| This journal is © The Royal Society of Chemistry 2017 |